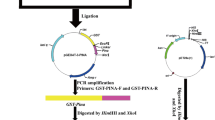
Abstract
Wheat puroindoline proteins, PINA and PINB, play key roles in determining wheat grain hardness as well as in defending the plant against pathogens. PINA has much greater membrane-binding property and antimicrobial activity because it contains more tryptophan residues in the unique tryptophan-rich domain (TRD). In order to obtain proteins with higher antimicrobial activity, mutants of PINA containing two or three copies of TRD, designated ABBC and ABBBC, respectively, were constructed and expressed in E. coli Rosetta-gami (DE3). Metal affinity chromatography was used to purify the soluble affinity-tagged recombinant proteins. The secondary structures of the recombinant proteins were predicted by the online program Protein Homology/analog Y Recognition Engine v2.0 and experimentally assessed using circular dichroism. Minimum inhibition concentration tests and fluorescence microscope analyses were employed to evaluate the antimicrobial activities of the mutants. The results showed that the purified recombinant ABBC was correctly folded and presented significantly higher antimicrobial activities against E. coli and S. aureus than wild-type PINA, suggesting its potential use as an antimicrobial agent. The results also confirmed that TRD is a determinant of the antimicrobial activity of PINA and demonstrated that it is feasible to enhance the antimicrobial activity of PINA by adding one copy of TRD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Two wheat endosperm-specific genes Puroindoline a and b (Pina and Pinb) (Blochet et al. 1993) represent the molecular-genetic basis of the Hardness (Ha) locus (Morris 2002) on chromosome 5DS in hexaploid wheat, and play key roles in determining wheat grain hardness (Giroux and Morris 1998). Moreover, puroindoline A (PINA) and puroindoline B (PINB) were also reported to have significant antimicrobial activities against bacteria and fungi (Capparelli et al. 2005, 2007). Indeed, transgenic crop over-expressing PINA and/or PINB showed significantly increased resistance to plant diseases (Krishnamurthy et al. 2001; Luo et al. 2008). These researches suggest that puroindolines (PINs) could be used as alternatives to conventional antibiotics to control plant and human diseases.
A critical characteristic of PINs is their ability to interact with polar lipids (Dubreil et al. 1997; Kooijman et al. 1997, 1998; Le Guerneve et al. 1998), which is the basis of the mechanism by which PINs determine grain hardness textures and exhibit antimicrobial activities (Bhave and Morris 2008). The lipid affinity of PINs depends on their tertiary structure, comprising four alpha helixes separated by loops and stabilized by five disulfide bonds (Le Bihan et al. 1996). Functional PINs rely heavily on the tryptophan-rich domain (TRD) localized on a surface loop between the first two helixes. Several lines of evidence (Evrard et al. 2008) showed that the TRDs are involved in the interaction of PINs with polar lipids. Having more Trp residues in TRD, PINA appears to penetrate lipid membranes more deeply than PINB (Kooijman et al. 1998). Results of several researches gave evidence that PINA has more antimicrobial activity than PINB (Sandras et al. 2009; Clifton et al. 2011). Moreover, a 13-residue synthetic peptide mimicking PINA TRD demonstrated antimicrobial activity in vitro (Jing et al. 2003). Consequently, the TRD might be the ‘antimicrobial domain’ of PINA.
The purpose of the present study is to construct mutants with better antimicrobial activities and to understand how the TRD influences the functions of PINA. PINA has a more pronounced antimicrobial activity than PINB, which makes it more suitable as a template for mutation. Mutations within the TRD, or altering the protein backbone, could cause the loss of PINA function (Feiz et al. 2009). We constructed mutants of PINA having two or three copies of TRDs, without disturbing the disulfide bonds or modifying any amino acid residues in the four alpha helixes, and tested their antimicrobial activities.
For convenience, here we designate the fragment of TRD as “B”, the fragment upstream as “A” and the fragment downstream as “C”. Then wild-type PINA is represented as “ABC” for short, the mutants containing two copies of TRD are designated as “ABBC”, and with three copies of TRD as “ABBBC”.
Materials and methods
Construction of ABBC and ABBBC mutants
Primers ForA 5′-CCCAAGCTTATGGAGGCCCTCTTCCTCAT-3′ (Hind III site underlined) and RevB 5′-GCGGATCCACCTCCCTTCCACCATTTCC-3′ (BamHI site underlined) were designed to amplify the fragment AB from 5′ start of fragment A to 3′ end of fragment B. Primers ForB 5′-CCCAGATCTTCAACGATGAAGGATTTCCC-3′ (BglII site underlined) and RevC 5′-CGGGATCCCCAGTAATAGCCAATAGTG-3′ (BamHI site underlined) were designed to amplify the fragment BC from 5′ start of fragment B to 3′ end of fragment C. After purification, two PCR products were cloned into pMD18-T vector, respectively, and used to transform Top10 competent cells. The plasmids were extracted and double-digested using restriction enzymes. Digested AB and BC fragments were incubated together with T4 DNA ligase at 16°C for 10 h.
Using the T4 ligation product as template and ForA, RevC as primers, PCR was carried out to amplify ABBC. The PCR product was cloned into pMD18-T vector and then transformed to Top10 competent cells. The positive clones were identified by PCR and confirmed by sequencing. Using ABBC as template, the fragments ABB and BBC were amplified, cloned and digested; ABBBC was constructed by linking the ABB and BC fragments.
Construction of E. coli expression vectors
Expression vector pET43.1a carries a Nus A fusion Tag at the N-terminal of the target protein. Two primers FnP 5′-GGAATTCGATGTTGCTGGCGGGGGT-3′ (EcoRI site underlined) and RnP 5′-TGCGGCCGCTCACCAGTAATAGCCAATA-3′ (NotI site underlined) were designed to remove the N-terminal cleavable peptides of Pina and of the mutant genes. After PCR amplification, the products and pET43.1a plasmid were double-digested, followed by gel extraction. Then Pina, ABBC and ABBBC fragments were ligated to the cohesive termini of the linearized pET43.1a. The ligation products were transformed to Top10 competent cells.
Expression and purification of fusion proteins
The expression plasmids designated pET43.1a-Pina/ABBC/ABBBC were introduced into E. coli strain Rosetta-gami (DE3). Positive clones were cultured to exponential phase (OD600 = 0.4–0.6) at 25°C in Terrific Broth (TB) medium (1.2% peptone, 2.4% yeast extract, and 0.4% glycerol, 17 mM KH2PO4 and 72 mM K2HPO4) containing 50 μg/mL ampicillin and 15 μg/mL kanamycin. The cultures were cooled to 14°C and induced by a final concentration of 0.5 mM IPTG at 14°C for 15 h. Bacterial cells were collected by centrifugation at 5,000g.
The purification of recombinant proteins was under native conditions, and all procedures employed were performed at 4°C. The bacteria pellet was resuspended in NTA buffer (20 mM Tris–HCl, 500 mM NaCl, 0.4% glycerol, 1 mM PMSF), and then treated by one shot high-pressure cell disruption instrument (Constant System) at 20 kPSI. After centrifugation, the supernatant was loaded onto a Ni–NTA affinity chromatography (Qiagen) column and washed sequentially by NTA buffer contained 20, 40, 60, 80, 100, 150 and 200 mM imidazole. The eluate was collected in 2 mL fractions and analyzed by SDS-PAGE. The fractions containing the target protein of the expected molecular weights were pooled and dialyzed versus 10 mM Tris–HCl buffer then ultrafiltered using an Amicon Ultra-15 centrifugal filter device (Millipore) with a NMWL of 30 kDa and a 4,000g centrifugation.
Cleavage and purification of the recombinant proteins
Purified recombinant fusion proteins were digested with recombinant thrombin for 5 h at 23°C according to the manufacturer’s instructions. Then the fusion tags and the enzyme were removed through two further steps of Ni–NTA affinity chromatography.
Secondary and tertiary structure prediction
Protein structure predictions were performed with Protein Homology/analog Y Recognition Engine v2.0 (Phyre2) (Kelley and Sternberg 2009) available on the web (http://www.sbg.bio.ic.ac.uk/Phyre2/); this suite is based on the work of the Structural Bioinformatics Group, Imperial College, London, and is supported by a BBSRC tools and resources grant. Phyre2 could predict the 3D structure adopted by a user-supplied protein sequence without additional configuration. Excluding the 28-residue long N-terminal cleavable peptides (Met1–Tyr28), amino acid sequences (Asp29 to the stop codon) of PINA (120 aa), ABBC (141 aa) and ABBBC (162 aa) were applied to Phyre2. The cleavable peptide comprises a 19-residue signal peptide (Met1–Ala19), then a peptide with cleavage site after the 28th residue (Gln20–Asp28). After alignment several templates (5 for PINA, 4 for ABBC and 4 for ABBBC) were selected by Phyre2 to model the 3D structures.
Circular dichroism spectroscopy
The circular dichroism (CD) spectra were measured at room temperature with a JASCO J-810 apparatus using a 1 mm path length cell. Each spectrum (190–240 nm) was obtained as the average of five times’ repeats. The protein samples were dissolved at the concentration of 10 μM in Tris–HCl buffer. The secondary structures of recombinant proteins were estimated using the online tool K2D2 (http://www.ogic.ca/projects/k2d2/).
Minimum inhibitory concentration assay
The minimum inhibitory concentration (MIC) against the bacterial strains E. coli ATCC 25922 and S. aureus ATCC 25923 was measured with a microplate reader. Bacteria were cultured in Bacto-Peptone medium until the exponential phase and collected by centrifugation. The resuspended cells were diluted to 2 × 106 CFU/mL and added to the wells of 96-well plates. Recombinant proteins in scalar concentrations were added to the wells and incubated overnight at 37°C. The data reported are the average of the experimental results of three repeats.
Fluorescence microscopy
Fluorescence microscopy analyses were carried out to evaluate the antimicrobial effect of the recombinant proteins. E. coli and S. aureus strains were cultured in Luria–Bertani (LB) medium until the exponential phase. Bacterial cells were collected and diluted to 2 × 106 CFU/mL then incubated with the recombinant proteins for 2 h. The treated cells were stained with 20 μM Hoechst 33342 and 15 μM propidium iodide (PI). Hoechst 33342 and PI staining were visualized by excitation with 350 and 488 nm UV light, respectively. Micrographs were captured with the CCD camera of a Nikon 80i fluorescence microscope. Cell counts were performed with the ImageJ software (http://rsbweb.nih.gov/ij/).
Results
Construction, expression and purification of PINA and its mutants
The PCR amplicons and the ligation products in the process of mutant construction were separated by agarose gel electrophoresis (as shown in Fig. 1). The lengths of the target fragments were as follows: AB 228 bp, BC 282 bp, ABB 291 bp, BBC 345 bp. The lengths of target mutant genes were as follows: Pina 447 bp, ABBC 510 bp, and ABBBC 573 bp.
Agarose gel electrophoresis of PCR amplified fragment AB (a), fragment BC (b), fragment ABB/BBC (c), Pina (d), ABBC (e), and ABBBC (f). a Lane M, DNA marker; lanes 1–4, fragment AB. b Lane M, DNA marker; lanes 1–4, fragment BC. c Lane M, DNA marker; lane 1, positive control 1 (fragment AB); lanes 2–4, fragment ABB; lane 5, positive control 2 (fragment BC); lanes 6–7, fragment BBC. d Lane M, DNA marker; lanes 1–3, Pina. e Lane M, DNA marker; lanes 1–3, ABBC. f Lane M, DNA marker; lanes 1–3, ABBBC. Experimental: in all case agarose concentration was 1.5%
The coding sequences of the N-terminal cleavable peptide of Pina and its mutants were removed by PCR to minimize the negative impact on the growth of E. coli strains. Protein expressions were induced with 0.5 mM IPTG for 15–20 h at 14°C. Low temperature used in preculture and induction significantly increased the yield of recombinant proteins, and most of the proteins expressed were soluble. The induction results and purification profile of PINA are shown in Fig. 2. Purified fusion proteins were digested with recombinant thrombin to cleave the tags.
SDS-PAGE analyses of intracellular soluble recombinant PINA, ABBC and ABBBC (a) and purification profile of PINA fusion protein (b). a Lane M, molecular weight marker; lane 1, lysate of induced Rosetta-gami (DE3) culture; lanes 2–4, lysate of induced Rosetta-gami (DE3) cultures containing pET43.1a-PINA, pET43.1a-ABBC and pET43.1a-ABBBC. b Lane M, molecular weight marker; lanes 1–2, lysate of induced Rosetta-gami(DE3) cultures containing recombinant PINA; lanes 3–4, purified recombinant PINA. Experimental: 4–15% polyacrylamide in Tris–glycine buffer
Secondary and tertiary structure prediction
The secondary and tertiary structures of three proteins were predicted with the online program Phyre2, which uses the alignment of hidden Markov models via HHsearch (Soding 2005). The secondary structure prediction by Phyre2 was the consensus of three methods including psiprep, jnet and sspro (shown in Fig. 3). The tertiary structure predicted by Phyre2 is shown in Fig. 4.
It can be clearly observed that in each protein the TRD (s) form (s) an extension of a surface loop. The basic globular conformation always consists of four helixes, which are not destroyed or disturbed in the artificial mutants, suggesting that the conformation predicted should be close to the real structure. In the case of PINA and ABBC, tryptophan residues in TRD (s) occupy the surface loop, as expected. In the case of ABBBC, the triplicate TRDs form a flexible extension with lacks support via disulfide bonds. In addition, Trp40, Trp82, Trp85 and Trp86 are shielded by adjacent residues and the rest of tryptophan residues in TRDs are dispersed in the protein backbone and might contribute less to the formation of a hydrophobic region.
Estimation of recombinant proteins’ secondary structure from circular dichroism spectroscopy
Circular dichroism spectroscopy was carried out to estimate the secondary structure of the purified recombinant proteins. The results are shown in Fig. 5. The proportions of secondary structural elements determined by CD and online tool K2D2 are shown in Table 1.
The proportions determined by CD show a difference among the three proteins. From PINA to ABBBC, the proportion of alpha helix increases significantly, while the proportion of random coil decreases.
Anti-microbial activity of recombinant PINA and its mutants
Minimum inhibitory concentration is defined as the lowest concentration of an antimicrobial agent that will inhibit the growth of a microorganism after overnight incubation. The MICs of recombinant PINA, ABBC and ABBBC against E. coli and S. aureus were evaluated and are listed in Table 2. Fluorescence microscopy analysis was carried out to observe the bacteria treated with the recombinant proteins. All recombinant proteins used in the analyses were at a concentration of 100 μg/mL for E. coli and 160 μg/mL for S. aureus. The fluorescence micrographs are shown in Figs. 6 and 7. The percentages of killed bacteria were calculated after colony count (as shown in Table 2).
Fluorescence micrographs of E. coli treated with recombinant PINA, ABBC, ABBBC. Cells were double-stained with Hoechst 33342 (blue for dead and live cells) and PI (red for dead cells). Negative control (untreated) (a, b); positive control (treated with 70% isopropanol) (c, d); treated with PINA (e, f); treated with ABBC (g, h); treated with ABBBC (i, j)
Fluorescence micrographs of S. aureus treated with recombinant PINA, ABBC, ABBBC. Cells were double-stained with Hoechst 33342 (blue for dead and live cells) and PI (red for dead cells). Negative control (untreated) (a, b); positive control (treated with 70% isopropanol) (c, d); treated with PINA (e, f); treated with ABBC (g, h); treated with ABBBC (i, j)
The MICs of recombinant ABBC against E. coli and S. aureus were lower than that of recombinant PINA (approximately 20–30%), which suggests that the antimicrobial ability of recombinant ABBC is enhanced. The MIC of recombinant ABBBC was higher than PINA (P < 0.05), which suggests that ABBBC has limited application value. The fluorescence microscopy analyses provide similar results.
Discussion
PINA has dual effects in determining grain texture and exhibiting antimicrobial activity. Evidence has shown that these different effects are mediated via the same TRD region, which can exhibit by itself antimicrobial activity. Although the mechanism by which PINA exhibits antimicrobial effect is not clear, over the past years several researchers have explored the application of PINA to control plant and human diseases (Capparelli et al. 2007; Luo et al. 2008; Krishnamurthy et al. 2001). However, there are no reports about the application of PINA artificial mutants. Using a PCR-based strategy we have successfully constructed, expressed and purified two artificial mutants of PINA (ABBC and ABBBC). The secondary and tertiary structure prediction suggests that the mutations may not affect the main backbone of the protein.
In this study, the pET43.1a vector was fused with 491 aa NusA fusion tag (Nus·Tag) to promote the solubility of the expressed proteins. The expression strain Rosetta-gami (DE3) has trxB and gor mutations, which enhance disulfide bond formation in E. coli cytoplasm, and also provides rare codon tRNAs. A low-temperature preculture and induction strategy was also used to increase substantially the yield of soluble, functional proteins.
Purification from Rosseta-gami (DE3) allowed for the characterization of PINA and its mutants by CD. The results show that recombinant PINA retains levels of secondary structure elements typical of a folded protein. This suggests that the recombinant proteins expressed in bacteria are correctly folded. Moreover, MIC tests and fluorescence microscopy analyses have fully confirmed the antimicrobial activities of the recombinant wild-type protein.
The decrease of antimicrobial activity in recombinant ABBBC might have resulted from the tertiary structure of the tripled TRD region. Tryptophan residues in TRD of native PINA form an extension of a surface loop, which is a close packing and relatively rigid structure stabilized by one pair of disulfide bonds. In the case of ABBBC, the tripled TRDs might be more flexible. It is possible that the close intra-molecular packing of the tryptophan residues in TRDs results in a conformational transition, likely from loops to helixes. This hypothesis is confirmed by the CD results, which show that ABBBC has the highest alpha helix and lowest random proportions amongst the three test proteins. Moreover, several tryptophan residues in TRDs do not occupy the surface of the loop but are shielded by adjacent residues.
In the case of ABBC, each TRD forms a rigid structure in the surface loop. All the hydrophobic and lipophilic tryptophan residues occupy the surface of the protein molecule forming two hydrophobic regions. These two TRDs are separated by several neutral residues instead of forming a single large hydrophobic region. There might be competition between the two regions for the interaction with the same target molecule, and they could also be subject to steric constraints when interacting with two lipid molecules. These effects might explain why the antimicrobial activity of recombinant ABBC is enhanced, but not doubled versus wild-type. The results anyhow suggest that ABBC would be a potential candidate as an antimicrobial agent.
Several studies have focused on the unique region of PINA known as the TRD. Our work provides new evidence for the involvement of TRD in PINA antibacterial activity. We also created an artificial puroindoline-like protein ABBC and proved that it has greater antibacterial activity than PINA against both gram-positive and gram-negative bacteria.
Bacteria gain drug resistance (Martinez and Silley 2010) due to a variety of reasons such as the abuse of antibiotics worldwide. Proteins such as ns-LTP present remarkable antimicrobial abilities (Gordon et al. 2005), suggesting a new approach to solve the problem of tolerance to antimicrobial drugs. It is believed that PINA kills bacteria by disturbing their lipid bilayer structure and destroying cell membranes. It is difficult for bacteria to gain tolerance by changing the lipid content of the plasma membrane, which opens interesting perspectives to the clinical application of PINs.
Because of the dual function of PINA in wheat, it is possible to modify wheat grain hardness texture as well as to extend the storage time of seeds through the over-expression of a single gene in transgenic plants. The antimicrobial activity in transgenic wheat of the mutants we describe in the present report and their effects in determining wheat seed texture should be evaluated in further studies.
References
Bhave M, Morris CF (2008) Molecular genetics of puroindolines and related genes: regulation of expression, membrane binding properties and applications. Plant Mol Biol 66(3):221–231
Blochet JE, Chevalier C, Forest E, Pebay-Peyroula E, Gautier MF, Joudrier P, Pezolet M, Marion D (1993) Complete amino acid sequence of puroindoline, a new basic and cystine-rich protein with a unique tryptophan-rich domain, isolated from wheat endosperm by Triton X-114 phase partitioning. FEBS Lett 329(3):336–340. doi:10.1016/0014-5793(93)80249-T
Capparelli R, Amoroso MG, Palumbo D, Iannaccone M, Faleri C, Cresti M (2005) Two plant puroindolines colocalize in wheat seed and in vitro synergistically fight against pathogens. Plant Mol Biol 58(6):857–867. doi:10.1007/s11103-005-8270-9
Capparelli R, Ventimiglia I, Palumbo D, Nicodemo D, Salvatore P, Amoroso MG, Iannaccone M (2007) Expression of recombinant puroindolines for the treatment of staphylococcal skin infections (acne vulgaris). J Biotechnol 128(3):606–614
Clifton LA, Sanders MR, Castelletto V, Rogers SE, Heenan RK, Neylon C, Frazier RA, Green RJ (2011) Puroindoline-a, a lipid binding protein from common wheat, spontaneously forms prolate protein micelles in solution. Phys Chem Chem Phys 13(19):8881–8888. doi:10.1039/c0cp02247k
Dubreil L, Compoint JP, Marion D (1997) Interaction of puroindolines with wheat flour polar lipids determines their foaming properties. J Agric Food Chem 45(1):108–116
Evrard A, Lagarde V, Joudrier P, Gautier MF (2008) Puroindoline-a and puroindoline-b interact with the Saccharomyces cerevisiae plasma membrane through different amino acids present in their tryptophan-rich domain. J Cereal Sci 48(2):379–386
Feiz L, Martin JM, Giroux MJ (2009) Creation and functional analysis of new Puroindoline alleles in Triticum aestivum. Theor Appl Genet 118(2):247–257
Giroux MJ, Morris CF (1998) Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Proc Natl Acad Sci USA 95(11):6262–6266
Gordon YJ, Romanowski EG, McDermott AM (2005) A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr Eye Res 30(7):505–515. doi:10.1080/02713680590968637
Jing WG, Demcoe AR, Vogel HJ (2003) Conformation of a bactericidal domain of puroindoline a: structure and mechanism of action of a 13-residue antimicrobial peptide. J Bacteriol 185(16):4938–4947
Kelley LA, Sternberg MJ (2009) Protein structure prediction on the web: a case study using the phyre server. Nat Protoc 4(3):363–371. doi:10.1038/nprot.2009.2
Kooijman M, Orsel R, Hessing M, Hamer RJ, Bekkers ACAPA (1997) Spectroscopic characterisation of the lipid-binding properties of wheat puroindolines. J Cereal Sci 26(2):145–159
Kooijman M, Orsel R, Hamer RJ, Bekkers ACAPA (1998) The insertion behaviour of wheat puroindoline-a into diacylgalactosylglycerol films. J Cereal Sci 28(1):43–51
Krishnamurthy K, Balconi C, Sherwood JE, Giroux MJ (2001) Wheat puroindolines enhance fungal disease resistance in transgenic rice. Mol Plant Microbe In 14(10):1255–1260
Le Bihan T, Blochet JE, Desormeaux A, Marion D, Pezolet M (1996) Determination of the secondary structure and conformation of puroindolines by infrared and Raman spectroscopy. Biochemistry 35(39):12712–12722. doi:10.1021/bi960869n
Le Guerneve C, Seigneuret M, Marion D (1998) Interaction of the wheat endosperm lipid-binding protein puroindoline-a with phospholipids. Arch Biochem Biophys 360(2):179–186
Luo L, Zhang JR, Yang GX, Li Y, Li KX, He GY (2008) Expression of puroindoline a enhances leaf rust resistance in transgenic tetraploid wheat. Mol Biol Rep 35(2):195–200
Martinez M, Silley P (2010) Antimicrobial drug resistance. Handb Exp Pharmacol 199:227–264. doi:10.1007/978-3-642-10324-7_10
Morris CF (2002) Puroindolines: the molecular genetic basis of wheat grain hardness. Plant Mol Biol 48(5):633–647
Sandras F, Pezolet M, Marion D, Grauby-Heywang C (2009) Raman study of the puroindoline-a/lysopamitoylphosphatidylcholine interaction in free standing black films. Langmuir 25(14):8181–8186
Soding J (2005) Protein homology detection by HMM–HMM comparison. Bioinformatics 21(7):951–960. doi:10.1093/bioinformatics/bti125
Acknowledgments
This research was financially supported by Genetically Modified Organisms Breeding Major Projects of China (Grant No. 2009ZX08002-013B, 2009ZX08002-006B, 2011ZX08002-004, 2011ZX08010-004), Key Projects of International Cooperation of Most of China (Grant No. 2009DFB30340) and Key Projects of S and T Research of MoE of China (Grant No. 109105). Authors are thankful to the Analytical and Testing Center of Huazhong University of Science and Technology, for technical support.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Y. Miao, L. Chen and C. Wang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Miao, Y., Chen, L., Wang, C. et al. Expression, purification and antimicrobial activity of puroindoline A protein and its mutants. Amino Acids 43, 1689–1696 (2012). https://doi.org/10.1007/s00726-012-1250-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-012-1250-x