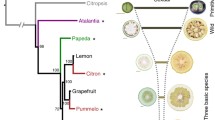
Key message
Review on citrus reproduction.
Abstract
Citrus is one of the most important and widely grown fruit crops. It possesses several special reproductive characteristics, such as nucellar embryony and self-incompatibility. The special phenomenon of nucellar embryony in citrus, also known as the polyembryony, is a kind of sporophytic apomixis. During the past decade, the emergence of novel technologies and the construction of multiple citrus reference genomes have facilitated rapid advances to our understanding of nucellar embryony. Indeed, several research teams have preliminarily determined the genetic basis of citrus apomixis. On the other hand, the phenomenon of self-incompatibility that promotes genetic diversity by rejecting self-pollen and accepting non-self-pollen is difficult to study in citrus because the long juvenile period of citrus presents challenges to identifying candidate genes that control this phenomenon. In this review, we focus on advances to our understanding of reproduction in citrus from the last decade and discuss priorities for the coming decade.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Citrus, including oranges, mandarins, lemons, and grapefruits, is some of our planet’s highest-yielding fruit crops and is distributed worldwide. The top ten citrus-producing countries are China, India, Nigeria, Brazil, Mexico, the USA, Spain, Egypt, Italy and Argentina. According to FAOSTAT, in 2014, the worldwide cultivation of citrus utilized more than 9.08 million hectares and yielded more than 139.79 million tons, with a gross production value of 66.44 billion US dollars. Most citrus species are xylophytas that are evergreen with juvenile periods that range from five to eight years. Although the long juvenile period hinders the breeding and research process, a series of unique reproduction features in citrus are attracting more and more interest from developmental biologists. During the last decade, research on reproduction in citrus has focused on three areas of reproductive development: gene-cytoplasmic interactions in mandarin that induce pollen sterility during gametogenesis (Zheng et al. 2014), production of seedless fruits in several pummelo and mandarin-like varieties with self-incompatibility (Ollitrault et al. 2007), and nucellar embryo development from somatic embryos derived from maternal nucellus tissue surrounding the sexual embryo sac. This last process is a kind of apomixis and is found in many citrus cultivars (Bicknell and Koltunow 2004).
Apomixis has been observed in more than 400 plant species spanning approximate 61 families, such as Gramineae, Compositae, Cruciferae, Roaceae, and Rutaceae (Ozias-Akins 2006). Except for apple and citrus, apomixis is rarely found in agricultural crop plants. The apomictic progeny inherit the mother’s genotype. Thus, apomixis can fix desirable genotypes for research and breeding (Spillane et al. 2001). Mechanisms of apomixis vary widely in woody plants and herbages. Mechanisms of apomictic reproduction are classified as either gametophytic or sporophytic according to their origin. In gametophytic apomixis, which is widely observed in grass species, the apomictic embryo develops from the female gametophytic structure through apospory or diplospory (Koltunow and Grossniklaus 2003). In adventitious embryony, typical in citrus and mango, somatic embryos initiate directly from nucellus or integument cells in the ovule. The offspring derived from nucellar embryony in Citrus possess the same genetic constitution as the female parent. The phenomenon of nucellar embryony hinders the formation of hybrid offspring and the progress of cross-breeding. However, it greatly benefits the production of offspring for true-to-type rootstock with good unity and yields virus-free seedlings. Therefore, investigating the mechanism of nucellar embryony in citrus will provide deeper insight into the processes controlling apomictic reproduction and facilitate the transfer of apomixis into other crops.
Another significant trait in citrus breeding is self-incompatibility (SI). SI is associated with parthenocarpy, which yields seedless citrus fruit (Gambetta et al. 2013; Honsho et al. 2009). Many commercial cultivars of citrus are self-compatible and seedy. However, seedlessness is an important commercial feature in many fruits. Recently, the self-incompatibility response was found to underpin seedlessness in a new seedless mutant of citrus—the mandarin cultivar ‘Wuzishatangju’ (Ye et al. 2009). The list of self-incompatible accessions in citrus is growing. Additionally, the bud mutants of cultivated varieties provide pairs of phenotypes that promote the study of self-incompatibility and provide a new approach for breeding seedless fruit in citrus. In addition to the normal outcrossing, apomixis and self-incompatibility are two major pathways of reproduction in angiosperms (Hörandl 2010). However, these mechanisms of reproduction remain poorly understood in citrus. This review mainly describes advances on the mechanisms of nucellar embryony and self-incompatibility, specifically focusing on advances in the mapping and cloning of genes controlling asexual reproduction and the identification of the potential S-locus of self-incompatibility.
Nucellar embryony in citrus
Many members of the genus Citrus and some closely related genera belonging to Rutaceae reproduce apomictically by nucellar embryony, including Zanthoxylum, Clausena, Aegle, Feroniella, Murraya, Ptelea, Ruta, and Triphasia (Carman, 1997). The presence of extra embryos from nucellar tissue gives rise to polyembryonic seeds. The closely related term polyembryony refers to the development of two or more embryos in one seed. A polyembryonic variety is commonly considered to be apomictic, as a monoembryonic variety is sexual or zygotic. However, the extra embryos are occasionally produced by the division of the zygote embryo, usually in polyploidy cultivars (Webber 1940; Aleza et al. 2010). Various polyembryonic resources in Citrus, Fortunella, and Poncirus provide excellent germplasm for studying this trait. We summarize the germplasm of citrus polyembryonic and monoembryonic cultivars, based on a survey by the Chinese Academy of Agricultural Sciences Institute of Citrus (Zhou and Ye 2010) and our own examination and previous reports (Table 1). The citrus relatives in Poncirus and Fortunella are mostly polyembryonic, except for F. margarita and F. japonica. In Citrus, most cultivars in C. paradise, C. sinensis, and C. reticulate are polyembryonic. However, several types of citrus with ancient origins always produce monoembryonic seeds, such as C. medica, C. grandis, C. mangshanensis, and Papeda. The exceptions are mainly derived from hybridization, such as ‘Kiyomi,’ which is derived from a cross between C. unshiu ‘Miyagawa wase’ and C. sinensis ‘Trovita’ orange—two polyembryonic cultivars (Nakano et al. 2008b). The diverse varieties of citrus germplasm provide rich resources for breeding and for studying the evolution of polyembryony in citrus.
The phenomenon of polyembryony was first reported by Leeuwenhoek in 1719, when he found that orange seeds each contain more than one embryo. Following this initial observation, numerous histological and genetic studies have advanced our understanding of this special phenomenon (Koltunow et al. 1995; Wakana and Uemoto 1987). The initial nucellar embryo cells have characteristic angular thick-walled cells with enlarged nuclei and a condensed cytoplasm. These cells are derived from one or two cell layers of the nucellus, which surround the embryo sac from the chalazal end to the micropyle prior to anthesis (Fig. 1) (Koltunow et al. 1995; Wakana and Uemoto 1987). The nucellar tissue acquires the potential for nucellar emrbryogenesis at the binucleate embryo sac stage, before the initial cells are evident (Koltunow et al. 1995). Subsequently, the initial cells become larger, rounded, thin walled, and vacuolated. Then, they divide and expand. The division and expansion of the embryonic cell groups is suspended as the zygotic embryo enters dominant period. As the zygotic embryo develops, these embryonic cell groups grow into the position of embryo sac and develop into mature nucellar embryos that frequently supersede the zygotic embryo (Koltunow 1993; Koltunow et al. 1995; Wakana and Uemoto 1987).
Overview of sexual and asexual seed development in citrus. Major processes of seed development are depicted during the formation of monoembryonic and polyembryonic seeds. Sexual reproduction pathways including meiosis, mitosis, and double fertilization occur in both monoembryonic and polyembryonic seeds. In apomictic ovules, nucellar cells may possess the ability to form nucellar embryos, which is histologically invisible, when the megaspore is divided into the binuclear embryo sac. Several nucellar embryo initial cells (purple) emerge and develop into adventitious embryos in the nucellus tissue (pink) surrounding the sexual embryo sac. In the depicted adventitious embryogenesis pathway, parts of the nucellar embryo initial cells begin to divide and launch embryogenesis when the zygote remains dormant after fertilization. Multiple adventitious embryos develop within the ovule forming a polyembryonic seed. The monoembryonic seed contains only the zygotic embryo
Genetic and molecular studies of nucellar embryogenesis
The genetic characteristics of apomixis are complex. Additionally, gametophytic and sporophytic apomixis may utilize distinct genetic mechanisms. During the last decade, several research groups have used segregating populations and molecular markers to develop several hypotheses that have substantially advanced our knowledge of the genetic basis and the locus controlling polyembryony in citrus (Table 2). Population construction and genetic mapping of polyembryony utilizes the ‘double pseudo-test cross’ theory and an F1 population because citrus plants have a long juvenile phase and high degrees of heterozygosity. Based on the ratios of phenotypes that Parlevliet and Cameron (1959) observed in several segregating populations, they proposed that the initiation of nucellar embryony is controlled by a single dominant gene in the Citrus genus and the Poncirus genus. Subsequently, Iwamasa et al. (1967) found that the one gene hypothesis could not explain the ratios of phenotypes in every segregating population. They proposed that duplicated genes or modified genes complicated the ratios of segregating phenotypes. Cameron and Soost (1979) proposed that two dominant heterozygous genes control polyembryony and that the genotype was P1p1 P2p2 in Poncirus. Based on an analysis of several populations and previously published results, Hong et al. (2001) hypothesized that two complementary dominant genes control apomixis in Citrus and Poncirus and that the dominant homozygous genotype A1A1 is lethal. Based on the ratios of segregating phenotypes, a unanimous conclusion on the genetic basis of this trait does not seem possible.
García et al. (1999) provided the first genetic analysis of nucellar embryony using molecular markers to analyze a hybrid population derived from C. volkameriana and P. trifoliate. They reported that apomixis in citrus is controlled by six quantitative trait loci that either positively or negatively affect apomixis. Based on these results, it appeared that different genetic mechanisms control the production of nucellar embryos and proportion of polyembryonic seeds. Furthermore, this research group investigated two complex rootstock populations and detected only three previously reported QTLs that control the type of embryony (Raga et al. 2012). Among these QTLs, Apo2 contributed to a greater degree than the other QTLs, consistent with the previous report by García et al. (1999). CR14,290 and TAA15 are two markers that were useful for the early selection of polyembryonic rootstocks in the progeny derived from C. reshni, C. aurantium, and C. volkameriana (García et al. 1999; Raga et al. 2012). Polyembryony in P. trifoliata was also investigated with a population derived from a cross between C. maxima and P. trifoliata using AFLP markers that were tightly linked to this trait (Roose and Kepiro 2010). These data also indicated that the proportion of polyembryonic seeds was probably dependent on distinct genes. Nakano et al. (2008a, b) constructed genetic and physical maps of the chromosomal region flanking the nucellar embryony locus using an F1 population derived from the monoembryonic cultivar C. unshiu × C. sinensis ‘Kiyomi’ backcrossed to its parent C. unshiu ‘Miyagawa wase.’ This work independently localized the polyembryony locus to an approximately 380-kb interval containing 70 genes in mandarin (Nakano et al. 2012).
Mechanisms that contribute to nucellar embryogenesis were suggested to depend on genes that are differentially expressed in closely related cultivars with different modes of reproduction. Thus, the expression of genes in the ovules of poly- and mono-embryonic cultivars was analyzed with suppression subtractive hybridization techniques and microarrays (Kumar et al. 2014; Nakano et al. 2013). These studies provided evidence that stress-responsive genes may contribute to nucellar embryony. A conjoint analysis of mRNA and microRNA was performed on the ovules from species with different reproductive modes prior to nucellar embryogenesis and during different stages of nucellar embryo initiation using two pairs of related citrus cultivars (Long et al. 2016). Results from these experiments indicated that the expression of the novel miRN23-5p is downregulated and its targets are upregulated in polyembryonic ovules. It was also shown that genes associated with stress responses were over-represented in polyembryonic cultivars, which is consistent with previous analyses.
Next-generation sequencing has fueled integrative projects on multiple citrus genomes to gain more insight into the development of citrus (Xu et al. 2013; Wang et al. 2017). The genetic locus responsible for citrus polyembryony was narrowed to an 80-kb region containing 11 candidate genes including CitRWP, which encodes a RWP-RK transcription factor that regulates cell differentiation during female gametophyte development and pattern formation in embryos (Wang et al. 2017; Jeong et al. 2011; Koi et al. 2016; Rovekamp et al. 2016; Waki et al. 2011). The expression of CitRWP is higher in polyembryonic citrus, probably because of a miniature inverted repeat transposable element (MITE) that is inserted into the promoter region of CitRWP. This MITE insertion cosegregated with polyembryony in 786 citrus accessions. This is the first study to provide strong evidence for a candidate gene responsible for the development of nucellar embryos in citrus. The reduced expression of a Marchantia polymorpha RWP-RK domain gene (MpRKD) leads to parthenogenetic-like egg cell divisions (Rovekamp et al. 2016). The PsASGR-BABY BOOM-like (PsASGR-BBML) gene in Pennisetum squamulatum induces parthenogenesis and produces haploid offspring in transgenic sexual pearl millet, rice, and maize (Conner et al. 2015, 2017). CitRWP seems to act in a different way than PsASGR-BBML because it is expressed only in the egg cell. It is still unclear how these genes induce cells to form embryos in the absence of fertilization. Nonetheless, it is obvious that significantly different developmental mechanisms induce parthenogenesis in haploid egg cells and embryogenesis in diploid nucellar cells. Therefore, CitRWP is probably functionally distinct from PsASGR-BBML.
Previous studies of polyembryony have revealed the preliminary genetic and molecular mechanism of polyembryony. However, when studying citrus, the long juvenile phase and the considerable space required for growing citrus trees constrain the size of populations. Additionally, the initial aim for the construction of these populations is always for breeding rather than for locating genes. These issues lead to population structures that are relatively complex, distorted segregation, and increased difficulty for genetic analysis and mapping. Based on genetic analyses of Poncirus and Citrus populations, the polyembryony trait of Citrus is probably controlled by one major gene. In contrast, other genes may regulate polyembryony in Poncirus because serious distortions were observed in the segregating Poncirus hybrid population and the genetic basis of these distortions is not known (Roose and Kepiro 2010). Additionally, the location of a quantitative trait that controls the proportion of apomictic seed is not known. Larger population sizes are required to mine these QTLs. Since the candidate gene in Citrus has been identified, it would seem important to test whether the same mechanism drives the development of polyembryony in Poncirus and Fortunella. It seems that polyembryony may have evolved from monoembryony because most species in the subgenus Metacitrus are polyembryonic and the species in Archicitrus are monoembryonic. This becomes problematic when considering the breadth of citrus relatives because Poncirus and Fortunella are generally polyembryonic. Further study of the evolution of polyembryony may lead us to the mechanism that led to the emergence of sporophytic apomixis during the domestication and evolution of citrus. The cosegregation of the MITE insertion in the promotor of CitRWP with polyembryony makes a strong case that the activities of transposable elements are coupled with these evolutionary events. High genome heterozygosity may also influence this phenomenon during domestication and evolution because of the feasibility of distant hybridization between different citrus species or relatives.
To unambiguously determine the contribution of the candidate gene CitRWP for nucellar embryony, it is necessary to knock out CitRWP in citrus. The CRISPR-Cas9 system may provide an excellent means for accomplishing this goal. The Hongkong kumquat, a wild citrus, should facilitate these experiments because it provides a short juvenile transformation platform (Zhang et al. 2009). The pathways that respond to CitRWP and the gene regulatory network that promotes the development of nucellar embryos will be explored using a delicate transcriptional approach, such as microdissection. Testing whether there are some distinct loci that interact with CitRWP in either Poncirus or Fortunella is also important to learn more about mechanisms that drive nucellar embryony. To understand the genetic basis of this process, several segregating populations using either Poncirus or Fortunella as parents should be constructed and analyzed. Fully understanding this process will not only provide information on how cell fate transitions take place during this particular reproductive process, but also provide some innovative and remarkable ideas for the utility of the mechanism of apomixis for the seed industry.
The S-RNase type of self-incompatibility
In contrast to apomixis, sexual reproduction promotes genetic diversity in citrus. Indeed, a species with higher rate of polymorphism has the ability to adapt to a wider range of environmental conditions. The SI system contributes to genetic diversity by preventing inbreeding and promoting outcrossing because this system recognizes and rejects pollen with same genotype (De Nettancourt 2001). This system includes two forms, the sporophytic SI (SSI) and the gametophytic SI (GSI). In SSI, the pollen S-genotype is determined by the diploid genotype of the parent. In contrast, in GSI, the genotype is determined by the haploid pollen. Currently, three different SI types have been described at the molecular level: the Brassicaceae type, Solanaceae type, and Papaveraceae type. The Solanaceae and Papaveraceae both use the GSI form of SI (Takayama and Isogai 2005).
The Solanaceae SI type is believed to be the most widely used mechanism of GSI, which is shared with two other families, Rosaceae and Plantaginaceae (McClure et al. 2011). This review will focus on the identification of the S-determinants specifying Solanaceae SI here. Classical genetic studies have established that this type of SI is usually controlled by a single polymorphic locus, termed the SI S-locus, which is organized in a haplotype fashion with at least two separate genes, the female determinant and the male determinant. These S-determinant genes are always tightly linked and also are expressed in a tissue-specific and developmentally controlled manner in the pistil and pollen. The female determinant of the Solanaceae-type SI was first identified in Nicotiana alata (Bredemeijer and Blaas 1981). A 32-kDa glycoprotein that segregated with S 2 -alleles was identified in the mature upper style segment (Anderson et al. 1986). Similar glycoproteins were identified among the S stylar proteins of tomato (Mau et al. 1986), potato (Kirch et al. 1989), and petunia (Broothaerts et al. 1990). Loss- and gain-of-function transformation experiments provided unambiguous evidence that these glycoproteins are the female determinant of recognition specificity in the style (Lee et al. 1994; Murfett et al. 1994). Because these glycoproteins share a homologous domain with the T2 and Rh ribonucleases from fungi (Horiuchi et al. 1988; Kawata et al. 1990) and were confirmed to be ribonucleases (McClure et al. 1989), they are named S-RNases. The Solanaceae type of SI is also called the S-RNase type. When the ribonuclease activity is abolished, the style fails to reject self-pollen (Huang et al. 1994). Moreover, McClure et al. (1990) reported that pollen RNAs were degraded by RNases after incompatible pollinations. Thus, it is believed that S-RNases serve as cytotoxins that inhibit the growth of pollen tubes by specifically degrading the RNA of incompatible pollen.
The S-locus contains another gene that specifically triggers the Solanaceae-type SI response that is expressed in the pollen as the male determinant. Earlier multiple attempts were made to identify this pollen S gene without success. These experiments utilized RNA differential display (McCubbin et al. 2000), subtractive hybridization (Dowd et al. 2000), and the yeast two-hybrid assay (Sims and Ordanic 2001). Lai et al. (2002) identified the first promising male determinant in Antirrhinum hispanicum using the bacterial artificial chromosome (BAC) approach. They found that this F-box gene located in the S2-haplotype of Antirrhinum was specifically expressed in pollen. Transformation experiments in Petunia inflata were used to demonstrate that the SLF (S-locus F-box protein) serves as the pollen S-determinant in the SI system (Sijacic et al. 2004). Since then, the polymorphic F-box genes were also identified within the S-locus regions of Prunus (Ushijima et al. 2004; Yamane et al. 2003) and Maleae (Sassa et al. 2007), which are referred to as SFB (S-haplotype-specific F-box protein) and SFBB (S-locus F-box brothers), respectively (McClure et al. 2004). SLF/SFB/SFBBs are thought to function as E3 ubiquitin ligases because they have a conserved F-box motif in the N terminus. E3 ubiquitin ligases interact with E2 ligases and ubiquitinate target proteins, which in many cases targets these proteins for degradation by the 26S proteasome pathway (Hua and Kao 2006; Huang et al. 2006). It remains to be clarified whether SLF ubiquitinates S-RNase. SLF was also proposed to ubiquitinate a general inhibitor gene (GI) which inhibits the activity of all S-RNases. These two different models will be discussed in the next paragraph. The observation that S-RNases are taken up by both self and non-self-pollen tubes provides evidence that SLF/SFB/SFBBs recognize S-RNases inside pollen tubes. Subsequently, cognate S-RNases function as cytotoxins that specifically degrade and arrest the growth of pollen tubes (Fig. 2a, b).
Two molecular models of the RNase-type self-incompatibility. a Non-self-recognition system. In the self-pollen tube, the self-SCFSLF complex is unable to recognize S-RNase. Therefore, S-RNase functions as cytotoxicity to degrade the RNA of pollen tube and induce the self-incompatibility reaction. In the non-self-pollen tube, the non-self-SCFSLF recognizes and polyubiquitinates S-RNase, resulting that S-RNase is degraded by the 26S proteasome. b Self-recognition system involved the general inhibitor (GI). The activity of S-RNase is inhibited by GI in pollen tube. Because of the inhibition, the RNA of non-self-pollen tube is able to escape the cytotoxicity of S-RNase and remain integrity. But the self-SCFSLF complex degrades GI by the ubiquitin proteasome system, which enables S-RNase to degrade RNA
RNAse-based SI in the xylophyta
There is no doubt that the identification of S glycoproteins in xylophyta is more difficult because the long juvenile period. They flower in 5–8 years. In contrast, Solanaceae can flower in few months. The long juvenile phase hinders the confirmation of phenotypes and the construction of populations. Nevertheless, considerable progress has been made in several species, particularly in Maleae and Prunus species. A self-compatible (SC) variety of Japanese pear (Pyrus pyrifolia), ‘Osa-Nijisseiki,’ was derived from the ‘Nijisseiki’ variety (Hirata 1989). The style proteins of the SI Japanese pear varieties with known S-genotypes and this pair of SI and SC pears were analyzed using isoelectric focusing–polyacrylamide gels and two-dimensional gel electrophoresis. As a result, a 30-kDa basic protein that segregated with the S-genotype was identified. This protein was expressed at much lower levels in the SC mutant than in the original SI variety (Sassa et al. 1992, 1993). Broothaerts et al. (1995) purified and identified a 29-kDa ribonuclease (S-RNase) from the pistil tissue of apple (Malus domestica). Subsequently, using the conserved sequences of the known S-RNases in Rosaceae, additional female S-RNases have been found in almond (Prunus dulcis) (Ushijima et al. 1998), plums (Prunus salicina) (Beppu et al. 2002), sweet cherry (Prunus avium) (Sonneveld et al. 2003), and apricot (Prunus armeniaca) (Romero et al. 2004).
Sequencing the chromosomal region of the S-haplotype was also found to be an effective way to identify the candidate male determinants in Rosaceae because the male and female S-determinants are tightly linked. An approximately 70-kb cosmid contig containing the Sc haplotype of almond was constructed that contains 12 open reading frames. But only one of them, SFB, was specifically expressed in the pollen (Ushijima et al. 2001, 2003). The function of the SFB gene was investigated in a pollen mutant from Prunus. Ushijima et al. (2004) found that the SFB gene in the SC mutant of Prunus lacks two hypervariable regions, HVa and HVb, and that the deletions of these hypervariable regions were responsible for loss of self-incompatibility. Researchers found that the S-locus from Maleae and Prunus are distinct and that the former contains multiple SFBs, referred to as SFBB (Matsumoto and Tao 2016). But the distinct evolution of the S-locus in Maleae and Prunus remains obscure. Minamikawa et al. (2010) identified 13 SFBB genes from the S 3 -haplotype and 12 from the S 9 -haplotype in apple. In Japanese pear, 10, 6, and 3 SFBB genes were isolated from the S 2- , S 4 -, and S 5 -haplotypes, respectively (Sassa et al. 2007).
Later, two distinct systems were proposed: the non-self-recognition system and the self-recognition system. A pool of F-box genes are confirmed to localize at the S-locus of Maleae, Solanaceae, and Plantaginaceae, which are assumed to adopted the non-self-recognition system. The pool of F-box proteins targets all non-self S-RNases in a collaborative action for proteolytic degradation by polyubiquitinating them. This mechanism allows the non-self-pollen tubes to escape the cytotoxic activity of the S-RNase. In contrast, each functional F-box fails to ubiquitinate the self S-RNase, which allows the S-RNases to degrade the RNAs of self-pollen tubes (De Franceschi et al. 2012; Kubo et al. 2010) (Fig. 2a). There is only one F-box in the Prunus S-locus. This SI type employs a self-recognition system to trigger the S-RNase cytotoxin. In this system, a general inhibitor (GI) is hypothesized to inhibit the activity of all S-RNases. When the F-box protein recognizes the self S-RNase-GI complex, it polyubiquitinates and degrades the GI. The degradation of the GI allows the S-RNases to function as cytotoxins and to inhibit the growth of self-pollen tubes (Matsumoto and Tao 2016) (Fig. 2b). Thus, studies in Rosaceae species (i.e., all woody species with some similar problems to citrus) have progressed well and provide a solid framework for studies in citrus.
Self-incompatibility in citrus
The SI varieties of citrus are mainly pummelo and mandarin (Ngo et al. 2001; Ngo et al. 2010; Yamamoto et al. 2006). The site of self-pollen tube rejection after pollination can serve as a good indicator of SI. In SSI, rejection generally occurs at or before pollen germination. In contrast, in GSI, the incompatible pollen is arrested within the style, rather than on the stigmatic surface (Newbigin et al. 1993). For example, in Nicotiana, the growth of the pollen tube is inhibited in the upper and lower parts of the transmitting tract (Lush and Clarke 1997). However, the GSI of poppies and grasses provides an exception to this rule (Lawrence 1975; Lewis 1956). ‘Shatian’ pummelo is a traditional cultivar of the Citrus genus that exhibits self-incompatibility. In self-pollination experiments with this cultivar, the pollen tubes penetrated the stigma and were rejected after growing through the top one-third of the style (Liang et al. 2017) (Fig. 3). The growth of self-pollen on ‘Comune’ clementine exhibited the same behavior (Distefano et al. 2009). The inhibition site for the self-pollen tubes of ‘Kagzi Kalan’ lemon was in the middle of the style (Kakade et al. 2017). Although the self-pollen tubes of ‘Wuzishatangju’ grew to the basal end of style, they became twisted and stopped growth once the tubes penetrated the ovary (Ye et al. 2009). Therefore, these studies are consistent with gametophytic rather than sporophytic control of the pollen rejection in citrus and this slower inhibition is also consistent with the proposed cytotoxic effect of S-RNases on pollen tubes.
The characteristics of self-incompatibility in citrus. Model for self-incompatibility in citrus. a Growth of a self-pollen tube with the genotype S1 or S2 is inhibited after penetrating the top one-third of the style. In contrast, a pollen tube with the S3 genotype can grow uninhibited to the bottom of style. b Self-pollinated (left) and cross-pollinated (right) styles were stained with aniline blue to show the growth of pollen tubes in incompatible and compatible pollinations, respectively. The arrows indicate the position of pollen tubes in style
It is vital to identify the S-haplotypes in citrus because linkage to the S-haplotypes is required to identify the candidate S genes. Although there is an ever-growing list of SI citrus accessions, the reported S-genotypes of these SI varieties are limited. The S-genotype of ‘Banpeiyu’ was defined as S1S2 (Ngo et al. 2010), which was used to produce self-fertilized seedlings (S1) with homozygous genotypes (Wakana et al. 2004). Homozygous S1 seedlings (S1S1 and S2S2) were identified by pollinated with ‘Banpeiyu’ (S1S2) pollen, and 55 pummelo accessions were separately pollinated with the pollen of S1S1 and S2S2 homozygous seedlings, indicating that 23/55 accessions had the S1 allele and 16 of them had the S2 allele (Kim et al. 2011). These studies provided a starting point for the identification of the S-determinants and the molecular mechanism of the SI response in citrus.
S-RNases share conserved sequence motifs and a similar topology with RNase T2, although their amino acid sequences have diverged substantially. A large number of S-genotypes in Rosaceae were identified based on these conserved motifs (Ushijima et al. 1998; Sonneveld et al. 2003). Many researchers tried to clone the homologous S genes from citrus using the same method, but none of the genes identified with this approach exhibited tissue-specific expression (Chai et al. 2011a, b; Miao et al. 2011a). By performing amino acid sequence similarity searches of the genomic databases of sweet orange (C. sinensis) and clementine mandarin (C. clementine), 16 genes encoding homologs of S-RNases were obtained. Among the 16 candidates, the amino acid sequence of only the protein encoded by CgRNS3 (Citrus grandis RNase 3) and the female determinant of SI were highly conserved. The CgRNS3 protein inhibited the growth of self-pollen tubes in an in vitro culture system, but after a heat treatment, this protein did not significantly inhibit the elongation of pollen tubes (Liang et al. 2017). We prepared multiple sequence alignments for the amino acid sequences of CgRNS3 and other reported S-RNases. We found that CgRNS3 fell into a single distance cluster. Thus, we conclude that citrus S-RNases have a considerably distant relationship to the S-RNases from Solanaceae and Rosaceae (Fig. 4). Thus, to some extent, an S-RNase from citrus, including the conserved sequence motifs, appears distinct from the known S-RNases.
The phylogenetic tree between CgRNS3 and other reported S-RNases. The tree was constructed using the neighbor-joining algorithm using the MEGA5.0, and the reliability was inferred from a bootstrap analysis of 1000 replicates. The accession numbers of these reported S-RNases are indicted in the study of Liang et al. (2017). CgRNS3 is indicated with the red color
In the past, constructing suppression subtractive hybridization (SSH) cDNA libraries provided an effective method for isolating differentially expressed genes. For this reason, two SSH libraries of self- and cross-pollinated styles from ‘Shatian’ pummelo were constructed. Among the 30 ESTs obtained from these experiments, one EST was homologous to the S1-haplotype sequence from Ipomoea trifida and the other was homologous to the S9-RNase from Malus domestica (Qin et al. 2008). To study the SC ‘Shatangju’ and its SI mutant ‘Wuzishatangju,’ mutant alleles of several genes were discovered using different SSH cDNA libraries (Miao et al. 2011b, 2013b). The expression levels of the gene encoding the S1 SI locus-linked pollen 3.15 gene (S1−3.15) (Miao et al. 2013c) and the gene encoding the ubiquitin-activating enzyme E1 (UBE1) (Miao et al. 2013a) in SC ‘Shatangju’ were higher in SC ‘Shatangju’ than in the SI mutant. This is an interesting finding, because male S-determinants are F-Box proteins which are involved in ubiquitination. However, further molecular characterization and expression analysis of these two genes provided evidence that they might be associated with the SC response in citrus, rather than serving as the key genes controlling SI (Miao et al. 2013a, c). Moreover, four days after pollination, the gene encoding the S-phase kinase-associated protein 1 (SKP1) was expressed at higher levels in the self-pollinated pistil than in the cross-pollinated pistils. The majority of pollen tubes became twisted after self-pollination in transgenic tobacco, suggesting that high expression of SKP1 has an effect on the growth of pollen tubes and is involved in the SI reaction of ‘Wuzishatangju’ (Li et al. 2015). These data indicate that most of these differentially expressed genes might function downstream of SI or perform functions that are related to the SC response.
After the introduction of next-generation sequencing technologies, RNA-seq became more popular than the construction of SSH cDNA libraries because RNA-seq provides a high-throughput approach for identifying differentially expressed genes. The transcript profiles of the styles from non-, self-, and cross-pollinated ‘Xiangshui’ lemon were determined to find candidate S genes using RNA-seq technology. Caruso et al. (2012) identified the differentially expressed genes in laser-microdissected stylar canal cells in ‘Comune’ and the SC mutant ‘Monreal.’ The transcriptomes of another pair of mutants were compared, the SI ‘Wuzishatangju’ and the SC ‘Shatangju’ (Ma et al. 2017). Liang et al. (2015) constructed a transcriptome dataset based from seven tissues from ‘Shatian’ pummelo, which provided a valuable resource for molecular biology studies in citrus. Moreover, the protein expression profiles of three developmental stages of ‘Hyuganatsu’ styles were obtained using two-dimensional gel electrophoresis and MALDI-TOF mass spectrometry (Uchida et al. 2012).
In summary, researchers used mainly two kinds of methods to identify the S-determinants of the citrus SI system in the past: homologous amplification and identification of differentially expressed genes. However, to date, there is no direct proof of a bona fide S-RNases in citrus. In xylophyta, SI and SC mutants are valuable to identify the S-determinants. The first female factor of Rosaceae SI was identified utilizing a mutated Japanese peer (Nakanishil et al. 1992). Several pairs of SI and SC mutants are also known in citrus, such as ‘Shatian’ pummelo and ‘Zigui shatian’ pummelo (Chai et al. 2011c), ‘Shatangju’ and ‘Wuzishatangju’ (Miao et al. 2011b), and ‘Comune’ and ‘Monreal’ (Caruso et al. 2012). However, a number of distinct mechanisms may lead to the loss of SI function, such as the deletion of a gene, a missense mutation that causes a single amino acid substitution that affects the female determinant (Li et al. 2009; Okada et al. 2008), an insertion in the coding region of the male determinant (Sonneveld et al. 2005), and changes in the non-S factors (Li et al. 2016). Thus, to identify S factors using SI and SC mutants, researchers should first determine whether the transformation from SI to SC is caused by a mutation that affects the function of the pollen or the pistil. Additionally, in vitro culture systems and antisense oligonucleotide technologies are the most effective means to test the function of candidate genes in xylophyta.
To date, our knowledge of SI in citrus is still less than that in the woody plants of Rosaceae. During the past twenty years, it was confirmed that many accessions of citrus adopt the SI system, especially in pummelo. With a variety of new molecular technologies and advanced sequencing technologies, it has been possible obtain many candidate genes related to SI, but most of these candidates appear to participate in the downstream reactions of SI and do not contribute to the S-determinants themselves. The failure of the homologous amplification experiments is consistent with significant differences between the conserved domains of S-RNase in citrus and known S-RNases. An alternative explanation might be that the S-RNase is not the female S-determinant. Thus, an important objective is to determine whether citrus is similar to other SI RNase systems in that a stylar RNase activity specifically accumulates to high levels or whether there is an SI mechanism in citrus that is distinct from the S-RNase-based mechanism. Franklin-Tong et al. (1991) showed that SI in Papaver rhoeas was different than in Solanaceae, because the level of ribonuclease activity in the mature stigmas of poppy is very low. There is no evidence for S-RNase activity in citrus yet, but experiments are underway to test this possibility. Although some SI accessions were identified in citrus, none of these accessions were used for reciprocal pollination experiments. This needs to be carried out in order to obtain some clearly specified S-haplotype materials to work on. Thus, in many cases, S-allele-associated proteins are the female S-determinants in styles and are possible to identify and isolate from different materials based on their S-haplotypes. These types of experiments will provide information on the processes of SI in citrus and lay a solid foundation for determining the molecular mechanism of SI in citrus.
Prospect of understanding the special reproductive traits of citrus
To accelerate the breeding efficiency for cultivating more citrus varieties with golden qualities and stable characters, it is essential to understand the mechanism of nucellar embryogenesis and self-incompatibility in citrus by utilizing new methods and technologies when they are developed. After the identification of the candidate gene CitRWP, priorities for the future study of nucellar embryony may include verifying the function of the candidate genes on developmental process and exploring related genes and pathways involved with this process. The study of self-incompatibility in citrus still focuses on the identification of S genes. It is essential to isolate the S-allele-associated proteins based on the genotypes of the self-incompatible accessions. Altogether, it is foreseeable that the engineering of apomixis and self-incompatibility in a broad range of crops will become a reality in the near future.
Author contribution statement
SQ Zhang, M Liang, and N Wang wrote the manuscript. Q Xu, XX Deng, and LJ Chai read and modified the manuscript.
References
Aleza P, Juarez J, Ollitrault P, Navarro L (2010) Polyembryony in non-apomictic citrus genotypes. Ann Bot 106:533–545
Anderson MA et al (1986) Cloning of cDNA for a stylar glycoprotein associated with expression of self incompatibility in Nicotiana alata. Nature 321:38–44
Beppu K, Yamane H, Yaegaki H, Yamaguchi M, Kataoka I, Tao R (2002) Diversity of S-RNase genes and S-haplotypes in Japanese plum (Prunus salicina Lindl.). J Hortic Sci Biotechnol 77:658–664
Bicknell RA, Koltunow AMG (2004) Understanding apomixis: recent advances and remaining conundrums. Plant Cell 16:S228–S245
Bredemeijer GMM, Blaas J (1981) S-specific proteins in styles of self-incompatible Nicotiana alata. Theor Appl Genet 59:185–190
Broothaerts WJ, Av Laere, Witters R, Preaux G, Decock B, Jv Damme, Vendrig JC (1990) Purification and N-terminal sequencing of style glycoproteins associated with self-incompatibility in Petunia hybrida. Plant Mol Biol 14:93–102
Broothaerts W, Janssens GA, Proost P, Broekaert WF (1995) cDNA cloning and molecular analysis of two self-incompatibility alleles from apple. Plant Mol Biol 27:499–511
Cameron JW, Soost RK (1979) Sexual and nucellar embryony in F1 hybrids and advanced crosses of Citrus with Poncirus. J Am Soc Hortic Sci 104:408–410
Carman JG (1997) Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol J Lin Soc 61(1):51–94
Caruso M, Merelo P, Distefano G, La Malfa S, Piero ARL, Tadeo FR, Talon M, Gentile A (2012) Comparative transcriptome analysis of stylar canal cells identified novel candidate genes implicated in the self-incompatibility response of Citrus clementina. BMC Plant Bio 12:20. https://doi.org/10.1186/1471-2229-12-20
Chai L, Ge X, Biswas MK, Deng X (2011a) Molecular analysis and expression of a floral organ-relative F-box gene isolated from ‘Zigui shatian’ pummelo (Citrus grandis Osbeck). Mol Biol Rep 38:4429–4436
Chai L, Ge X, Xu Q, Deng X (2011b) CgSL2, an S-like RNase gene in ‘Zigui shatian’ pummelo (Citrus grandis Osbeck), is involved in ovary senescence. Mol Biol Rep 38:1–8
Chai LJ, Ge XX, Biswas MK, Xu Q, Deng XX (2011c) Self-sterility in the mutant ‘Zigui shatian’ pummelo (Citrus grandis Osbeck) is due to abnormal post-zygotic embryo development and not self-incompatibility. Plant Cell Tiss Organ Cult 104:1–11
Conner JA, Mookkan M, Huo H, Chae K, Ozias-Akins P (2015) A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc Natl Acad Sci USA 112:11205–11210
Conner JA, Podio M, Ozias-Akins P (2017) Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes. Plant Reprod 30(1):41–52
De Franceschi P, Dondini L, Sanzol J (2012) Molecular bases and evolutionary dynamics of self-incompatibility in the Pyrinae (Rosaceae). J Exp Bot 63:4015–4032
De Nettancourt D (2001) Incompatibility and incongruity in wild and cultivated plants, vol 3. Springer, Berlin
Deidda P, Chessa I (1982) A three-gene hypothesis for the inheritance of nucellar embryony in Citrus. Riv Ortoflorofruttic Ital 66(6):431–436
Distefano G, Caruso M, La Malfa S, Gentile A, Tribulato E (2009) Histological and molecular analysis of pollen-pistil interaction in clementine. Plant Cell Rep 28:1439–1451
Dowd PE, McCubbin AG, Wang X, Verica JA, Tsukamoto T, Ando T, Kao T-H (2000) Use of Petunia inflata as a model for the study of Solanaceous type self-incompatibility. Ann Bot 85:87–93
Franklin-Tong VE, Atwal KK, Howell EC, Lawrence MJ, Franklin FCH (1991) Self-incompatibility in Papaver rhoeas: there is no evidence for the involvement of stigmatic ribonuclease activity. Plant Cell Environ 14:423–429
Gambetta G, Gravina A, Fasiolo C, Fornero C, Galiger S, Inzaurralde C, Rey F (2013) Self-incompatibility, parthenocarpy and reduction of seed presence in ‘Afourer’ mandarin. Sci Hortic 164:183–188
García R, Asíns MJ, Forner J, Carbonell EA (1999) Genetic analysis of apomixis in Citrus and Poncirus by molecular markers. Theor Appl Genet 99:511–518
Hirata N (1989) Self-compatible mutant in Japanese pear. Gamma Field Symp 28:71–80
Hong QB, Xiang SQ, Chen KL, Chen LG (2001) Two complementary dominant genes controlling apomixis in genus Citrus and poncirus. Acta Genet Sin 28:1062–1067
Honsho C, Kotsubo M, Fukuda Y, Hamabata Y (2009) Reproductive characteristics for self-compatibility and seedlessness in ‘Nishiuchi Konatsu’, a bud mutation of Hyuganatsu (Citrus tamurana hort. ex Tanaka). HortScience 44:1547–1551
Hörandl E (2010) The evolution of self-fertility in apomictic plants. Sex Plant Reprod 23:73–86
Horiuchi H et al (1988) Primary structure of a base non-specific ribonuclease from Rhizopus niveus. J Biochem 103:408–418
Hua Z, Kao TH (2006) Identification and characterization of components of a putative petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell 18:2531–2553
Huang S, Lee HS, Karunanandaa B, Kao TH (1994) Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell 6:1021–1028
Huang J, Zhao L, Yang Q, Xue Y (2006) AhSSK1, a novel SKP1-like protein that interacts with the S-locus F-box protein SLF. Plant J 46:780–793
Iwamasa M, Ueno I, Nishiura M (1967) Inheritance of nucellar embryony in Citrus. Bull Hortic Res Stn Jpn, Ser B 7:1–8
Jeong S, Palmer TM, Lukowitz W (2011) The RWP-RK factor GROUNDED promotes embryonic polarity by facilitating YODA MAP kinase signaling. Curr Biol 21:1268–1276
Kakade V, Dubey AK, Sharma RM, Malik SK (2017) Gametophytic self-incompatibility causes seedlessness in ‘Kagzi Kalan’ lemon (Citrus limon). J Jpn Soc Hortic Sci Biotechnol 92:303–312
Kawata Y, Sakiyama F, Hayashi F, Kyogoku Y (1990) Identification of two essential histidine residues of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem 187:255–262
Kim J-H, Mori T, Wakana A, Ngo BX, Sakai K, Kajiwara K (2011) Determination of self-incompatible Citrus cultivars with S1 and/or S2 alleles by pollination with homozygous S1 seedlings (S1S1 or S2S2) of ‘Banpeiyu’pummelo. J Jpn Soc Hortic Sci 80:404–413
Kirch HH, Uhrig H, Lottspeich E, Salamini F, Thompson RD (1989) Characterization of proteins associated with self-incompatibility in Solanum tuberosum. Theor Appl Genet 78:581–588
Koi S et al (2016) An evolutionarily conserved plant RKD factor controls germ cell differentiation. Curr Biol 26:1775–1781
Koltunow AM (1993) Apomixis: embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5:1425–1437
Koltunow AM, Grossniklaus U (2003) APOMIXIS: a developmental perspective. Annu Rev Plant Biol 54:547–574
Koltunow AM, Soltys K, Nito N, McClure S (1995) Anther, ovule, seed, and nucellar embryo development in Citrus sinensis cv. Valencia. Can J Bot 73:1567–1582
Kubo K, Entani T, Takara A, Wang N, Fields AM, Hua Z et al (2010) Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330:796–799
Kumar V, Malik SK, Pal D, Srinivasan R, Bhat SR (2014) Comparative transcriptome analysis of ovules reveals stress related genes associated with nucellar polyembryony in citrus. Tree Genet Genomes 10:449–464
Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y (2002) An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol 50:29–42
Lawrence MJ (1975) The genetics of self-incompatibility in Papaver rhoeas. Proc R So B Biol Sci 188:275–285
Lee HS, Huang S, Kao TH (1994) S proteins control rejection of incompatible pollen in Petunia inflata. Nature 367:560–563
Lewis D (1956) Incompatibility and plant breeding Genetics in plant breeding. Brook-haven Symp Biol 9:89–100
Li MF, Li XF, Han ZhH, Shu HR, Li T (2009) Molecular analysis of two Chinese pear (Pyrus bretschneideri Rehd.) spontaneous self-compatible mutants, Yan Zhuang and Jin Zhui. Plant Biol 11:774–783
Li P et al (2015) CrWSKP1, an SKP1-like gene, is involved in the self-incompatibility reaction of “Wuzishatangju” (Citrus reticulata Blanco). Int J Mol Sci 16:21695–21710
Li W et al (2016) Molecular and genetic characterization of a self-compatible apple cultivar, ‘CAU-1’. Plant Sci 252:162–175
Liang M, Yang XM, Li H, Su SY, Yi HL, Chai LJ, Deng XX (2015) De novo transcriptome assembly of pummelo and molecular marker development. PLoS One 10:e0120615
Liang M et al (2017) Genome-wide identification and functional analysis of S-RNase involved in the self-incompatibility of citrus. Mol Genet Genomics 292:325–341
Long JM et al (2016) Genome-scale mRNA and small RNA transcriptomic insights into initiation of citrus apomixis. J Exp Bot 67:5743–5756
Lush WM, Clarke AE (1997) Observations of pollen tube growth in Nicotiana alata and their implications for the mechanism of self-incompatibility. Sex Plant Reprod 10:27–35
Ma Y, Li Q, Hu G, Qin Y (2017) Comparative transcriptional survey between self-incompatibility and self-compatibility in Citrus reticulata Blanco. Gene 609:52–61
Matsumoto D, Tao R (2016) Distinct self-recognition in the Prunus S-RNase-based gametophytic self-incompatibility system. Hortic J 85:289–305
Mau S-L et al (1986) Style proteins of a wild tomato (Lycopersicon peruvianum) associated with expression of self-incompatibility. Planta 169:184–191
McClure BA, Hairing V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE (1989) Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342(6252):955–957
McClure BA, Gray JE, Anderson MA, Clarke AE (1990) Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 347:757–760
McClure BA (2004) S-RNase and SLF determine S-haplotype-specific pollen recognition and rejection. Plant Cell 16:2840–2847
McClure BA, Cruz-García F, Romero C (2011) Compatibility and incompatibility in S-RNase-based systems. Ann Bot 108:647–658
McCubbin AG, Wang X, T-h Kao (2000) Identification of self-incompatibility (S-) locus linked pollen cDNA markers in Petunia inflata. Genome 43:619–627
Miao HX, Qin YH, Teixeira da Silva JA, Ye ZX, Hu GB (2011a) Cloning and expression analysis of S-RNase homologous gene in Citrus reticulata Blanco cv. Wuzishatangju. Plant Sci 180:358–367
Miao HX, Qin YH, Teixeira Da Silva JA, Ye ZX, Hu GB (2011b) Isolation and differential expression analysis of self-compatibility-related genes from mature pistils of ‘Shatangju’ mandarin (Citrus reticulata Blanco). J Hortic Sci Biotechnol 86:575–582
Miao H-x, Ye Z-x, Hu Y-h, Qin G-b (2013a) Identification of differentially expressed genes in 72 h styles from self-incompatible Citrus reticulata. Sci Hortic 161:278–285
Miao HX, Qin YH, Ye ZX, Hu GB (2013b) Molecular characterization and expression analysis of ubiquitin-activating enzyme E1 gene in Citrus reticulata. Gene 513:249–259
Miao HX, Ye ZX, da Silva JAT, Qin YH, Hu GB (2013c) Identifying Differentially Expressed Genes in Pollen from Self-Incompatible “Wuzishatangju” and Self-Compatible “Shatangju”. Mandarins Int J Mol Sci 14:8538–8555
Minamikawa M, Kakui H, Wang S, Kotoda N, Kikuchi S, Koba T, Sassa H (2010) Apple S locus region represents a large cluster of related, polymorphic and pollen-specific F-box genes. Plant Mol Biol 74:143–154
Murfett J, Atherton TL, Mou B, Gassert CS, Mcclure BA (1994) S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature 367:563–566
Nakanishil T et al (1992) Isoelectric focusing analysis of stylar proteins associated with self-incompabibility alleles in Japanese pear. J Jpn Soc Hortic Sci 61:239–248
Nakano M et al (2008a) Marker enrichment and construction of haplotype-specific BAC contigs for the polyembryony genomic region in Citrus. Breed Sci 58:375–383
Nakano M, Shimizu T, Kuniga T, Nesumi H, Omura M (2008b) Mapping and haplotyping of the flanking region of the polyembryony locus in Citrus unshiu Marcow. J Jpn Soc Hortic Sci 77:109–114
Nakano M et al (2012) Characterization of genomic sequence showing strong association with polyembryony among diverse Citrus species and cultivars, and its synteny with Vitis and Populus. Plant Sci 183:131–142
Nakano M, Kigoshi K, Shimizu T, Endo T, Shimada T, Fujii H, Omura M (2013) Characterization of genes associated with polyembryony and in vitro somatic embryogenesis in Citrus. Tree Genet Genomes 9:795–803
Newbigin E, Anderson MA, Clarke AE (1993) Gametophytic self-incompatibility systems. Plant Cell 5:1315–1324
Ngo BX, Wakana A, Park SM, Nada Y, Fukudome I (2001) Pollen tube behaviors in self-incompatible and self-compatible Citrus cultivars. J Fac Agric Kyushu Univ 45:443–457
Ngo BX, Wakana A, Kim JH, Mori T, Sakai AK (2010) Estimation of self-incompatibility S genotypes of Citrus cultivars and plants based on controlled pollination with restricted number of pollen grains. J Fac Agric Kyushu Univ 55:67–72
Okada K et al (2008) Deletion of a 236 kb region around S 4 -RNase in a stylar-part mutant S sm4-haplotype of Japanese pear. Plant Mol Biol 66:389–400
Ollitrault P, Luro F, Yamamoto M (2007) Seedlessness and ploidy manipulations. In: Khan IA (ed), Citrus Genetics, Breeding and Biotechnology, CAB International, UK, pp 197–218
Ozias-Akins P (2006) Apomixis: developmental characteristics and genetics. Crit Rev Plant Sci 25:199–214
Parlevliet JE, Cameron JW (1959) Evidence on the inheritance of nucellar embryony in citrus. Proc Am Soc Hortic Sci 74:252–260
Qin XM, Xiong J, Yang J, Wan S, Wei SL (2008) Construction and analysis of suppression subtractive hybridization library related to gametophytic self-incompatibility in style of Citrus grandis var. shatinyu. Hort J Trop Subtrop Bot 16:425–429
Raga V, Bernet GP, Carbonell EA, Asins MJ (2012) Segregation and linkage analyses in two complex populations derived from the citrus rootstock Cleopatra mandarin. Inheritance of seed reproductive traits. Tree Genet Genomes 8:1061–1071
Romero C, Vilanova S, Burgos L, Martinez-Calvo J, Vicente M, Llacer G, Badenes ML (2004) Analysis of the S-locus structure in Prunus armeniaca L. Identification of S-haplotype specific S-RNase and F-box genes. Plant Mol Biol 56:145–157
Roose ML, Kepiro JL (2010) AFLP markers closely linked to a major gene essential for nucellar embryony (apomixis) in Citrus maxima × Poncirus trifoliata. Tree Genet Genomes 6:1–11
Rovekamp M, Bowman JL, Grossniklaus U (2016) Marchantia MpRKD regulates the gametophyte-sporophyte transition by keeping egg cells quiescent in the absence of fertilization. Curr Biol 26:1782–1789
Sassa H, Hirano H, Ikehashi H (1992) Self-incompatibility-related RNases in styles of Japanese pear (Pyrus serotina Rehd.). Plant Cell Physiol 33:811–814
Sassa H, Hirano H, Ikehashi H (1993) Identification and characterization of stylar glycoproteins associated with self-incompatibility genes of Japanese pear, Pyrus serotina Rehd. Mol Gen Genet 241:17–25
Sassa H et al (2007) S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics 175:1869–1881
Sijacic P et al (2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429:302–305
Sims TL, Ordanic M (2001) Identification of a S-ribonuclease-binding protein in Petunia hybrida. Plant Mol Biol 47:771–783
Sonneveld T, Tobutt KR, Robbins TP (2003) Allele-specific PCR detection of sweet cherry self-incompatibility (S) alleles S1 to S16 using consensus and allele-specific primers. Theor Appl Genet 107:1059–1070
Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP (2005) Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17:37–51
Spillane C, Steimer A, Grossniklaus U (2001) Apomixis in agriculture: the quest for clonal seeds. Sex Plant Reprod 14:179–187
Takayama S, Isogai A (2005) Self-incompatibility in plants. Annu Rev Plant Biol 56:467–489
Uchida A et al (2012) Comprehensive analysis of expressed proteins in the different stages of the style development of self-incompatible ‘Hyuganatsu’ (Citrus tamurana hort. ex Tanaka). J Jpn Soc Hortic Sci 81:150–158
Ushijima K, Sassa H, Tao R, Yamane H, Dandekar AM, Gradziel TM, Hirano H (1998) Cloning and characterization of cDNAs encoding S-RNases from almond (Prunus dulcis): primary structural features and sequence diversity of the S-RNases in Rosaceae. Mol Gen Genet 260:261–268
Ushijima K, Sassa H, Tamura M, Kusaba M, Tao R, Gradziel TM et al (2001) Characterization of the S-locus region of almond (Prunus dulcis): analysis of a somaclonal mutant and a cosmid contig for an S haplotype. Genetics 158:379–386
Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao Ryutaro, Hirano H (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15:771–781
Ushijima K, Yamane H, Watari A, Kakehi E, Ikeda K, Hauck NR et al (2004) The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J 39:573–586
Wakana A, Uemoto S (1987) Adventive embryogenesis in Citrus. I. The occurrence of adventive embryos without pollination or fertilization. Am J Bot 74:517–530
Wakana A, Ngo BX, Fukudome I, Kajiwara K (2004) Estimation of the degree of self-incompatibility reaction during flower bud development and production of selffertilized seeds by bud pollination in self-incompatible Citrus cultivars. J Fac Agric Kyushu Univ 49:307–320
Waki T, Hiki T, Watanabe R, Hashimoto T, Nakajima K (2011) The Arabidopsis RWP-RK protein RKD4 triggers gene expression and pattern formation in early embryogenesis. Curr Biol 21:1277–1281
Wang X et al (2017) Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat Genet 49:765–772
Webber JM (1940) Polyembryony. Bot Rev 6:575–598
Xu Q et al (2013) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45:59–66
Yamamoto M, Kubo T, Tominaga S (2006) Self- and cross-incompatibility of various citrus accessions. J Jpn Soc Hortic Sci 75:372–378
Yamane H, Ikeda K, Ushijima K, Sassa H, Tao R (2003) A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol 44:764–769
Ye W et al (2009) Seedless mechanism of a new mandarin cultivar ‘Wuzishatangju’ (Citrus reticulata Blanco). Plant Sci 177:19–27
Zhang J, Tao N, Xu Q, Zhou W, Cao H, Xu J, Deng X (2009) Functional characterization of Citrus PSY gene in Hongkong kumquat (Fortunella hindsii Swingle). Plant Cell Rep 28(11):1737
Zheng BB, Fang YN, Pan ZY, Sun L, Deng XX, Grosser JW, Guo WW (2014) iTRAQ-based quantitative proteomics analysis revealed alterations of carbohydrate metabolism pathways and mitochondrial proteins in a male sterile cybrid pummelo. J Proteome Res 13:2998–3015
Zhou K, Ye Y (2010) Fruit tree of China: volume of citrus. China Agricultural Press, Beijing
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (Nos. 31772259, 31630065, 31521092) and the Special Fund for Agro-scientific Research in the Public Interest (201303093). We also thank Prof. Robert M. Larkin and Prof. Pengwei Wang for help with English language editing.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Tetsuya Higashiyama.
A contribution to the special issue ‘Plant Reproduction Research in Asia’.
Rights and permissions
About this article
Cite this article
Zhang, S., Liang, M., Wang, N. et al. Reproduction in woody perennial Citrus: an update on nucellar embryony and self-incompatibility. Plant Reprod 31, 43–57 (2018). https://doi.org/10.1007/s00497-018-0327-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-018-0327-4