Abstract
Antimicrobial proteins, and especially antimicrobial peptides (AMPs) hold great promise in the control of animal and plant diseases with low risk of pathogen resistance. The two puroindolines, a and b, from wheat control endosperm softness of the wheat caryopsis (grain), but have also been shown to inhibit the growth and kill various bacteria and fungi, while showing little toxicity to erythrocytes. Puroindolines are small (~ 13 kDa) amphipathic proteins with a characteristic tryptophan-rich domain (TRD) that is part of an 18 or 19 amino acid residue loop subtended by a disulfide bond. This review presents a brief history of the puroindolines, their physical–chemical characteristics, their interaction with lipids and membranes, and their activity as antimicrobial proteins and AMPs. In this latter context, the use of the TRDs of puroindoline a and b in puroindoline AMP function is reviewed. The activity of puroindoline a and b and their AMPs appear to act through similar but somewhat different modes, which may involve membrane binding, membrane disruption and ion channel formation, and intra-cellular nucleic acid binding and metabolic disruption. Natural and synthetic mutants have identified key elements of the puroindolines for antimicrobial activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction and historical background
Plants and animals are in a constant battle against pathogenic microbes. Human intervention in this struggle has relied heavily on synthetic compounds such as antibiotics and fungicides. However, the ability of microbes to develop resistance to these compounds necessitates alternative strategies. One such strategy is the use of antimicrobial proteins and peptides (AMPs). AMPs are small peptides, typically of < 100 amino acid residues. Most are amphipathic and cationic (Avci et al. 2018; Shagaghi et al. 2018; Zasloff 2002); many specifically target features of the microbial cell membrane that are different from multicellular plants and animals (Zasloff 2002). This review describes how the puroindoline proteins and specific portions of the proteins (peptides) act as antimicrobial agents.
Puroindolines were first reported in 1990 (Blochet et al. 1991). Their discovery brought together two divergent fields of study. The first involved wheat (Triticum ssp.) kernel texture (grain hardness). Kernel texture is the single most important trait governing the milling, flour quality and food applications of wheat (Heinze et al. 2016; Murray et al. 2016). Although quantitative in nature, a single genetic locus determines the majority of texture variation, which falls into three major phenotypic classes (Morris 2002). This locus, referred to as Hardness (Ha), exhibits simple Mendelian inheritance (Morris et al. 1999). The first two kernel texture classes are ‘soft’ and ‘hard’ in T. aestivum L., commonly known as bread wheat. The third texture class is comprised of the very hard kernel, T. turgidum subsp. durum. In brief, durum wheat formed through the natural hybridization of two wild diploid grasses, both of which possessed a Ha locus and had soft kernels. However, the two Ha loci were lost. Since the Ha locus confers the soft kernel phenotype, the grains of durum wheat devoid of any Ha loci are extremely hard (Bhave and Morris 2008a). Approximately 10,000 years ago, a second polyploidation event occurred involving durum wheat and a wild diploid grass, Aegilops tauschii (Li et al. 2008; Massa and Morris 2004, 2006). In this instance, the Ha locus of Ae. tauschii was retained, thus conferring soft kernel texture to bread wheat. All hard wheats have arisen through mutation since the original polyploidization event (Giroux and Morris 1997, 1998; Morris and Bhave 2008a). This fact has bearing on the subsequent discussion of antimicrobial effects of the puroindolines.
Early work on kernel texture and the soft, hard and durum wheat classes identified a small ~ 13 kDa protein associated with the surface of isolated wheat starch (Greenwell and Schofield 1986). This ‘protein’, termed ‘friabilin’, was eventually resolved to be comprised of three individual components, two of which were puroindolines (Morris et al. 1992, 1994). Friabilin was shown to be associated with starch granule surface lipids, and was quantitatively expressed according to gene dosage (Bettge et al. 1995). It would eventually be shown that the two major components of friabilin were puroindoline a and b (Fig. 1, Supplementary Fig. 1), and that they comprised the molecular-genetic basis of wheat kernel texture.
Native protein sequences of puroindoline a and puroindoline b from wheat. Sequence 1, puroindoline a wild-type Pina-D1a; 2, puroindoline b wild-type Pinb-D1a; 3, puroindoline b G46S Pinb-D1b (‘Pin-bH’ of Clifton et al. 2007a, b, 2008); 4, puroindoline b W44R Pinb-D1d (‘Pin-bS’ of Clifton et al. 2007a, b, 2008). The prepro signal peptide (C-terminal) and cleavable N-terminal residues are underlined. The tryptophan-rich domains are double underlined. Cysteine residues are in red. Mutant amino acids are highlighted
The second field of study involved amphiphilic lipid binding proteins that were associated with the functionality (baking quality) of wheat flour. A key aspect of this research was the isolation of proteins from wheat flour using Triton X-114, which can undergo a temperature dependent phase transition, thereby producing ‘detergent rich’ and ‘detergent depleted’ fractions. The report of Blochet et al. (1991) included the N-terminal amino acid sequence of three of these unique Triton X-114 isolated wheat proteins. Subsequent research has demonstrated that two of these proteins were puroindoline a and b, with the third being a similar protein (Grain softness protein-1), which also resides at the Ha locus and is related evolutionarily (Chantret et al. 2005; Morris et al. 2013; Massa et al. 2004, 2006).
These two foregoing fields of study coalesced around the protein sequence of Blochet et al. (1991), Morris et al. (1994), and others (see Fig. 1 in Morris 2002). Blochet et al. (1993) would subsequently provide the complete amino acid sequence of one of these proteins, as well as the name, ‘puroindoline’. With the isolation of a second protein, this first specific protein would later be designated puroindoline a. Gautier et al. (1994) cloned and reported full-length cDNA sequences for both puroindoline a and b. From this point, research on puroindolines rapidly advanced.
Eventually it would be shown that puroindoline a and b from soft kernel wheat represent the ‘wild-type’ sequences, contributed by Ae. tauschii (Massa et al. 2004, 2006; Giroux and Morris 1997, 1998) (Fig. 1, Supplementary Fig. 1). Further, it was demonstrated that all hard kernel bread wheats possessed a mutation in puroindoline a or b (Morris and Bhave 2008), which disrupts the co-operative softening of the puroindolines. For example, if puroindoline a was absent, the kernel texture was intermediate between soft wheat and durum wheat (Morris and Bhave 2008; Bhave and Morris 2008a). Many of the observations related to puroindolines and kernel texture have some parallel to their antimicrobial activity (Bhave and Morris 2008b).
Puroindoline chemistry and structure
Puroindolines share many features of other antimicrobial proteins, especially those in wheat and related taxa. Figure 1 shows the amino acid sequence of the puroindolines. A key feature of these types of proteins is their conserved cysteine backbone, in the case of the puroindolines, 10 cysteines each (Fig. 2). Puroindolines have an uninterrupted ORF of 444 nt, 148 amino acid residues (~ 13 kDa), with cleavable N- and C-terminal signal and propeptides (Gautier et al. 1994), for mature proteins of 115–120 residues (Fig. 1). Minor C-terminal variants of PINA were reported by Blochet et al. (1993) and Gautier et al. (1994).
General structure of puroindoline a and b, and related proteins. The conserved cysteine backbone is shown in boxes, with the number of intervening amino acid residues indicated. Adapted from Gautier et al. (1994). Cysteines colored blue sub-tend the tryptophan rich domain and form a disulfide bond
Puroindoline a and b, although paralogous, share only 60% identity at the amino acid level and 70% identity at the nucleic acid level (coding region) (Fig. 1). In vitro at pH 7.0, they have a similar secondary structure comprised of about 30% α-helix (bundle of four helices, about 13–14 residues each), 30% β-sheets, and 40% unordered structure (Le Bihan et al. 1996). This secondary structure changes, however, when puroindolines interact with lipids and membranes (Bottier et al. 2008). Their pIs have been estimated from 10.5 to 11, with net charges of + 5 (PINA) and + 8 (PINB) (Blochet et al. 1993; Gautier et al. 1994). The most notable feature of puroindoline a and b are their tryptophan-rich domains (TRDs) (Fig. 1). Generally, these are defined as WRWWKWWK (PINA) and WPTKWWK (PINB), although the two sub-tending cysteines, which form a disulfide bond, create extended loops of 18 or 19 residues for each protein (Fig. 2).
Interaction with lipids
Observations associated with friabilin
Unbeknownst to Greenwell and Schofield (1986) when they first reported the discovery of friabilin, it was their method of starch isolation that essentially produced the observed phenomenon. A minor fraction of the total seed friabilin was associated with the surface of the starch granules and qualitatively related to the kernel texture (Jolly et al. 1993, 1996; Greenblatt et al. 1995). Later, Greenblatt et al. (1995) would show that friabilin could be extracted from starch using aqueous propan-2-ol and NaCl (90:10, propan-2-ol:water, 100 mM NaCl), indicating both hydrophobic and ionic interactions were involved. Neither propan-2-ol nor NaCl was effective alone. Of interest, bound glycolipids and phospholipids were also qualitatively associated with the starch granule surface in the same relationship as kernel texture and friabilin. It could not be fully resolved exactly how the friabilin-lipid-starch interaction occurred. Nevertheless, as will be discussed next, a key feature of puroindolines is their ability to interact with certain lipids under certain conditions.
Finnie et al. (2010) provided a more detailed analysis using near-isogenic wheat lines that differed only for puroindoline gene/protein sequences (Fig. 1) (Morris and King 2008). They found the same pattern of interaction of puroindolines with the lipids as reported earlier (Greenblatt et al. 1995). In whole grain, bound polar lipids were present at approximately 3200 nmol/g, and did not differ among the four wheat puroindoline haplotypes. When isolated starch was examined, a dramatic difference was observed, both in reduction and in differential: ~ 476 nmol/g for the wild-type puroindolines (soft kernel texture), and a range of 39 to 111 nmol/g for the mutant haplotypes. The lowest amount of bound polar lipids was present in the mutant line that lacked puroindoline a; the two puroindoline b mutants were intermediate, again suggesting the possibility of co-operative binding. The predominant lipids were DGDG and MGDG (Kim et al. 2012) (Table 1), representing 82% of bound polar lipids present on the surface of wild-type starch.
Direct interaction of puroindolines and TRDs with lipids
Wilde et al. (1993) were the first to study the interaction of purified puroindoline lipids and found that puroindoline bound approximately five LPC molecules in a co-operative manner. Dubreil et al. (1997) found that puroindoline a was capable of binding tightly to both wheat phospholipids and glycolipids. In contrast, puroindoline b interacted tightly only with negatively charged phospholipids, and formed loose lipoprotein complexes with glycolipids. Wheat phospholipids were 60% NAPE whereas wheat glycolipids were mainly neutral galactolipids (MGDG, DGDG).
Kooijman et al. (1997) were perhaps the first to directly show that the tryptophan loop was involved in binding lipids. Lipids could be bound as monomers, without the need for micelle formation. In addition to hydrophobic interactions of the tryptophan residues with the lipid, an electrostatic interaction between the negative head group of the lipid with the tryptophan loop enhanced the strength of binding. They also appear to be the first to use a synthetic TRD peptides (Fig. 1 and Tables 2 and 3). In their subsequent report, Kooijman et al. (1998) used an interfacial film comprised of MGDG to demonstrate that PINA and the PINA TRD both had a strong interaction with the monolayer. PINA initially interacted with the surface, and then inserted itself into the monolayer. It should be kept in mind that the TRD was a linear peptide, whereas the TRD is restrained as a loop in the native protein (Figs. 1, 2).
Le Guernevé et al. (1998) extended this work by showing that the interactions of PINA with model phospholipid bilayers and micelles depended on the headgroup, acyl chain length, ionic environment, and lipid to protein concentration. Although PINA interacted with zwitterionic phospholipids, the interaction with negatively charged headgroups was much stronger. Interaction with phosphatidylcholine was weak and was with the surface of the bilayer. With phosphatidylglycerol, not only was the interaction much stronger, but PINA partially penetrated the bilayer, disrupting the acyl chain packing. Dubreil et al. (2003) reported additional details on the interaction of PINA with monolayers comprised of the zwitterionic DPPC and anionic DPPG. PINA interacted strongly with both, but much more so with DPPG.
Beyond these informative lipid interaction studies, Charnet et al. (2003) showed how PINA could form ion channels in model lipid bilayers comprised of PC:PE 7:3. Channels also formed with the inclusion of PS (PC:PE:PS 7:3:1), although the incorporation of PINA was slower (see below). The studies of Llanos et al. (2004, 2006) indicated that PINA formed single channel cationic pores in giant liposomes with selectivity K+ > Na+ > > Cl−. Bottier et al. (2008) used monolayers of the galactolipids MGDG and DGDG to study the interaction of the puroindolines with lipids. MGDG and DGDG are the primary components of amyloplast membranes. In MGDG, PINA forms a dense interconnected network with no aggregation. PINB, however, penetrates the monolayer film and forms round domains with diameters ~ 100–200 nm. In the case of DGDG, PINA forms isolated domains of one to several micrometers, whereas PINB again forms patches of ~ 100–200 nm, but also larger domains of irregular form.
Clifton, Sanders and co-workers have published a highly informative series of papers on how puroindoline a and b interact with lipids, with each other, and with themselves. A particularly useful aspect of this research was the use of naturally occurring puroindoline b mutants (Fig. 1). Clifton et al. (2007b) used condensed monolayers of zwitterionic DPPC and anionic DPPG to compare the interaction of puroindoline b wild-type with two natural mutants (identified as G46S and W44R; amino acid residue, its position, followed by the mutant residue) (Fig. 1). They observed a stronger interaction of PINB wild-type compared to the mutant forms, with PINB G46S ranked next, and then PINB W44R. However, wild-type PINB showed a greater selectivity for DPPG than the other two mutant forms. Based on surface pressure changes, PINB wild-type penetrated the lipid monolayer to the greatest extent, followed by PINB G46S, and significantly less by PINB W44R. The authors extended their results to suggest that the mutant PINBs G46S and W44R would have reduced antimicrobial activities compared to wild-type PINB. Of note, both mutations occur in the TRD, and both are associated with hard kernel texture (Morris and King 2008).
In subsequent reports, Clifton et al. (2007a,2008) studied the interaction of PINA with PINB wild-type and the previously described two mutant forms. In addition to PINA + PINB combinations, PINA:PINB ratios of 3:1, 1:1 and 1:3 were examined. Of the individual puroindolines, PINA seemed to interact more with the surface of the DPPG monolayer, whereas PINB wild-type penetrated more deeply into the monolayer. When combined with PINA, the mutant PINB proteins penetrated the monolayer significantly less than PINA + PINB wild-type. PINA + PINB wild-type penetration was similar to PINB wild-type alone. Clifton et al. (2011b) reported that PINA interacted mainly with the head-group of the condensed DPPG monolayer, to form a 33 Å thick layer below the lipid film. The ranking of changes in surface pressure to the DPPC monolayer (equated as the degree of puroindoline insertion) was PINB wild-type > PINA > and PINB mutant W44R (Sanders et al. 2013). When DPPE and DPPG were used alone and in various monolayer combinations, the greatest pressure change was with PINB wild-type and a 9:1 DPPG:DPPE, although 100% DPPG was nearly the same (Sanders et al. 2016). When the same experiments were conducted with PINB W44R mutant, the 9:1 lipid mixture had the greatest effect, but less than with PINB wild-type (10.8 vs. 8.9 mN/m). However, whereas PINA decreased pressure of most of the DPPG and DPPE mixtures by ~ 7 mN/m, changes in pressure due to PINB W44R were much less, and with the 100% DPPE monolayer, the change was nearly zero, indicating different effective insertion of PINB wild-type versus the mutant protein, and different interactions based on lipid mixture (Sanders et al. 2016). Sanders et al. (2017) examined model eukaryotic membranes, including cholesterol and sphingomyelin in DPPG + DPPC monolayers. Again, PINB wild-type was compared to the PINB W44R with a modified TRD (Fig. 1). The mutant PINB protein was less responsive to changes in lipid monolayer composition, and although it was able to absorb to the monolayer surface, it did not penetrate the layer as effectively a PINB wild-type. One additional report from this group (Clifton et al. 2011a) showed that PINA spontaneously self-assembled into prolate ellipsoidal micelles of ca. 112 Å (major axis), which were comprised of 38 PINA molecules. These micelles were stable over wide ranges of pH and temperature. As a final comment, these studies observed an evocative relationship between reduced lipid interaction/monolayer penetration of the mutant PINB proteins that correlated with the soft-to-hard kernel texture change in wheat.
Alfred et al. (2013b) constructed large unilamellar vesicles (LUVs) from DOPC:DOPG 1:3 to mimic bacterial membranes or DOPC alone to mimic mammalian membranes (Table 1). At a typical MIC (‘minimum inhibitory concentration’ = the lowest concentration able to completely prevent bacterial growth) of 16 μg/mL against E. coli, PINA TRD (Table 2) increased leakage and changed the morphology of DOPC:DOPG LUVs, but did not completely lyse them; PINA had no effect on the DOPC LUVs. PINB had no effect. Results with PINA P35S was similar to the PINA wild-type TRD sequence. The authors concluded that “The results indicate that the peptides [PINA TRDs] disrupted the negatively charged phospholipid vesicles but did not cause complete lysis… and were ineffective against zwitterionic lipid vesicles.”
Lastly, Keller (2018) described an in silico bioinformatic approach to identifying lipid binding regions of cereal proteins. This approach identified three binding sites each in PINA and PINB. The author mentioned that the number of lipid binding regions was different from the actual number of lipid molecules that could be bound.
Antimicrobial activity
Native puroindoline proteins in situ
In contrast to kernel texture, few studies have examined the antimicrobial role of native puroindolines in situ in wheat. Zhang et al. (2016) reportedly found PINB in the embryos of a soft wheat variety but not a hard variety. This simple observation was suggested to relate antimicrobial activity and hence response of grain to storage pathogens. The results and this line of logic were not particularly convincing given that (1) their sequence had only 12% coverage with PINB, (2) the results were completely confounded with the two varieties, and (3) no storage treatment/microbe challenge was reported.
Tripathi et al. (2013) found that PINB transcripts were present from 0 to 12 h after inoculation of wheat with Tilletia indica (karnal bunt) in both a resistant and a susceptible variety, but then they were not detected at 24 and 48 h after inoculation. This pattern of gene expression did not seemingly have any direct relationship to bunt resistance, and the “zero” time sampling was not precisely indicated. Of note, the basidiospores were introduced with a syringe so a wound response could not be ruled out. Of some relevance, Kiszonas et al. (2019) recently reported that durum wheat possessing the puroindolines chromosome translocation (Morris et al. 2011) were highly resistant to dwarf bunt (Tilletia controversa).
Puroindoline proteins expressed trans-genetically
Some of the more compelling results related to the antimicrobial activity of the puroindolines against plant pathogens come from transgenic studies. The first involved a series of three studies on transgenic apple (Malus x domestica) cvs. Galaxy and Ariane, and resistance to apple scab (Venturia inaequalis) (Tables 4, 5). Galaxy is scab-susceptible, whereas Ariane carries a resistance gene (Vf) to scab. Chevreau et al. (2001) reported that PINB at 200 μg/mL significantly inhibited scab mycelium growth in vitro. Subsequently Chevreau et al. (2004) showed that when PINB coding sequence was expressed under the control of the CaMV35S promoter, transgenic plants inoculated in a growth chamber had a significant reduction in scab pathogenicity. The responses were clearly an interaction between pathogen strain and host resistance: with the common V. inaequalis strain 1, there was no decrease in susceptibility, whereas with the more virulent strain 6, there was a significant reduction in susceptibility. Conversely, cv. Ariane (resistant to strain 1), showed significant reduction in susceptibility to strain 6. In both Galaxy and Ariane, there was a significant negative correlation between PINB expression and susceptibility to V. inaequalis strain 6. In their third report, Faize et al. (2004) used the same cvs. Galaxy and Arianne expressing the PINB transgene and challenged with the same two races of scab. PINB had an effective MIC at 200 μg/mL against V. inaequalis mycelium growth in vitro. PINB reduced scab in transgenic susceptible cv. Galaxy against strain 6, but had no effect against strain 1. In resistant cv. Ariane transgenic lines with PINB, resistance to strain 6 was observed (Ariane is normally susceptible to stain 6). The quantitative amount of expressed PINB was correlated with disease reduction across individual transgenic trees. These studies are noteworthy in that they demonstrate the usefulness of PIN expression in a distantly related, woody taxon.
Krishnamurthy et al. (2001) found that transgenic rice (Oryza sativa) plants expressing PINA, PINB or PINA + PINB had improved resistance to rice blast (Magnaporthe grisea) and sheath blast (Rhizoctonia solani) (Tables 4 and 5). Rice blast symptoms were delayed and disease severity was reduced. The combination of PINA + PINB was better than either protein alone. Similarly, sheath blast severity was reduced in plants expressing PINA and PINB; but again the combination of both was better. The authors concluded that the PINs inhibited fungal growth, but not infection. Leaf extracts from transgenic plants reduced fungal growth in vitro.
The next study to examine the role of puroindolines in pathogen resistance did not use recombinant PIN proteins, but rather the puroindoline gene promoters which were expressed trans-genetically in planta in rice (Evrard et al. 2007; cf. Simeone et al. 2006) (Tables 4, 5). Promoter activity was induced by wounding/inoculating the plants with rice blast. The longest promoter sequence for PINA (1214 bp upstream ATG start codon), and a truncated 390-bp sequence were sufficient for expression in seeds, whereas a truncated 214-bp promoter was not. In transgenic plants inoculated with rice blast, the PINA promoter found to be not seed specific, but could induce expression in roots, flowers and leaves, although the highest expression was in seeds. PINB was observed to be seed specific, consistent with Digeon et al. (1999). Further, the PINB promoter did not respond to wounding or pathogen attack.
As described above, durum wheat lacks puroindolines, but have been introduced through transformation. Luo et al. (2008) produced transgenic durum wheat varieties expressing PINA, which showed enhanced resistance to leaf rust (Puccinia triticina) (Tables 4, 5). Transgenic plants showed delayed appearance of infection after inoculation and more rapid disappearance of disease symptoms after fungicide treatment. Similar to Krishnamurthy et al. (2001), they concluded that PINA did not prevent infection completely, but slowed fungal growth.
The next in planta expression of PINs was that of Zhang et al. (2011). Here, PINA and PINB were introduced trans-genetically into Zea mays (maize) (Tables 4, 5). PINs significantly increased tolerance to Chochliobolus heterostrophus (Southern corn leaf blight), averaging a 42% reduction in symptoms. Similar to rice (Krishnamurthy et al. 2001; Luo et al. 2008), the initial fungal infections were similar in PIN positive and PIN negative transgenic plants, the increased tolerance to disease was due to inhibition of the fungal growth.
Native puroindoline proteins tested in vitro
The next topic is the use of native (wheat derived) puroindoline proteins and their antimicrobial activity in vitro. Dubreil et al. (1998) was the first to study the antimicrobial activity (Tables 4, 5) of puroindolines. Five fungi were tested (Alternaria brassicola, Ascochyta pisi, Botrytis cinereal, Fusarium culmorum, and Verticillium dahlia) with PINA and PINB. In vitro results showed that the IC50 (concentration for 50% growth inhibition) was greater than or equal to 100 μg/mL for PINA and 20–70 μg/mL for PINB against A. brassicola, A. pisi, F. culmorum, and V. dahliae; the PINs were not effective against B. cinerea. In combinations, the PINs showed some synergy against F. culmorum. In contrast to the studies of apple scab, above, pear (Pyrus communis) fire blight (Erwinia amylovora) was not affected by PINA at concentrations up to 200 μM in vitro (Mourgues et al. 1998).
In a series of four papers, Capparelli and co-workers (Capparelli et al. 2005, 2006, 2007; Palumbo et al. 2010) examined the effect of PINA and PINB against various microbes. The first (Capparelli et al. 2005) included Gram-positive and -negative bacteria in vitro: Agrobacterium tumefaciens, Clavibacter michiganensis, E. coli, Erwinia carotovora, Pseudomonas syringae phaseoli, and Staphylococcus aureus (Tables 4, 5). Against all bacteria, the MIC was the same for PINA and PINB, 30–50 μg/mL. Synergism was noted when the two PINs were used together against E. coli: PINB at 2/3 MIC + 1 μg/mL PINA produced 100% inhibition. At 10X MIC (300 μg/mL), both PINA and PINB killed bacteria to nearly undetectable levels. Their subsequent studies (Capparelli et al. 2006, 2007; Palumbo et al. 2010) used recombinant PINs and are presented below in the next section.
Chugh et al. (2015) isolated PINA and PINB from soft wheat grain and used them in in vitro assays against E. Coli, Serratia marcenscens and S.Aureus (Tables 4, 5). PINs at 1 mg/mL produced roughly 40–50% inhibition growth of all three microbes.
Recombinant puroindoline proteins in vitro
Although a few studies used native PINs isolated from wheat grain or flour, for various reasons the following studies employed recombinant proteins. Capparelli et al. (2006) produced recombinant PINA and PINB, which were tested in vitro against E. coli, S. aureus and S. epidermis (Tables 4, 5) at 0.125–40 μg and 106 cells. The MIC was essentially the same as the native PINs, 30 μg/mL (Capperelli et al. 2005). PINB alone at tenfold dilution killed 70% of E. coli, whereas PINA had little effect at this concentration. Differences were observed among microbe response: much higher concentration of PINs was required to kill S. epidermis (5 μg/106 cells, PINB killed 56% of cells) compared to E. coli (1.25 μg/106 cells, killed 100% of the cells). In their third paper, Capparelli et al. (2007) produced correctly folded recombinant PINA and PINB which showed similar antimicrobial activity as native proteins on S. epidermis (PINA and PINB each had a MIC90 of 30 μg/mL). Bactericidal activity of recombinant PINA was 125 μg/mL and PINB was 42 μg/mL. In a second assay, PINA and PINB killed all S. Epidermis cells at 20 μg/mL, and could kill intracellular Staphylococci by entering infected mouse monocyte macrophage cells. PINA or PINB at tenfold higher concentrations (1250 μg/mL PINA, 400 μg/mL PINB) used for extracellular bacteria reduced intracellular bacteria by 3 log units within 3 h.
Sorrentino et al. (2009) transformed tobacco (Nicotiana tabacum) cell cultures with recombinant PINA and PINB constructs. The PINB construct was present in the total soluble protein fraction of the cell culture, and inhibited E. coli growth by 90%, and produced both bacteriostatic and bactericidal activities. The effective PINB concentrations were estimated at 15 and 30 μg/mL.
Miao et al. (2012) constructed PINA with 1 (wild-type), 2 or 3 copies of the TRD (21 AA each) (Fig. 1, Tables 4, 5). MIC of the three proteins was assessed against E. coli and S. aureus. One additional TRD decreased MIC 20–30% (MIC = 70 μg/mL E. coli, 120 μg/mL S. aureus) whereas two additional TRDs were less effective with an increased MIC compared to the wild-type. Bactericidal activity was also highest with two TRDs, and reduced with three TRDs. Circular dichroism analysis indicated that the three TRD form had increased alpha helix, with several Trp residues shielded as opposed to exposed at the surface of the loop.
Niknejad et al. (2016) tested purified PINA and PINB recombinant proteins against Colletotrichum graminicola, Drechslera brizae (post-harvest rot), E. coli, Fusarium oxysporum, Rhizoctonia cerealis, R. solani (sheath blight), and S. aureus. Both PINs retained antimicrobial activity. MICs were as low as 32 μg/mL for PINA and PINB against E.coli, and 32–250 μg/mL for both proteins against R. cerealis, R. solani, and S. aureus. PINs exhibited low activity (MIC ≥ 500 μg/mL) against C. graminicola, D. brizae, and F. oxysporum.
Recombinant puroindoline proteins in situ
Evrard et al. (2008) created recombinant PINs (Fig. 1, Tables 2, 3) point mutations in the TRD, and then used those constructs to transform Saccharomyces cerevisiae in a complementation assay to determine which residues were key to allowing PINs to interact with the plasma membrane (Tables 4, 5). In this yeast two-hybrid system, each PIN was able to function individually. Using this system, they showed that PINA residues W41 and W44 were essential for PINA interaction with the yeast plasma membrane. For PINB, it was found that tryptophan residues K42 and/or K45 were involved in PINB membrane interaction. They suggested that likely ionic interactions were also likely (cf. Greenblatt et al. 1995). PINB interaction required that the yeast be grown on glucose but not galactose, indicating that the composition of the plasma membrane was critical. Dubreil et al. (1997) had shown that PINA was capable of binding both phospholipids and glycolipids in vitro, but that PINB interacted tightly only with negatively charged phospholipids.
Palumbo et al. (2010) used recombinant PINA and PINB (see Capparelli et al. 2007) against Listeria monocytogenes in situ (mice, Mus musculus) in infected mice (Tables 4, 5). Highest bacteria counts were in the liver after 7 days of infection. PINA and PINB individually injected at 5 mg/mouse inhibited bacteria growth completely (in vitro PINA and PINB were bactericidal). When combined with lactoferrin and lysozyme, PINA at 59 μg/mouse and PINB at 19 μg/mouse completely inhibited growth of L. monocytogenes. PINA and PINB individually significantly reduced the expression level of pro-inflammatory cytokines, acute phase protein, and T lymphocyte antigens. PINA and PINB exhibited prophylactic activity against L. monocytogenes at 1.25 and 0.312 mg/mouse, respectively. Activity was defined as a 3 log reduction in cfu in livers. However, when administered concurrently with lactoferrin, complete sterilization was achieved with 14 μg PINA, 5 μg PINB, and 78 μg lactoferrin per mouse.
TRDs of puroindoline proteins in vitro
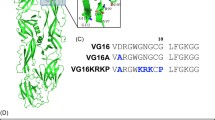
This section covers the testing of various TRDs in vitro against microorganisms. Jing et al. (2003) tested the 13-AA TRD of PINA and the 12-AA TRD of PINB against S. aureus and E. coli (Tables 2, 3, 4, 5). The PINA MIC against E. coli was 7 μM and against S. Aureus, 16 μM. PINB was not found to be effective with an MIC > 200 μM, and was not considered further. PINA disrupted phospholipid unilamellar vesicles. Leakage was greatest when the proportion of negatively charged headgroups of phospholipid increased. These results were consistent with PINA perturbing negatively charged bacterial membranes versus mammalian cells (i.e. very low hemolytic activity, see below). They indicated that PINA resides at the interface of membranes and does not penetrate deeply nor form pores, although the location of PINA in the membrane will vary with charge and structure of the lipid polar headgroup.
Phillips et al. (2011) produced nine synthetic peptides based on the PINA and PINB TRDs. All began with F34 (PINA 13 AA residues) or F35 (PINB 12 AA residues) (Tables 2, 3, 4, 5). PINA wild-type TRD exhibited an MIC of 16 μg/mL against E. coli and S. aureus. For Rhizoctonia cerealis and R. solani, the MICs were 32 and 64 μg/mL, respectively. MICs against Collectotrichum graminicola, Drechslera brizae, and Fusarium oxysprorum ≥ 250 μg/mL. Changing the sequence to P35S, increased activity slightly against the bacteria (MICs of 13 μg/mL); other changes decreased activity, but not dramatically. Wild-type PINB was not particularly effective against the bacteria and C. graminicola, D. brizae, and F. Oxysprorum (> 250 μg/mL). None of the altered sequences improved activity. PINB wild-type was most effective against the two Rhizoctonia species (MIC of 64 μg/mL). But again, sequence alterations did not improve efficacy.
Alfred et al. (2013a) extended the work of Phillips et al. (2011) to examine the effects of PINA and PINB TRD peptides on wheat rusts, Puccinia triticina (leaf rust) and P. stiiformis f. tritici (stripe rust) (Tables 2, 3, 4, 5). PINA, PINA R39G, and PINB adversely affected the morphology of stripe rust spores, whereas PINA and PINB inhibited spore germination. PINA and PINB when sprayed onto the leaves of stripe rust infected plants showed a moderate reduction in the number of uredia, as did PINB when sprayed on leaves 5 days prior to spore infection.
Alfred et al. (2013b) observed inhibition of yeast (Saccharomyces cerevisiae) cell growth by PINA, PINA P35S, PINA W → F (all three Ws), and PINB TRDs (Tables 2, 3) at MICs of 125 or 250 μg/mL. As assessed by propidium iodide (PI) uptake and fluorescence, PINA and PINA P35S at 64 μg/mL caused 100% of the cells to take up PI. PINA W → F was less effective, and PINB had no effect. SEM corroborated the morphological changes to the cell membranes due to TRDs. The various PINA TRDs also showed complete inhibition of DNA migration in gel retardation assays, indicating strong binding. DNA binding was related to the net charge of the TRDs, with the strongest binding at + 3. The DNA binding assay agreed well with the filamentous growth of E. coli.
Haney et al. (2013) synthesized TRD sequence variants of PINB (Table 3) and determined their MIC against E. coli and S. Aureus. Whereas PINB wild-type had MICs of > 125 μg/mL, altered PINB TRD sequences had MICs as low as 3–15 μg/mL (E. Coli) and < 3 μg/mL (S. aureus). As a comparison, wild-type PINA had MICs of 5 and 20 μg/mL for E. coli and S. Aureus, respectively (Jing et al. 2003). All of these peptides were considered to have low mammalian toxicity. Based on a number of methods, the authors concluded that the PINB TRDs have a strong binding constant for nucleic acids and that their primary antimicrobial mode of action is intra-cellular as opposed to membrane disruption.
Shagagahi et al. (2016) used PINA and PINB wild-type, and PINA P35S TRD peptides (Tables 2, 3) against Bacillus subtillis, Pseudomonas aeruginosa, Listeria monocytogenes, and L. innocua (Tables 4, 5). MICs for PINA were 8 μg/mL against B. subtillis, L. monocytogenes, and L. innocua, and 64 μg/mL against P. aeruginosa. PINB P35S was equally or more highly effective than PINA. With the exception of B. Subtillis (MIC of 64 μg/mL), wild-type PINB MICs were > 250 μg/mL. This study was unique in that it showed that PIN TRDs could inhibit the initial cell attachment of planktonic cells at the MIC level. The PIN TRDs were also effective against preformed biofilms. Lastly, this study showed that PIN TRDs effectively reduced viability of B. Subtillis spores by 2–3 log. In their follow-up report (Shagagaghi et al. 2017), they examined the mode of action of PINA TRD and Candida albicans (Tables 2, 3, 4, 5). At PNA concentrations of 0.5 × and 1 × MIC (64 and 125 μg/mL, respectively), pores formed on the membrane surface, and viable cells were reduced by 85% after 30 min, with complete (> 99%) kill after 60 min. They reported that PINA arrested cell proliferation at the S-phase, thus inhibiting normal cellular processes. Low levels (8 μg/mL) of fluor-labelled PINA was seen entering the cells within 30 s and locate to the nucleus. After 20–40 min, PINA accumulated at the cell membrane, which was eventually disrupted allowing PI to enter via pores.
Boden et al. (2018) immobilized PINA TRD (Table 2) to 2 μm carboxylated polystyrene beads, and in conjunction with smaller (0.11 μm) poly(methyl methacrylate) particles, produced ‘binary colloidal crystal’ monolayers. This coating significantly decreased the viability of adherent E. coli cells, which were morphologically deflated due ostensibly to leakage of cellular contents.
Interaction with animal cells
Not all studies involving puroindolines and their TRDs have focused exclusively on antimicrobial properties. In this section, those studies involving animal cells are reviewed. These studies have generally had one of two objectives: (1) the examination of the effect of puroindolines on membranes and cell function, or (2) the establishment of the MIC (in this context, the “safety” of PINs). Naturally, the two are directly related. Mattei et al. (1998) found that PINA induced swelling in frog (Xenopus) myelinated axons at 10 and 100 μM, but not at 1 μM (Table 6). They suggested that the effect was due to increased internal osmolality due to PINA forming pores in the membrane such that external Na+ and water entered the cell. Charnet et al. (2003) examined PINA and PINB singly (20–50 μM each) and found the formation of ion channels in the membranes of Xenopus oocytes at 50 μM. Ion passage was restricted to cations, with selectivity in the order of Cs+ > K+ > Na+ > Li+ > choline = tetraethylammonium. Ca2+ inhibited pore formation. However, this pore formation was voltage dependent. No effect of PINA was observed on resting oocytes, whereas pores formed in hyperpolarized cells. Similarly, Llanos et al. (2006) found that PINA at 1 μM had no effect on the membrane potential of mouse muscle fibers, whereas at 10 μM, PINA caused hyperpolarization of the muscle membrane, likely due to loss of K+ through cation channels. At 50 μM or above, neuroblastoma cells swelled and formed blebs.
Regarding the “safety” of PINs, and as has been discussed throughout, the distinct compositional differences of microbial vs. Plant and animal cells is directly related to the way in which PINs interact with the lipids comprising their membranes. Jing et al. (2003) found that to achieve 50% hemolysis (HD50) of erythrocytes, the concentration of PINA and PINB TRDs had to be greater than > 1000 μg/mL (an actual HD50 value was not provided). Capparelli et al. (2007) reported no hemolytic activity of recombinant PINA or PINB at 150 and 50 µg/mL, respectively. Phillips et al. (2011) reported that hemolysis by PINA and PINB required > 500 μg/mL, and Niknejad et al. (2016) found little hemolytic activity against erythrocytes at the MIC effective against E. coli (32–64 μg/mL). Again, actual HD50 values were not provided. Haney et al. (2013) synthesized PINB TRDs with altered sequences (Table 3); all of these peptides were considered to have low mammalian toxicity (erythrocyte HD50 > 300 or 600 μg/mL).
Concluding remarks
Humans have an unrelenting challenge to protect themselves, their crops, livestock and pets from pathogenic microbes. In this endeavor, antimicrobial proteins and peptides (AMPs) from higher organisms will likely play an important role as they seemingly have millions of years of evolutionary advantage over bacteria and fungi due to differences in the composition of their membranes. Consequently, many antimicrobial proteins and AMPs from higher organisms such as the puroindolines and their TRDs of wheat are ‘natural antibiotics’ with anti-fungal, bactericidal and bacteriostatic properties. This review has documented the studies related to the examination of the puroindolines, their mutant forms and their tryptophan-rich domains (TRDs) with regards to how they interact with lipids, natural and artificial membranes, and a long list of microbes (Table 5). The efficacy of the puroindolines and their TRDs show a complex range of interactions. These interactions undoubtedly reflect to some extent the various experimental procedures, nevertheless, puroindolines clearly have the ability (and potential) to disrupt bacterial membranes and bacterial growth and reproduction, decrease fungal spore growth and reproduction, while simultaneously, having low toxicity to mammalian cells. As such, this area of research (and its eventual practical application) will undoubtedly continue and expand. Specifically, when and how to express puroindolines and TRDs transgenetically, how to make TRDs more potent (e.g. sequence variants), how to scale up the production of puroindolines and TRDs for pathogen control, and what microbes to target will be essential areas of future work.
References
Alfred RL, Palombo EA, Panozzo JF, Bariana H, Bhave M (2013) Stability of puroindoline peptides and effects on wheat rust. World J Microbiol Biotechnol 29:1409–1419
Alfred RL, Palombo EA, Panozzo JF, Bhave M (2013) The antimicrobial domains of wheat puroindolines are cell-penetrating peptides with possible intracellular mechanisms of action. PLoS ONE 8:e75488
Avci FG, Akbulut BS, Ozkirimli E (2018) Membrane active peptides and their biophysical characterization. Biomolecules 8:77
Bettge AD, Morris CF, Greenblatt GA (1995) Assessing genotypic softness in single wheat kernels using starch granule-associated friabilin as a biochemical marker. Euphytica 86:65–72
Bhave M, Morris CF (2008) a) Molecular genetics of puroindolines and related genes: allelic diversity in wheat and other grasses. Plant Mol Biol 66:205–219
Bhave M, Morris CF (2008) b) Molecular genetics of puroindolines and related genes: regulation of expression, membrane binding properties and applications. Plant Mol Biol 66:221–231
Blochet J-E, Kaboulou A, Compoint J-P, Marion D (1991) Amphiphilic proteins from wheat flour: specific extraction, structure and lipid binding properties. In: Bushuk W, Tkachuk R (eds) Gluten Proteins 1990. AACCI, St. Paul, MN, pp 314–325
Blochet J-E, Chevalier C, Forest E, Pebay-Peyroula E, Gautier M-F, Joudrier P, Pezolet M, Marion D (1993) Complete amino acid sequence of puroindoline, a new basic and cysteine-rich protein with a unique tryptophan-rich domain, isolated from wheat endosperm by Triton X-114 phase partitioning. FEBS Lett 329:336–340
Boden A, Bhave M, Wang P-Y, Jadhav S, Kingshott P (2018) Binary colloidal crystal layers as platforms for surface patterning of puroindoline-based antimicrobial peptides. ACS Appl Mater Interfaces 10:2264–2274
Bottier C, Gean J, Desbat B, Renault A, Marion D, Vie V (2008) Structure and orientation of puroindolines into wheat galactolipid monolayers. Langmuir 24:10901–10909
Capparelli R, Amoroso MG, Palumbo D, Iannaccone M, Faleri C, Cresti M (2005) Two plant puroindoline colocalize in wheat seed and in vitro synergistically fight against pathogens. Plant Mol Biol 58:857–867
Capparelli R, Palumbo D, Iannaccone M, Ventimiglia I, Di Salle E, Capuano F, Salvatore P, Amoroso MG (2006) Cloning and expression of two plant proteins: similar antimicrobial activity of native and recombinant form. Biotechnol Lett 28:943–949
Capparelli R, Ventimiglia I, Palumbo D, Nicodemo D, Salvatore P, Amoroso MG, Iannaccone M (2007) Expression of recombinant puroindolines for the treatment of staphylococcal skin infections (Acne vulgaris). J Biotechnol 128:606–614
Chantret N, Salse J, Sabot F, Rahman S, Bellec A, Laubin B et al (2005) Molecular basis of evolutionary events that shaped the Hardness locus in diploid and polyploidy wheat species (Triticum and Aegilops). Plant Cell 17:1033–1045
Charnet P, Molle G, Marion D, Rousset M, Lullien-Pellerin V (2003) Puroindolines form ion channels in biological membranes. Biophys J 84:2416–2426
Chevreau E, Dupuis F, Ortolan C, Parisi L (2001) Transformation of apple for durable scab resistance: expression of a puroindoline gene in susceptible and resistant (Vf) genotypes. Proceedings of 4th international symposium in vitro culture and horticulture breed 560:323–326
Chevreau E, Faize M, Dupuis F, Sourice S, Parisi L (2004) Combination of a transgene-mediated defense mechanism with a natural resistance gene increases apple scab resistance. Acta Hort 663:447–452
Chugh V, Kaur K, Singh D, Kumar V, Kaur H, Dhaliwal HS (2015) Molecular characterization of diverse wheat germplasm for puroindoline proteins and their antimicrobial activity. Turkish J Biol 39:359–369
Clifton LA, Green RJ, Frazier RA (2007) Puroindoline-b mutations control the lipid binding interactions in mixed Puroindoline-a:Puroindoline-b systems. Biochemistry 46:13929–13937
Clifton LA, Lad MD, Green RJ, Frazier RA (2007) Single amino acid substitutions in puroindoline-b mutants influence lipid binding properties. Biochemistry 46:2260–2266
Clifton LA, Green RJ, Hughes AV, Frazier RA (2008) Interfacial structure of wild-type and mutant forms of puroindoline-b bound to DPPG monolayers. J Phys Chem B 112:15907–15913
Clifton LA, Sanders MR, Castelletto V, Rogers SE, Heenan RK, Neylon C, Frazier RA, Green RJ (2011) Puroindoline-a, a lipid binding protein from common wheat, spontaneously forms prolate protein micelles in solution. Phys Chem Chem Phys 13:8881–8888
Clifton LA, Sanders MR, Hughes AV, Neylon C, Frazier RA, Green RJ (2011) Lipid binding interactions of antimicrobial plant seed defence proteins: puroindoline-a and B-purothionin. Phys Chem Chem Phys 13:17153–17162
Digeon J-F, Guiderdoni E, Alary R, Michaux-Ferriere N, Joudrier P, Gautier M-F (1999) Cloning of a wheat puroindoline gene promoter by IPCR and analysis of promoter regions required for tissue-specific expression in transgenic rice seeds. Plant Mol Biol 39:1101–1112
Dubreil L, Compoint J-P, Marion D (1997) Interaction of puroindolines with wheat flour polar lipids determines their foaming properties. J Agric Food Chem 45:108–116
Dubreil L, Gaborit T, Bouchet B, Gallant DJ, Broekaert WF, Quillien L, Marion D (1998) Spatial and temporal distribution of the major isoforms of puroindolines (puroindoline-a and puroindoline-b) and nonspecific lipid transfer protein (ns-LTP1e1) of Triticum aestivum seeds. Relationships with their in-vitro antifungal properties. Plant Sci 138:121–135
Dubreil L, Vie V, Beaufils S, Marion D, Renault A (2003) Aggregation of puroindoline in phospholipid monolayers spread at the air-liquid interface. Biophysical J 85:2650–2660
Evrard A, Meynard D, Guiderdoni E, Joudrier P, Gautier M-F (2007) The promoter of the wheat puroindoline-a gene (PinA) exhibits a more complex pattern of activity than that of the PinB gene and is induced by wounding and pathogen attack in rice. Planta 225:287–300
Evrard A, Lagarde V, Joudrier P, Gautier M-F (2008) Puroindoline-a and puroindoline-b interact with the Saccharomyces cerevisiae plasma membrane through different amino acids present in their tryptophan-rich domain. J Cereal Sci 48:379–386
Faize M, Sourice S, Dupuis F, Parisi L, Gautier M-F, Chevreau E (2004) Expression of wheat puroindoline-b reduces scab susceptibility in transgenic apple (Malus x domestica Borkh.). Plant Sci 167:347–354
Finnie SM, Jeannotte R, Morris CF, Faubion JM (2010) Variation in polar lipid composition among near-isogenic wheat lines possessing different puroindoline haplotypes. J Cereal Sci 51:66–72
Gautier M-F, Aleman M-E, Guirao A, Marion D, Joudrier P (1994) Triticum aestivum puroindolines, two basic cysteine-rich seed proteins: cDNA sequence analysis and developmental gene expression. Plant Mol Biol 25:43–57
Giroux MJ, Morris CF (1997) A glycine to serine change in puroindoline b is associated with wheat grain hardness and low levels of starch surface friabilin. Theor Appl Genet 95:857–864
Giroux MJ, Morris CF (1998) Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Proc Natl Acad Sci USA 95:6262–6266
Greenblatt GA, Bettge AD, Morris CF (1995) Relationship between endosperm texture and the occurrence of friabilin and bound polar lipids on wheat starch. Cereal Chem 72:172–176
Greenwell P, Schofield JD (1986) A starch granule protein associated with endosperm softness in wheat. Cereal Chem 63:379–380
Haney EF, Petersen AP, Lau CK, Jing W, Storey DG, Vogel HJ (2013) Mechanism of action of puroindoline derived tryptophan-rich antimicrobial peptides. Biochem Biophys Acta 1828:1802–1813
Heinze K, Kiszonas AM, Murray JC, Morris CF, Lullien-Pellerin V (2016) Puroindoline genes introduced into durum wheat reduce milling energy and change milling behavior similar to soft common wheat. J Cereal Sci 71:183–189
Jing W, Demcoe AR, Vogel HJ (2003) Conformation of a bactericidal domain of puroindoline a: structure and mechanism of action of a 13-residue antimicrobial peptide. J Bacteriol 185:4938–4947
Jolly CJ, Rahman S, Kortt AA, Higgins TJV (1993) Characterisation of the wheat Mr 15 000 “grain-softness protein” and analysis of the relationship between its accumulation in the whole seed and grain softness. Theor Appl Genet 86:589–597
Jolly CJ, Glenn GM, Rahman S (1996) GSP-1 genes are linked to the grain hardness locus (Ha) on wheat chromosome 5D. Proc Natl Acad Sci USA 93:2408–2413
Keller RCA (2018) Identification of potential lipid binding regions in cereal proteins and peptides with the use of bioinformatics. J Cereal Sci 80:128–134
Kim K-H, Feiz L, Martin J-M, Giroux MJ (2012) Puroindolines are associated with decreased polar lipid breakdown during wheat seed development. J Cereal Sci 56:142–146
Kiszonas AM, Higginbotham R, Chen XM, Garland-Campbell K, Bosque-Perez NA, Pumphrey M, Rouse MN, Hole D, Wen N, Morris CF (2019) Agronomic traits in durum wheat germplasm possessing puroindoline genes. Agron J doi: 10.2134/agronj2018.08.0534.
Kooijman M, Orsel R, Hessing M, Hamer RJ, Bekkers ACAPA (1997) Spectroscopic characterisation of the lipid-binding properties of wheat puroindolines. J Cereal Sci 26:145–159
Kooijman M, Orsel R, Hamer Bekkers ACAPA (1998) The insertion behaviour of wheat puroindoline-a into diacylgalactosylglycerol films. J Cereal Sci 28:43–51
Krishnamurthy K, Balconi C, Sherwood JE, Giroux MJ (2001) Wheat puroindolines enhance fungal disease resistance in transgenic rice. Mol Plant-Microbe Interactions 14:1255–1260
Le Bihan T, Blochet J-E, Desormeaux A, Marion D, Pezolet M (1996) Determination of the secondary structure and conformation of puroindolines by infrared and Raman spectroscopy. Biochemistry 35:12712–12722
Le Guernevé C, Seigneuret M, Marion D (1998) Interaction of the wheat endosperm lipid-binding protein puroindoline-a with phospholipids. Archives Biochem Biophys 360:179–186
Li W, Huang L, Gill BS (2008) Recurrent deletions of puroindoline genes at the grain hardness locus in four independent lineages of polyploidy wheat. Plant Physiol 146:200–212
Llanos P, Henriquez M, Minic J, Elmorjani K, Marion D, Riquelme G, Molgo J, Benoit E (2004) Neuronal and muscular alterations caused by two wheat endosperm proteins, puroindoline-a and alpha1-purothionin, are due to ion pore formation. Eur Biophys J 33:283–284
Llanos P, Henriquez M, Minic J, Elmorjani K, Marion D, Riquelme G, Molgo J, Benoit E (2006) Puroindoline-a and α1-purothionin form ion channels in giant liposomes but exert different toxic actions on murine cells. FEBS J 273:1710–1722
Luo L, Zhang J, Yang G, Li Y, Li K, He G (2008) Expression of puroindoline a enhances leaf rust resistance in transgenic tetraploid wheat. Mol Biol Rep 35:195–200
Massa AN, Morris CF (2006) Molecular evolution of the puroindoline-a, puroindoline-b and grain softness protein-1 genes in the tribe Triticeae. J Mol Evol 63:526–536
Massa AN, Morris CF, Gill BS (2004) Sequence diversity of puroindoline-a, puroindoline-b and the grain softness protein genes in Aegilops tauschii Coss. Crop Sci 44:1808–1816
Mattei C, Elmorjani K, Molgo J, Marion D, Benoit E (1998) The wheat proteins puroindoline-a and α1-purothionin induce nodal swelling in myelinated axons. NeuroReport 9:3803–3807
Miao Y, Chen L, Wang C, Wang Y, Zheng Q, Gao C, Yang G, He G (2012) Expression, purification and antimicrobial activity of puroindoline A protein and its mutants. Amino Acids 43:1689–1696
Morris CF (2002) Puroindolines: the molecular genetic basis of wheat grain hardness. Plant Mol Biol 48:633–647
Morris CF, Bhave M (2008) Reconciliation of D-genome puroindoline allele designations with current DNA sequence data. J Cereal Sci 48:277–287
Morris CF, King GE (2008) Registration of hard kernel puroindoline allele near-isogenic line hexaploid wheat genetic stocks. J Plant Reg 2:67–68
Morris CF, Greenblatt GA, Malkawi HI (1992) Enhanced electrophoretic detection and isolation of friabilin, a starch granule protein. Cereal Chem 69:467–468
Morris CF, Greenblatt GA, Bettge AD, Malkawi HI (1994) Isolation and characterization of multiple forms of friabilin. J Cereal Sci 21:167–174
Morris CF, DeMacon VL, Giroux MJ (1999) Wheat grain hardness among chromosome 5D homozygous recombinant substitution lines using different methods of measurement. Cereal Chem 76:249–254
Morris CF, Simeone MC, King GE, Lafiandra D (2011) Transfer of soft kernel texture from Triticum aestivum to durum wheat Triticum turgidum ssp. durum. Crop Sci 51:114–122
Morris CF, Geng H, Beecher BS, Ma D (2013) A review of the occurrence of Grain softness protein-1 genes in wheat (Triticum aestivum L.). Plant Mol Biol 83:507–521
Mourgues F, Brisset M-N, Chevreau E (1998) Activity of different antibacterial peptides on Erwinia amylovora growth, and evaluation of the phytotoxicity and stability of cecropins. Plant Sci 139:83–91
Murray JC, Kiszonas AM, Wilson JD, Morris CF (2016) Effect of soft kernel texture on the milling properties of soft durum wheat. Cereal Chem 93:513–517
Niknejad A, Webster D, Bhave M (2016) Production of bioactive wheat puroindoline proteins in Nicotiana benthamiana using a virus-based transient expression system. Protein Expr Purif 125:43–52
Palumbo D, Iannaccone M, Porta A, Capparelli R (2010) Experimental antibacterial therapy with puroindolines, lactoferrin and lydozyme in Listeria monocytogenes-infected mice. Microbes Infection 12:538–545
Phillips RL, Palombo EA, Panozzo JF, Bhave M (2011) Puroindolines, Pin alleles, hordoindolines and grain softness proteins are sources of bactericidal and fungicidal peptides. J Cereal Sci 53:112–117
Sanders MR, Clifton LA, Neylon C, Frazier RA, Green RJ (2013) Selected wheat seed defense proteins exhibit competitive binding to model microbial lipid interfaces. J Agric Food Chem 61:6890–6900
Sanders MR, Clifton LA, Frazier RA, Green RJ (2016) Role of lipid composition on the interaction between a tryptophan-rich protein and model bacterial membranes. Langmuir 32:2050–2057
Sanders MR, Clifton LA, Frazier RA, Green RJ (2017) Tryptophan to arginine substitution in puroindoline-b alters binding to model eukaryotic membrane. Langmuir 33:4847–4853
Shagaghi N, Alfred RL, Clayton AHA, Palombo EA, Bhave M (2016) Anti-biofilm and sporicidal activity of peptides based on wheat puroindoline proteins. J Pept Sci 22:492–500
Shagaghi N, Bhave M, Palombo EA, Clayton AHA (2017) Revealing the sequence of interactions of PuroA peptide with Candida albicans cells by live-cell imaging. Sci Rep 7:43542
Shagaghi N, Palombo EA, Clayton AHA, Bhave M (2018) Antimicrobial peptides: biochemical determinants of activity and biophysical techniques of elucidating their functionality. World J Microbiol Biotechnol 34:62
Simeone M, Gedye KR, Mason-Gamer R, Gill BS, Morris CF (2006) Conserved regulatory elements identified from a comparative puroindoline gene sequence survey of Triticum and Aegilops diploid taxa. J Cereal Sci 44:21–33
Sorrentino A, Iannaccone M, Palumbo D, Capparelli R, Porta R, Mariniello L (2009) Tobacco (Nicotiana tabacum cv. Bright Yellow 2) cells as an effective bioreactor for the production of puroindolines. Biotechnol Appl Biochem 53:193–199
Tripathi A, Aggarwal R, Yadav A (2013) Differential expression analysis of defense-related genes responsive to Tilletia indica infection in wheat. Turkish J Biol 37:606–613
Wilde PJ, Clark DC, Marion D (1993) Influence of competitive adsorption of a lysopalmitoylphosphatidylcholine on the functional properties of puroindoline, a lipid-binding protein isolated from wheat flour. J Agric Food Chem 41:1570–1576
Zasloff M (2002) Antimicrobial peptides of multicellular organisms. Nature 415:389–395
Zhang J, Martin JM, Balint-Kurti P, Huang L, Giroux MJ (2011) The wheat puroindoline genes confer fungal resistance in transgenic corn. J Phytopathol 159:188–190
Zhang S-B, Zhai H-C, Lv Y-Y, Cai J-P, Wang J-S (2016) Proteomic analysis reveals the fungal resistance of soft wheat during storage. J Stored Products Res 69:195–198
Acknowledgements
The assistance of Stacey Sykes and Shawna Vogl are gratefully acknowledged. Funding was provided by the USDA ARS CRIS Project 2090-43440-007-00D.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Morris, C.F. The antimicrobial properties of the puroindolines, a review. World J Microbiol Biotechnol 35, 86 (2019). https://doi.org/10.1007/s11274-019-2655-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-019-2655-4