Abstract
F-box proteins are a large family of eukaryotic proteins that contained a conserved motif of approximately 40 amino acids. They play an important role in the processing of degradation of cellular regulatory proteins. In this study we isolated a full-length of cDNA encoding a putative F-box protein from Citrus grandis Osbeck CV ‘Zigui shatian’ pummelo and designated as CgF-box. The cDNA sequence of CgF-box was 920 bp containing a 585 bp open reading frame encoding a precursor protein of 194 amino acid residues. The deduced protein comprised a conserved F-box domain at the position from the 40th to 84th amino acids. Cluster analysis suggested that CgF-box was more closely related to the grape F-Box protein. Southern hybridization verified CgF-box existed in the genome as multiple copies. The expression analysis revealed that the expression level of CgF-box gene remarkably increases during the flower developmental process of ‘Zigui shatian’ pummelo, such as high level of expression was noted in style, petal and anther, on the other hand low level of expression was found in ovary and leaf. For further verifying the different expression in different tissue of this gene, in situ hybridization was conducted, strong expression signal could be observed in the style, stigma and anther, low even no signal was observed in ovary. According to their findings we can conclude that CgF-box was not only involved in flower maturation, but also showed different roles in different tissue.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In plants, like other living organisms, there is a key regulatory mechanism to control protein turnover, called ubiquitin (Ub)/26S proteasome pathway. It is responsible for selective degradation of most intracellular proteins by ubiquitination these proteins [1]. There are three key enzymes involving in the ubiquitination process, including: Ub-activating enzyme (E1), Ub-conjugating enzyme (E2) and Ub-protein ligases (E3). The most important is the Ub E3 ligases that catalyze the attachment of poly Ub chains to target proteins for their subsequent degradation by the 26S proteasome. Among the E3 families, Skp1p-cullin-F-box (SCF) protein is a major class as is well understands. As one of SCF multi-protein complex component, the F-box protein performs the crucial role of conferring specificity to the complex for appropriate targets [2–5].
F-box proteins are a large family of proteins found in various eukaryotes, including yeasts, nematodes, plants, fishes, human and mammals [6–11]. These proteins share a conserved F-box domain approximately 40 amino acid motif at their N terminus, which interacts with Skp1 subunit another component of SCF multi-protein complex. Because it was first identified at the N-terminal region of cyclin F, named F-Box protein [12]. Since the discovery of the first F-box protein (Cyclin F) from human [12], numerous F-box proteins have been identified by the presence of a well-conserved N-terminally F-box domain [13]. Very interestingly, different species show different number of F-box genes [3, 12, 13]. It was reported that there were 4, 337, 24, and 38 F-box genes in budding yeast, nematode, fruit-fly, and human respectively [13]. In plants, at least 692, 337 and 779 F-box genes have been identified in the Arabidopsis thaliana, popular and rice genomes respectively [14]. The huge number of F-box genes exists in many different species, which makes the F-box superfamily as one of the largest in plants.
Plants have acquired a large number of F-box genes most likely because of their roles in protein ubiquitination and degradation; it implicates protein degradation as a prevalent developmental control mechanism in plants. Recently many researchers have proved that F-box proteins regulate a variety of biological processes, such as lateral root development [15], hormonal responses [16–18], senescence [19], and pathogen resistance [20] and so on. Although much number of F-box genes and diverse functions were revealed in many living organisms, however, till now there is very little information about F-box gene in citrus species, especially flower relative F-box gene. F-box proteins related with flower have been found to function in two fields; one is involved in the regulation of a complex set of development events during floral development, including floral meristem and floral organ identity [21–23], the other is participated in plant self-incompatibility [24–28]. In citrus, self-incompatibility is also an important mechanism as it can lead to seedless fruits when coupled with parthenocarpy. Thus, researches have been conducted to determine self-incompatibility by pollination test in major cultivars and many accessions, such as pummelo [29]. Till now, it is unknown whether the S-locus-linked F-box protein existed or not in gametophytic incompatibility mechanism of pummelo. Because very little information about flower related F-box gene can be found in citrus, it becomes very emergently to uncover the F-box gene information in pummelo. In this study, we used ‘Zigui shatian’ pummelo as plant material, which was a natural self-compatible mutant from ‘Shatian’ pummelo [30], and constructed a Full-length cDNA library using the total RNA from seven development stages of flowers. An EST contained full-length open reading frame (ORF) was found to encode F-box protein, designated as CgF-box. Further expression analysis revealed that CgF-box is not only involved in flower maturation, but also showed different roles in different tissue.
Materials and methods
Sample collection
‘Zigui shatian’ pummelo was cultivated in Zigui county, Hubei, China; Seven flower developing stages of ‘Zigui shatian’ pummelo were collected according to the length of flower bud (1) 0.3–0.5 cm, (2) 0.7–0.9 cm, (3) 1.1–1.2 cm, (4) 1.5–1.7 cm, (5) 1.9–2.1 cm, (6) 2.4–2.6 cm, (7) 2.9–3.0 cm for cDNA library construction. The tissues of ovaries, petals, styles and anthers were collected separately from the 10 days before anthesis stage flower; the leaf was collected from middle position of tree, and it was growing on new germination shoot. Other species were cultivated in the National Center of Citrus Breeding located at Huazhong Agricultural University, Wuhan, China.
cDNA library construction and sequencing of the cDNA insert
Total RNAs of seven different developing stage flower of ‘Zigui shatian’ pummelo were isolated according to the protocol Liu et al. [31]. A cDNA library was constructed using the SMART cDNA Library Construction Kit (Clontech, USA). The insert fragment sizes of the positive recombinants were analyzed by PCR amplification using the vector-specific M13 primers. BLAST analysis of the EST sequences revealed that a CgEST838 contained the F-box motif. This EST was selected for further analysis of the CgF-box gene of ‘Zigui shatian’ pummelo.
Multiple sequence alignment and phylogenetic analysis
The full-length cDNA sequence was used to search homologous sequences via BLASTX in NCBI (http://www.ncbi.nlm.nih.gov/BLASTX/). Sequences of other plant RNase proteins were retrieved from Gen Bank database. Sequence alignment was carried out using Clustal W program, and a phylogenetic relationship tree was constructed with the MEGA 4 program by Neighbor-Joining method [32].
DNA extraction and Southern blot
Genomic DNA was isolated from the leaves of six citrus cultivars, i.e. (1) Zigui shatian pummelo (Citrus grandis); (2) Citrus ichangensis; (3) Cocktail grapefruit (C. paradisi); (4) Sour orange (C. aurantium); (5) Red tangerine (C. reticulata); (6) Bendizao tangerine (C. reticulata) by the cetyltrimethylammonium bromide (CTAB) method [33]. 20 μg genomic DNA was digested with HindIII and EcoRI, blotted onto Nytran Super Charge nylon membrane. The DNA gel blot was hybridized with digoxigenin-dUTP labeled full ORF cDNA probes, and the subsequent procedures were performed according to the manufacturer’s instructions (Roche, Switzerland).
Semi-quantitative reverse transcription PCR
Total RNA was extracted from seven stage of flower development and others ovaries, petals, styles leaf and stamen which were collected separately from fully opened flowers. Semi-quantitative reverse transcription PCR (RT-PCR) analysis was conducted with specific primers derived from CgF-box as well as actin as a control. The primers of gene specific primers are 5′-CTGCTTCTCAGATTGGCATTTG-3′ (forward) and 5′-GATACTCTGTCCACTGAGAGTG-3′ (reverse). Actin is 5′-CCAAGCAGCATGAAGATCAA-3′ (forward) and 5′-ATCTGCTGGAAGGTGTGAG-3′ (reverse). The PCR programs were as follows: 94°C for 4 min, 28 cycles of amplification (94°C for 30 s, variable annealing temperature depending on specific primer set for 50 s, and 72°C for 2 min) followed by a final extension at 72°C for 5 min. The RT-PCR products from each tissue were subject to electrophoresis on 1.5% agarose gels. The experiments were repeated three times with the similar results and one of them was presented.
Quantitative real-time PCR reaction
The same materials mentioned above by RT-PCR were used. The total RNA yield and quality were determined spectrophotometrically at wave lengths of 230, 260 and 280 nm, and the integrity of total RNA were verified by running samples on 1% agarose gels. First strand cDNA was synthesized using RevertAidTM First Strand cDNA Synthesis Kit (Fermentas) according to the guidelines provided by the manufacturer (Roche, Switzerland). Real-time PCR was performed with the ABI 7500 Real-Time System (PE Applied Biosystems, Foster City, CA, USA). The target gene products were amplified with primers (forward: 5′-GAGAAACATCGTGGATCAACCA-3′ and reverse: 5′-ACTAAGCTCTAAGTACGAATCAAACTCATT-3′). These primers were designed with the Primer Express software (Applied Biosystems, Foster City, CA, USA) and following the manufacturer’s guidelines for primer design. The levels of gene expression were analyzed with ABI 7500 Sequence Detection System Software (PE Applied Biosystems, Foster City, CA, USA) and normalized with the results of actin (primer, forward: 5′-CCAAGCAGCATGAAGATCAA-3′ and reverse: 5′-ATCTGCTGGAAGGTGCTGAG-3′). The primers for the target gene and actin were diluted in the SYBER GREEN PCR Master Mix (PE Applied Biosystems) and 10 μl of the reaction mix were added to each well. Reactions were performed by an initial incubation at 50°C for 2 min and at 95°C for 1 min, and then cycled at 95°C for 15 s and 60°C for 1 min for 40 cycles. Output data generated by the instrument on-board software Sequence Detector Version 1.3.1 (PE Applied Biosystems) were transferred to a custom-designed Microsoft Excel macro for analysis. Real-time quantitative RT-PCR was performed in four replicates for each sample.
RNA in situ hybridization and detection
Different tissues of ‘Shatian pummelo’ were fixed overnight in FAA (100% formalin: 100% acetic acid: 50% ethanol, in proportions of 5:5:90 in volume) at 4°C for 12 h, and dehydrated through ethanol to 70% for long-term storing. Fixed tissues were embedded in wax and sectioned at 10 μm with a rotary microtome. The sense and antisense probes were transcribed with full cDNA fragments cloned in pGEM-T vector (Promega, USA) and were labeled with the digoxigenin-UTP by SP6 or T7 RNA polymerase in vitro transcription kit according to the methods described in the manufacturer’s technical manual (Roche, Switzerland). Detection was carried out with anti-DIG/alkaline phosphatase (Roche, Switzerland), and the hybridization signal detection was as previously described by Hu et al. [34].
Results
Cloning and identification of CgF-box gene from ‘Zigui shatian’ pummelo
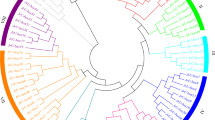
A full-length cDNA of CgF-box gene was identified from a cDNA library. The library was constructed from the seven flower development stages of ‘Zigui shatian’ pummelo. The identified CgF-box gene was 920 bp, it contained a 585 bp ORF and encoding a peptide of 194 amino acids (Fig. 1b), with a calculated molecular mass of 22 kDa and an isoelectric point of 4.74 (http://cn.expasy.org/tools/protparam.html). Further, BlastX (NCBI) result indicated that the deduced amino acid sequence of CgF-box had a moderate degree of homology with other F-box subunit proteins from various plant species viz., Ricinus communis (79%), Populus trichocarpa (76%), Vitis vinifera (75%), A. thaliana (60%), and Oryza sativa Japonica Group (51%). F-box conserved domain also found in the N-terminal of CgF-box protein sequence (Fig. 1a). In order to estimate the phylogenetic relationship of CgF-box gene with other F-box gene of higher plants, a neighbor-joining phylogenetic tree was constructed based on amino acid sequences of CgF-box and other 15 homologous amino acid sequences from higher plants and the results showed that CgF-box was closer to the F-box gene of V. vinifera (Fig. 1c).
a Sequence alignment of CgF-box with other F-box gene. Multiple sequence alignment was performed using the Clustal W program. b Nucleotide sequence of CgF-box cDNA from ‘Zigui shatian’ pummelo and its deduced amino acid sequence. The start and stop codons are in bold.c A phylogenetic tree of plant F-box derived from the alignment of 11 F-box gene using the Neighbor-joining method in MEGA 4.0. The position of pummelo was indicated by the box. The Genbank Accession No. of aligned sequences are as follows: Ricinus communis (XP_002511947); Populus trichocarpa (XP_002301389); Vitis vinifera (XP_002276444); Arabidopsis thaliana (Q9M310); Oryza sativa Indica Group (EEC71213); Zea mays (ACR34152); Sorghum bicolor (XP_002456082); Malus x domestica (ACI13687); Lotus japonicus (AAN87351); Solanum pimpinellifolium (ACI24355)
Genomic organization of CgF-box
To investigate the genomic organization of CgF-box gene, DNA blot analysis was carried out using genomic DNA of ‘Zigui shatian’ pummelo and other citrus relatives. The result reveal that not only more than one HindIII fragment existed in all of tested cultivar under the condition of high stringency, but also in condition of low stringency using EcoRI digestion (Fig. 2). Because there was only one restriction site in the full-length ORF region of the CgF-box gene which was uses as the probe for hybridization about HindIII and there was no EcoRI restriction site in the ORF region, so it indicated that there were possible closely related genes sharing sequence similarity to CgF-box in genomic sequence. This result implied that CgF-box appears to be a member of big family in citrus genome.
Southern blot analysis of CgF-box gene in citrus genome. DNA was digested with HindIII (a) and EcoRI (b) restriction enzymes. Hybridization to DIG-probe generated with primers specific to CgF-box. Samples were: 1 Zigui shatian pummelo (Citrus grandis); 2 Citrus ichangensis; 3 Cocktail grapefruit (C. paradisi); 4 Sour orange (C. aurantium); 5 Red tangerine (C. reticulata); 6 Bendizao tangerine (C. reticulata)
CgF-box expression in ‘Zigui shatian’ pummelo
To detect the CgF-box gene expression during seven flower development stages of ‘Zigui shatian’ pummelo, we extracted the RNA from the whole flower of each development stage, and the RNA was used for further qRT-PCR and RT-PCR experiment. The result showed that the CgF-box gene expression level was high in later developmental stage, and there was significantly increase during this process (Fig. 3). Then further expression of CgF-box gene was studied in different tissues by qRT-PCR and RT-PCR. The material included mature of petals, style, anther, leaf and ovary. The result showed that different tissue may show different expression pattern. The high level of expression of CgF-box was observed in style, petal and anther, low even no expression appeared in the ovary and leaf (Fig. 4). To further dissect the tissue distribution of CgF-box mRNA in different tissues, and confirm the qRT-PCR and RT-PCR expression of different tissues, especially the expression pattern between style, anther and ovary, in situ hybridization was performed. An anti-sense probe of CgF-box was employed to detect its transcripts in the anther, stigma, style, and ovary. The signal of CgF-box distributed uniformly in the stigma and style (Fig. 5d, f), but stronger signal was located in vascular tissues of anther (Fig. 5b). Moreover, the CgF-box exhibited very low even no expression at ovary (Fig. 5g). These results were consisting with qRT-PCR and RT-PCR expression of different tissues.
Detection of CgF-box transcript level by quantitative real-time PCR and RT-PCR in seven flower developing stages of ‘Zigui shatian’ pummelo; Seven developing stages of flower according to the length of flower bud 1 0.3–0.5 cm, 2 0.7–0.9 cm, 3 1.1–1.2 cm, 4 1.5–1.7 cm, 5 1.9–2.1 cm, 6 2.4–2.6 cm, 7 2.9–3.0 cm
Detection of CgF-box transcript level by quantitative real-time PCR and RT-PCR in different tissues from ‘Zigui shatian’ pummelo. The tissues of ovaries, petals, styles and anthers were collected separately from the 10 days before anthesis stage flower; the leaf was collected from middle position of tree, and it was growing on new germination shoot
In situ hybridization analysis of CgF-box gene expressed in different parts of the flower. Left pictures (a, c, e, g) were control under the bright light after hybridization with the sense probe for CgF-box; b, d, f, and h under bright light after hybridization with the anti-sense probe for CgF-box. a, b for anthers from the anthesis flower. Ovaries were from full opened flowers (g, h). Stigmas (c, d) and styles (e, f) both from the anthesis flower. The dark color showed the hybridization signal which was indicated by arrows. Bar represented 200 μm (a, b) and 500 μm (c–h), respectively
Discussion
In this study, we cloned an F-box gene named CgF-box from ‘Zigui shatian’ pummelo. The deduced amino acid sequence of the cDNA fragment showed similarity to the others of F-box proteins. Moreover, sequence comparison of CgF-box with homologs from other plant species revealed that one conserved region called F-box motif existed among these proteins in the N-terminal region (Fig. 1a). In many diverse organisms, F-box proteins are predicted to contain various protein–protein interaction domains at their C terminus. For example, in mammals, F-box proteins contained WD40 repeat domains, LRR domains, and other domains [35]. In Arabidopsis, many different domains existed including LRR, kelch repeats, FBD, WD40, PAS/PAC, ring finger, tubby (TUB), and PPR [3, 36]. In rice, it was estimated that more than 30 putative statistically significant motif could be identified [37]. By searching the Pfam motif database, no putative domain or repeat without F-box domain was matched with CgF-Box. According to the result of rice F-Box clarification, we thought that our CgF-box showed similarity structure belonging to the FBX subfamily of rice [37]. Large number of F-box proteins exists in different living organisms; it implies the same situation as in pummelo. Further, Genomic hybridization was carried out using ORF of CgF-box as probe and result showed multiple copy of bands (Fig. 2), which absolutely proved F-box gene family existed in pummelo.
Previously we cloned another core component and subunit of the SCF (SKP1-CUL1-F-box protein)-type E3 Ub ligases in pummelo, SKP1 (S-phase Kinase-associated Protein 1) gene. It was up-regulated during flower development in pummelo [38]. In the SCF complex, SKP1 will directly interact with F-box forming function unit together with CUL1 [39]. Herein, CgF-box was also up-regulated during the flower development process in pummelo. The expression pattern of SKP1 and CgF-box was coincident with each other. Both of results suggest that Ub-dependent proteolytic pathway plays an important role and participates in the flower maturation process of pummelo.
‘Shatian’ pummelo shows typical self-incompatible characteristic which is very similar with the gametophytic self-incompatibility (GSI) system of Rosaceae [40]. In Rosaceae, the S-RNase gene was first isolated and identified as female component of the S-locus [41, 42]. We thought that some molecular mechanism could be also existed in pummelo. So we cloned an S-like RNase gene in pummelo previously. Further expression analysis verified that it played key role in ovary senescence process not self-incompatibility [26]. In Rosaceae, the male component associated with SI had been detected and functionally verified, named SLB/SLF (S-haplotype-specific F-box/S-locus F-box) [43]. To be SLF gene which should has some unique features, one of the important features is specific expression in the mature pollen [44]. For testing whether CgF-box is potential candidate of SLF or not in pummelo, we conducted RT-PCR and Real-time PCR experiment, both of these results showed that the expression of CgF-box was not pollen specific. So we concluded that CgF-box did not control the self-incompatibility mechanism in pummelo. Further analysis of expression of CgF-box gene in different tissue showed that it expressed in style petal and anther at highest level, but in ovary and leaf it expressed at the lowest level. These findings implied that CgF-box played different roles in different tissue. Further work needed to be carried out to understand its detailed function.
In conclusion, this is the first time to report cloning gene containing F-box conserved domain from ‘Shatian’ pummelo as well as citrus species. Expression analysis results suggested the gene not only involved in flower development process of ‘Zigui shatian’ Pummelo, but also played different roles in different tissue. Here, we provided the preliminary molecular information and function of CgF-box gene. These results constituted an important step for further research on the role of CgF-box in the flower development, especially for further cloning F-box family member, which is showing mature pollen specific expression possibility involved in pummelo self-incompatibility.
Abbreviations
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction.
- ORF:
-
Open reading frame.
References
Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55:555–590
Patton E, Willems AR, Tyers M (1998) Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet 14:236–243
Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99:11519–11524
Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479
Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533
Xiao W, Jang JC (2000) F-box proteins in Arabidopsis. Trends Plant Sci 5:454–457
Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW (1999) A family of mammalian F-box proteins. Curr Biol 9:1180–1182
Zeng L, Gu SH, Li Y, Wang WH, Huang Y, Ye X, Xu J, Zhao EP, Ji CN, Ying K, Xie Y, Mao YM (2004) cDNA cloning and expression analysis of a novel human F-box domain containing gene. Mol Biol Rep 31:51–57
Kawakami K, Amsterdam A, Shimoda N, Becker T, Mugg T, Shima A, Hopkins N (2000) Proviral insertions in the zebrafish hagoromo gene, encoding an F-box/WD40-repeat protein, cause stripe pattern anomalies. Curr Biol 10:463–466
Li Y, Yang SL, Tang ZL, Cui WT, Mu YL, Chu MX, Zhao SH, Wu ZF, Li K, Peng KM (2010) Expression and SNP association analysis of porcine FBXL4 gene. Mol Biol Rep 37:579–585
Wang ZW, Li XY, Tang ZL, Yang SL, Ying ZZ, Fu T, Fan B, Mu YL, Ao H, Li K (2010) Molecular characterization and association analysis of FBXO40 with partial hematological indexes in pig. Mol Biol Rep 37(7):3393–3400
Bai C, Richman R, Elledge SJ (1994) Human cyclin F. EMBO J 13:6087–6098
Kipreos ET, Pagano M (2000) The F-box protein family. Genome Biol 1: Rev 3002
Xu GX, Ma H, Nei M, Kong HZ (2009) Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci USA 106:835–840
Coates JC, Laplaze L, Haseloff J (2006) Armadillo-related proteins promote lateral root development in Arabidopsis. Proc Natl Acad Sci USA 103:1621–1626
Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF (TIR1)-dependent degradation of AUX/IAA proteins. Nature 414:271–276
Gao Y, Zhao Y, Li TT, Liu Y, Ren CX, Wang ML (2010) Molecular cloning and expression analysis of an F-box protein gene responsive to plant hormones in Brassica napus. Mol Biol Rep 37:1037–1044
Qiao H, Chang KN, Yazaki J, Ecker JR (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23(4):512–521
Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13:1779–1790
Luo Y, Lv GL, Wu WT, Chen SN, Cheng ZQ (2010) Analysis of genome expression in the response of Oryza granulata to Xanthomonas oryzae pv oryzae. Mol Biol Rep 37:875–892
Samach A, Klenz JE, Kohalmi SE, Risseeuw E, Haughn GW, Crosby WL (1999) The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J 20:433–445
Ingram GC, Doyle S, Carpenter R, Schultz EA, Simon R, Coen ES (1997) Dual role for FIMBRIATA in regulating floral homeotic genes and cell division in Antirrhinum. EMBO J 16:6521–6534
Ni W, Xie D, Hobbie L, Feng B, Zhao D, Akkara J, Ma H (2004) Regulation of flower development in Arabidopsis by SCF complexes. Plant Physiol 134:1574–1585
Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H (2003) Structural and transcriptional analysis of the self incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15(3):771–781
Yamane H, Ikeda K, Ushijima K, Sassa H, Tao R (2003) Pollen-expressed gene for a novel protein with an F-box motif that very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol 44(7):764–769
Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao TH (2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429(6989):302–305
Qiao H, Wang H, Zhao L, Zhou J, Huang J, Zhang Y, Xue Y (2004) The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16(3):582–595
Qiao H, Wang F, Zhao L, Zhou J, Lai Z, Zhang Y, Robbins TP, Xue Y (2004) The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 16(9):2307–2322
Masashi Y, Tatsuya K, Shigeto T (2006) Self- and cross-incompatibility of various citrus accessions. J Jpn Soc Hortic Sci 75(5):372–378
Chai LJ, Ge XX, Biswas MK, Xu Q, Deng XX (2010) Self-sterility in the mutant ‘Zigui shatian’ pummelo (Citrus grandis Osbeck) is due to abnormal post-zygotic embryo development and not self-incompatibility. Plant Cell, Tissue Organ Cult. doi: 10.1007/s11240-010-9793-6
Liu YZ, Liu Q, Tao NG, Deng XX (2006) Efficient isolation of RNA from fruit peel and pulp of ripening navel orange (Citrus sinesis Osbeck). J Huazhong Agric Univ 25(3):300–304
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Cheng YJ, Guo WW, Yi HL, Pang XM, Deng XX (2003) An efficient protocol for genomic DNA extraction from Citrus species. Plant Mol Biol Rep 21:177a–177g
Hu ZY, Tong Z, Wei H, Yi HL, Deng XX (2006) Mitochondrial gene expression in stamens is differentially regulated during male gametogenesis in Citrus unshiu. J Hortic Sci Biotechnol 81:565–569
Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW (2004) Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 18:2573–2580
Kuroda H, Takahashi N, Shimada H, Seki M, Shinozaki K, Matsui M (2002) Classification and expression analysis of Arabidopsis F-box containing protein genes. Plant Cell Physiol 43:1073–1085
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice: genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143:1467–1483
Chai LJ, Biswas MK, Ge XX, Deng XX (2010) Isolation, characterization, and expression analysis of an SKP1-like gene from ‘Shatian’ Pummelo (Citrus grandis Osbeck). Plant Mol Biol Rep 28:569–577
Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P (2006) F-box proteins everywhere. Curr Opin Plant Biol 9:631–638
Xue MN, Chen TT, Yang XH (1995) Observations on self and cross-compatibility in Shatinyu. Acta Hortic Sin 22(2):127–132
Sassa H, Nishio T, Kowyama Y, Hirano H, Koba T, Ikehashi H (1996) Self-incompatibility (S) alleles of the Rosaceae encodes members of a distinct class of the T2/S ribonuclease super family. Mol Gen Genet 50:547–557
Wang Y, Wang X, Skirpan AL, Kao TH (2003) S-RNase mediated self-incompatibility. J Exp Bot 54:115–122
Chai LJ, Ge XX, Xu Q, Deng XX (2010) CgSL2, an S-like RNase gene in ‘Zigui shatian’ pummelo (Citrus grandis Osbeck), is involved in ovary senescence. Mol Biol Rep. doi: 10.1007/s11033-010-0070-x
Kao TH, Tsukamoto T (2004) The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16:S72–S83
Acknowledgments
This research was financially supported by the National Basic Research Program of China (2011CB100600) and the National Natural Science Foundation of China (Nos. 30830078, 30921002).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chai, L., Ge, X., Biswas, M.K. et al. Molecular analysis and expression of a floral organ-relative F-box gene isolated from ‘Zigui shatian’ pummelo (Citrus grandis Osbeck). Mol Biol Rep 38, 4429–4436 (2011). https://doi.org/10.1007/s11033-010-0571-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0571-7