Abstract
Key message
The proposed height–diameter model applicable to many tree species in the multi-layered and mixed stands across Czech Republic shows a high accuracy in the height prediction. This model can be useful for estimating forest yield and biomass, and simulation of the vertical stand structures.
Abstract
We developed a generalized nonlinear mixed-effects height–diameter (H–D) model applicable to Norway spruce (Picea abies (L.) Karst.), European beech (Fagus sylvatica L.) and other conifer and broadleaved tree species using the modelling method that includes dummy variables accounting for species-specific height differences and random component accounting for within- and between-sample plot height differences and randomness in the data. We used two large datasets: the first set (model fitting dataset) originated from permanent research sample plots and second set (model-testing dataset) originated from the Czech national forest inventory (NFI) sample plots. The former dataset comprises 224 sample plots with 29,390 trees and the latter dataset comprises 14,903 sample plots with 382,540 trees, each representing wide variabilities of tree size, ecological zone, growth condition, stand structure and development stage, and management regime across the country. Among the four versatile growth functions evaluated as base functions with diameter at breast height (DBH) included as a single predictor, the Chapman-Richards function showed the most attractive fit statistics. This function was then extended through the integration of other predictor variables, which better describe the stand density (stand basal area), stand development and site quality (dominant height), competition (ratio of DBH to quadratic mean DBH), that would act as modifiers of the original parameters of the function. The mixed-effects H–D model described a large part of the variations in the H–D relationships (\(R_{{{\text{adj}}}}^{2}\) = 0.9182; RMSE = 2.7786) without substantial trends in the residuals. Testing this model against model-testing dataset confirmed the model’s high accuracy. The model may be used for estimating forest yield and biomass, and therefore will serve as an important tool for decision making in forestry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tree height and diameter at breast height (DBH) are important in forest inventories and timber management, as these variables are necessary for estimating stand volume and biomass. Tree height data modeled from DBH can be employed as an input variable in various forest models, together with DBH, such as growth and yield models, site productivity models, crown models, biomass models, and carbon budget models. These models serve as important tools in forest management decision making. Compared to the DBH measurement, measuring height is more tedious, time-consuming, costly, and has relatively larger errors, as it is affected by visual obstruction due to uneven landforms and crowded stands. Tree height is often measured for few trees per sample plot and height of the remaining trees is imputed using height–diameter model (H–D model) (MacPhee et al. 2018; Sharma and Breidenbach 2015; Vargas-Larreta et al. 2009). The H–D models are used to minimize the cost associated with inventories and to reduce the problems associated with height measurement errors (MacPhee et al. 2018). A realistic description of stand growth and stem volume, and simulation of stand structure and dynamics is possible only with use of the stand-specific H–D models (Mehtätalo et al. 2015). The H–D models may be used to evaluate site productivity (Duan et al. 2018; Fu et al. 2018; Vanclay 1994).
Based on the number of predictors used, there can be two types of H–D models: model that expresses height as a function of DBH (Fang and Bailey 1998; Robinson and Wykoff 2004), and model that includes DBH and other variables describing stand characteristics (Calama and Montero 2004; Paulo et al. 2011; Temesgen and Gadow 2004). The former model is known as a local model and appropriate for homogenous stands, while the latter model is known as a generalized model and appropriate for forests comprising heterogonous stands. The H–D relationship differs from one stand to another, and even within the same stand due to different competitive effects among trees (Clutter et al. 1983). Furthermore, site quality, stand structure, and development stage also significantly affect the H–D relationships (Calama and Montero 2004; Liu et al. 2017; Saud et al. 2016; Sharma and Parton 2007; Sharma and Breidenbach 2015; Schmidt et al. 2011; Vargas-Larreta et al. 2009). The height prediction accuracy can be increased through the integration of variables describing these stand characteristics into the model. In addition to this, an unstructured random component describing natural variability of tree heights, caused by stochastic factors must be included into the H–D models using the mixed-effects modelling approach. This allows for sample plot-specific height prediction, and enables the inclusion of correlations among the hierarchically structured random effects that account for correlations of the height measurements per sample plot (Pinheiro and Bates 2000; West et al. 1984; Vargas-Larreta et al. 2009). In contrast, a fixed-effect H–D model, also known as an ordinary least square (OLS) regression model, only provides unbiased mean height prediction, even under the unbalanced sampling design, which may not be a precise mean height for each sample plot. The OLS regression violates the assumptions of independent errors and leads to biased parameter estimates and variances, causing invalidation of the hypothesis tests (Gregoire et al. 1995; Pinheiro and Bates 2000; Fu et al. 2017a).
A large dataset acquired with a high measurement accuracy is necessary to develop the H–D model with robust parameter estimates. This dataset is often made available from forestry research projects, which would have established permanent sample plots covering all possible growth conditions and silviculture treatments. Measurement accuracy of the individual tree attributes on these sample plots may be relatively higher than that on other types of inventory sample plots, for example, national forest inventory (NFI) sample plots. The NFI may provide database for modelling H–D relationships; however, this database is not intended for modelling growth and other individual tree attributes, but for monitoring forests in general (Lawrence et al. 2010; Sharma et al. 2011). All empirical models including H–D models are developed based on the assumptions that all predictor variables are free from systematic errors. If they have such errors, the error-in-variable modelling approach (Kangas 1998; Sharma et al. 2011) must be applied, which is computationally complex. Developing forest models using research sample plot data and testing against NFI data is often carried out (Sharma et al. 2017c). Testing the models against NFI dataset increases credibility and confidence about the model, as NFI data cover wider environmental variabilities and growth conditions. In recent years, the NFI data have been used to develop various forest models despite their errors (Adame et al. 2008; Bollandsås and Næsset 2009; Crecente-Campo et al. 2010; Huuskonen and Miina 2007; Mehtatalo 2005; Monserud and Sterba 1996; Hasenauer and Monserud 1996; Nanos et al. 2004; Sharma and Breidenbach 2015; Sharma and Brunner 2017; Sharma et al. 2011, 2012).
Developing accurate H–D models for the multi-layered mixed forests is more challenging, as H–D relationships vary more in these stands than in a single species and/or a single-layered stands due to the effects of species-specific differences and vertical stand structural differences that also causes increased competitive interactions among trees (Eerikäinen 2009; Sharma et al. 2016b; Temesgen et al. 2014; Vargas-Larreta et al. 2009). When adequate measurements are not available to develop species-specific H–D models, a single model applicable to several tree species can be developed based on the species-specific data pooled together using the dummy variable modelling approach. This study aims (1) to develop a generalized mixed-effects H–D model applicable to several tree species using permanent research sample plot data with application of the dummy variable modelling approach, and (2) to evaluate the prediction performance of the H–D model using an independent dataset acquired from the Czech NFI sample plots. Variables describing effects of stand characteristics, dummy variables describing effects of species-specific differences, and random component describing the effects of sample plot-level variability on the H–D relationships were included into the model. The proposed model can be used for precise predictions of the tree height for several tree species from a minimum set of predictors that can be easily derived from forest inventory database.
Materials and methods
Data materials
Two different datasets were used in this study: one originating from permanent research sample plots (model fitting dataset, hereafter named as a training dataset) and another originating from the Czech NFI sample plots (model-testing dataset, hereafter named as a validation dataset). Dataset differs from each other in terms of sampling design, measurement methods and accuracy, statistical characteristics, and extent of the coverage of tree population. We briefly describe each of these datasets in the following sub-sections.
Training dataset
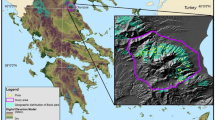
This dataset originates from 224 permanent research sample plots, hereafter termed as sample plots, which are squared-shaped and size varies from 2500 to 4900 m2. Sample plots are located in various parts of the Czech Republic (Fig. 1a), covering most of the forested parts of the country. All sample plots were established by considering criteria, such as canopy structure, status of regeneration, piles of dead wood, and stand development stage and site quality. It was assumed that sample plots represented the full range of variabilities in site quality, stand development stage, stand density, species mixture, stand structure, and management regime. Sample plot locations varied from 240 to 1370 m a.s.l., where mean climate characteristics and growing season length largely vary [i.e., annual temperature (4–9.5 °C), mean annual precipitation (500–1450 mm), and growing season length (45–180 days)]. Most of the studied stands, especially European beech and other broadleaved tree species stands originated from natural regeneration and about 20% Norway spruce and 60% Scots pine stands originated from plantation (Sharma et al. 2017d). About 70% stands aged between 20 and 165 years were left for spontaneous development where minimum selection felling was done, and this included salvage cutting and sanitary interventions, e.g., extraction of the trees affected by bark beetles and diseases. Management of the rest of studied forests included a mainly shelter wood selection system (minimum share of clear-cut), which involved formation of the 5% gap (Sharma et al. 2017b). Measurements from those sample plots, which were substantially damaged by natural epidemics such as air pollution, wind, bark beetles, fungal pathogens and diseases in 1980s (Vacek et al. 2013), were excluded. Description of the study sites is also available in the literature (Sharma et al. 2017a, 2016a, b, d; Vacek et al. 2016). In total, 23 tree species were included into the training dataset, comprising 25% monospecific sample plots (Scots pine 7%, Norway spruce 8% European beech 8% and others 2%) and 75% mixed species sample plots. Definition of the monospecific stands considered the inclusion of all individuals other than a species of interest if they had over-bark diameter at breast height (DBH, 1.3 m above ground) ≥ 4 cm (Sharma et al. 2016c). All individuals with DBH ≥ 2 and height ≥ 1.5 m were measured on each sample plot. Following the inventory protocols developed by Forest Management Institute (FMI 2003), all dendrometric measurements were made between April 2007 and March 2016. However, no repeated measurements were involved, meaning that no temporal variation was included into the data. DBH and total height were measured with precisions of 1 mm and 0.1 m, respectively. The numbers of height sample trees by species used in this study are presented in Appendix-T1.
Validation dataset
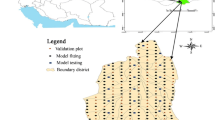
The validation dataset originates from the Czech NFI sample plots. Laying out of the NFI sample plots (Fig. 1b) involves two main steps: first, squared-shaped basic plots (inventory squares) were located in each 2 × 2 km grid, which were generated randomly for the territory of the Czech Republic. Second, two circular plots (of size 500 m2 and radius 12.62 m, hereafter, termed as sample plots) were located within each squared-shaped plot. The center of the first sample plot was located by generator of the random numbers either in the center of an inventory square or in its vicinity (0–3600), however, no further than 300 m. The center of the second circular plot was located by generator of the random numbers in the vicinity (0–3600) of the center of the first sample plot in a distance equaling 300 m. All sample plots were located by following the Field-Map technology of the IFER-Monitoring and Mapping Solutions Ltd (Šmelko and Merganič 2008). Locations of the NFI sample plot vary from 120 to 1405 m a.s.l. The first inventory was carried out between 2001 and 2004 and second inventory between 2011 and 2015. All trees with DBH ≥ 7 cm were measured on each sample plot. We used all measurements from both inventory cycles, which resulted in a selection of 14,903 sample plots with 382,540 trees, representing large variabilities in site quality, stand structure and development stage, stand density, species mixture, silvicultural tending, and other management interventions and natural disturbances. All tree and stand characteristics were measured following the inventory protocols developed by Forest Management Institute (FMI 2003). Details of sampling design and measurement methods applied to the NFI sample plots are reported in the literature (FMI 2007; Kučera 2016; Sharma et al. 2017c) or on the website (http://www.nil.uhul.cz). Conifer tree species predominate with a share of 58.9% (Norway spruce 44.1%), while broadleaved tree species occupy 41.1% (European beech 10.3%). The numbers of height sample trees by each species used in this study are presented in Appendix-T2. Graphs of the total height versus DBH in the training and validation datasets are shown in Fig. 2.
Data analysis
Tree and stand variables
Variables describing stand characteristics (e.g., site quality and stand development stage, stand density) would have significant influences on the H–D relationships (Calama and Montero 2004; Castedo-Dorado et al. 2006; Crecente-Campo et al. 2010; Sharma and Parton 2007). We evaluated many measured stand variables for their potential effects on the H–D relationships. However, we could not use site index as a site quality measure in our H–D model, as this could not be estimated due to lack of stand age in our data. Instead, we used dominant height (HDOM, m), which was calculated using methods suggested by Sharma et al. (2011, 2016a), to describe the combined effects of stand development and site quality (Fu et al. 2013, 2017c; Sharma et al. 2016a, c, 2017a, b, d). For mixed stands, where numbers of dominant trees of species or species group were inadequate as per the definition (i.e., 250 largest trees per hectare), height measurements of the largest trees per sample plot regardless of species were used to calculate HDOM. We assumed that HDOM in such a mixed stand would also reflect stand development stage. We also calculated dominant diameter (DDOM—an average DBH of dominant trees, cm). Both DDOM and HDOM are considered independent of thinning, except in the case of thinning from above. Most of forest stands in the Czech Republic were thinned from below. We evaluated the potential contributions of stem crowding (N, ha−1) and stand basal area (BA, m2 ha−1) to the H–D model. We evaluated other sample plot-level measures, such as total DBH (DBHsum, cm), arithmetic mean DBH (AMD, cm), arithmetic mean height (AMH, m), and quadratic mean DBH (QMD, cm), DBH difference between the thickest and thinnest trees (DBHrange, cm), height difference between the tallest and shortest trees (Hrange, m), and tallest tree per sample plot (HTALL, m). We also evaluated the tree-centred competition measures, such as ratio of DBH to QMD (dq) and basal area of trees larger in diameter than a subject tree (BAL, m2 ha−1) for their potential contributions to the H–D model. All aforementioned variables were calculated using all living trees per sample plot regardless of species. Summary statistics including mean, minimum, maximum, and standard deviation of the main tree and stand variables in the training and validation datasets are presented in Table 1.
Selection of a base model
The relationship between height and diameter of a tree through time is a nonlinear, because growth of the height with respect to diameter increases rapidly in earlier age and slowly in the latter age, thereby exhibiting a sigmoidal pattern (Clutter et al. 1983; Lei and Parresol 2001). Thus, selection of a functional form for modelling H–D relationship should not be restricted to the ease-of-fit to the data, but also should consider characteristics of a chosen function, i.e., sigmoidal pattern that has an inflection point, monotonic increment, and asymptote (Castedo-Dorado et al. 2005; Lei and Parresol 2001). We considered only those growth functions, which possess these important characteristics, in our preliminary analyses, and they are Chapman-Richards function (Chapman 1961; Richards 1959), Weibull function (Yang et al. 1978), Schnute function (Schnute 1981), and Korf function (Zeide 1989). These functions have frequently been used for modelling H–D relationships (e.g., Peng 2001; Zhang 1997; Sharma and Zhang 2004; Sharma and Parton 2007; Li et al. 2015) and modelling other individual tree attributes (Burkhart and Tomé 2012; Carmean and Lenthall 1989; Goelz and Burk 1992; Huang and Titus 1994; Lei and Parresol 2001; Tewari and Kishan Kumar 2002). We fitted the functions using OLS method, and following the form of the Chapman-Richards function (Eq. 1) was found the most appropriate to our data, as it showed the least residual variations (i.e., smallest sum of squared errors). Henceforth, this model is termed as a base model.
where Hij, and DBHij are total height (m) and diameter at breast height (cm) of the jth tree on the ith sample plot, respectively, b1,b2,b3 are parameters to be estimated, and \({\varepsilon _{ij}}\) is an error term. In the context of H–D modelling, it is a common practice to force the H–D curve to pass through (0, 1.3) to prevent the negative estimate of height for small trees, and therefore 1.3 was added to avoid the illogical estimates when DBH would approach zero.
Extension of a base model
The H–D relationship is significantly influenced by other tree and stand characteristics, such as tree health and vigor, site quality and stand development stage, and stand density or competition, and species mixture in a stand (Calama and Montero 2004; Sharma and Zhang 2004). Considering this, we evaluated several variables (Table 1) for their potential contributions to description of the variations in the H–D relationships. Evaluation was based on whether variables were suited to the model fitting procedure beginning with graphical exploration of the data and examination of the correlation statistics (Fu et al. 2017a; Sharma and Breidenbach 2015; Uzoh and Oliver 2008). The interaction effects between predictor variables and their transformations (logarithmic, inverse, and square) were also evaluated. We identified HDOM, BA, and dq as more significantly contributing predictors than others based on the results from the stepwise variable selection procedures (Montgomery et al. 2001). The model provided the greatest fitting improvement to the data when b1 of Eq. 1 was modeled as a function of HDOM, BA, and dq as below:
where HDOM = dominant height (m); BA = stand basal area (m2 ha−1); dq = ratio of DBH to quadratic mean DBH. For the purpose of simplicity (i.e., ease-of-fit to the data, and easy application of the developed H–D model), two individual species (Norway spruce, European beech) and two species groups were considered from a total of 23 tree species recorded from research sample plot inventory (see Appendix-T1). Following the principles of modelling indicator variables (Bates and Watts 1988; Sharma et al. 2012), we formed three dummy variables (S1, S2, S3) for four tree species to describe the effect of species-specific differences on the H–D relationships.
Species | S1 | S2 | S3 |
|---|---|---|---|
Norway spruce (TS1) | 0 | 0 | 0 |
Other conifers (TS2) | 1 | 0 | 0 |
European beech (TS3) | 0 | 1 | 0 |
Other broadleaved (TS4) | 0 | 0 | 1 |
The effect of species-specific differences was best described when b1 of a base model (Eq. 1) was expressed as a linear function of dummy variables as below:
Our database contained multiple sampling units (multiple species, multiple trees and measurements within each sample plot), and therefore we formulated the mixed-effects H–D model using Eq. 1 (with Eq. 2a, 2b included) through inclusion of the sample plot random effects. The objective of inclusion of the random effects into the fixed-effect model is to secure a higher prediction accuracy (Pinheiro and Bates 2000; Sharma et al. 2017d; Vargas-Larreta et al. 2009). Various mixed-effects model variants formed with all possible combinations of the random effects and three fixed parameters in Eq. 1 were fitted to the data. However, convergence with the smallest Akaike information criterion (AIC) was found only with the random effects added to b1 and b3 in Eq. 1 (with Eq. 2a, 2b included). The mixed-effects model formulation steps are available in the standard textbooks (Pinheiro and Bates 2000; Vonesh and Chinchilli 1997), and therefore, only final form of our mixed-effects H–D model is presented here (Eq. 3) that best described the variations of the H–D relationships in our data:
where Sk is tree species or species group (k = 1, 2, 3); bk (k = 1, 2, 3) and αk (k = 1,2,...,7) are parameters to be estimated, and \({\varepsilon _{ij}}\) is an error term; and all other abbreviations are the same as in Eq. 1, 2a, and 2b. In this equation, vectors of error \({\varepsilon _{ij}}\) and sample plot random effects (ui1, ui2) are defined by \(~{\varvec{\varepsilon}_{\varvec{i}}}\sim {\varvec{N}}\left( {0,~{\varvec{R}}} \right)~{\text{and}}~~{{\varvec{u}}_{\varvec{i}}}\sim {\varvec{N}}\left( {0,~{\varvec{D}}} \right)\), respectively, meaning that error vector εi is assumed to have a normal distribution with zero mean and within-sample plot variance–covariance matrix R, defined by Eq. 4.
A vector ui of the random effects (ui1, ui2) in Eq. 3 was assumed to have a multivariate normal distribution with zero mean and sample plot variance–covariance matrix D, defined by Eq. 5.
In Eq. 4, σ2 is a scaling factor for error dispersion (Gregoire et al. 1995) and it is equivalent to the residual variance of the estimated H–D model and common to all sample plots. A matrix Гi accounts for within-sample plot autocorrelations of the residual errors, but this was assumed to be an identity matrix, Ii, because our data lacked the repeated measurements. A matrix Gi accounts for a within-sample plot variance heteroscedasticity, and its diagonal elements are provided by variance function introduced into the H–D model. Since our preliminary analyses showed a within-sample plot heteroskedasticity in the residuals even after inclusion of the random effects, we added a power variance function of the following formulation (Eq. 6) to the mixed-effects H–D model (Eq. 3). This function reduced the heteroscedasticity most effectively among three-variance functions evaluated, such as exponential, power, and constant plus power functions (Fu et al. 2013; Pinheiro and Bates 2000).
where \(\phi\) is a parameter to be estimated; \({\hat {H}_{ij}}~\) is the estimated height of jth tree on the ith sample plot using fixed part of the mixed-effects model (Eq. 3); and σ2 is the same as defined in Eq. 4.
Model estimation and evaluation
We estimated the generalized mixed-effects H–D model with the restricted maximum likelihood in the SAS macro NLINMIX (SAS Institute Inc. 2012) using the expansion-around-zero method (Littell et al. 2006). However, we estimated all base models with OLS method using PROC NLIN in SAS (SAS Institute Inc. 2012). We used most common goodness-of-fit statistics to evaluate the fitted model alternatives, and they are root mean squared error (RMSE) that analyses precision of estimation, coefficient of determination (R2) that reflects total variability described by model considering total numbers of the parameters to be estimated, and AIC. The AIC is based on minimizing the Kullback–Lieber distance and it imposes the penalty for the number of parameters of the model (Akaike 1972; Burnham and Anderson 2002). The formulae of these three statistical measures are available in the statistical textbooks, e.g.. Montgomery et al. (2001). The R2 for mixed-effects model is often expressed into two different coefficient of determinations: marginal coefficient of determination (\(R_{{\text{m}}}^{{\text{2}}}\)) and conditional coefficient of determination (\(R_{{\text{c}}}^{2}\)). The former is concerned with variance described by fixed-effect factors, but latter is concerned with variance described by both fixed and random effect factors (Nakagawa and Schielzeth 2013; Sharma et al. 2017b). We also analyzed the residual graphs and simulated H–D curves overlaid on the measured data. Unless otherwise specified, we used 1% level of significance in all analyses. We tested the H–D model using NFI dataset. We examined the sample plot-specific H–D curves produced with a calibrated mixed-effects H–D model using the random effects that were estimated with the empirical best linear unbiased prediction (EBLUP) method (Pinheiro and Bates 2000).
Calibration of the mixed-effects model and subject-specific height prediction
Tree height can be predicted with or without using random effects in the mixed-effects H–D model. A prediction with the estimated random effects included is known as calibration or localization of the mixed-effects model (Pinheiro and Bates 2000). This requires the prior measurements of a response variable (tree height) for every sample plot in addition to the information of other predictors of the model. Height measurements of any number of trees per sample plot may be used for estimation of the random effects and adjusted to a fixed part of the mixed-effects H–D model. We calibrated the mixed-effects H–D model using the random effects estimated from the randomly selected trees that varied from one to four, depending on the numbers of trees of a particular species or species group available on the plot. We applied the following EBLUP equation (Pinheiro and Bates 2000) to predict the sample plot-specific random effects using PROC IML in SAS (SAS Institute Inc. 2012).
where ui is a random effect vector accounting for sample plot variations of the mean H–D relationships for the ith sample plot. The values of \({{\varvec{R}}_{\varvec{i}}}~{\text{and }}{\varvec{D}}~\) were obtained from Eq. 4, 5, respectively. A vector \(~{\varvec{\varepsilon}_{\varvec{i}}}\) containing residual errors was obtained from fixed part of the mixed-effects H–D model. The elements of a designed matrix Zi, were obtained from partial derivatives of Eq. 3 with respect to its fixed parameters (b1 and b3) (Calama and Montero 2004; Castedo-Dorado et al. 2005; Crecente-Campo et al. 2010; Pinheiro and Bates 2000; Sharma and Parton 2007) as shown below.
where ln is a natural logarithm, all other abbreviations and symbols are the same as in Eqs. 1, 2a, 2b and 3. Since our objective of developing the mixed-effects H–D model was for subject-specific height prediction, we evaluated the prediction performance of the model in both training and validation datasets using the following statistical measure (Huang et al. 2009; Sharma et al. 2017d):
where \({\bar {e}_i}\) is a mean prediction error for tree heights on the ith sample plot, \({H_{ij}}~\) and \({\hat {H}_{ij}}~\) are measured and predicted heights for the jth tree on the ith sample plot, \({\bar {H}_{ij}}~\) is a mean of measured height for the jth tree on the ith sample plot, and ni is the number of observations for the ith sample plot. In addition to this, we also examined other common prediction statistics, such as RMSE and R2, and sample plot-specific H–D curves produced with a calibrated model.
Results
The base model described less than 81% variations in the H–D relationship for each species or species group (\(R_{{{\text{adj}}}}^{2}\) = 0.81: Norway spruce; 0.74: other confers; 0.79: European beech; 0.74: other broadleaved) while applying the OLS method to estimate its parameters. When covariate predictors: dominant height (HDOM), stand basal area (BA), ratio of DBH to quadratic mean DBH (dq) were added to base model, there was a substantial improvement on the model fits (\(R_{{{\text{adj}}}}^{2}\) = 0.89: Norway spruce; 0.81: other confers; 0.88%: European beech; 0.78%: other broadleaved). Model fits were further improved when sample plot random effect parameters (ui1, ui2) and power variance function (Eq. 6) were included into the model, i.e., model described the largest part of the variations in the H–D relationship for each species or species group (92.4%: Norway spruce; 88.3%: other confers; 91.6%: European beech; 84.4%: other broadleaved). Since our main interest of developing H–D model is for sample plot-specific prediction, only results of the mixed-effects model are presented in Table 2. All parameter estimates of the mixed-effects model (Eq. 3) were highly significant, even p value of the least significant estimate of α3 was less than 0.0002, and all estimated values and signs are biologically plausible and interpretable. Differences of the parameter estimates of dummy variables indicated clear differences in the H–D relationships among tree species or species groups. The reduction of unexplained variance (i.e., mean squared errors, σ2) in the mixed-effects model relative to its OLS model variant was about 44%. A larger value of ui1 indicated that b1 in Eq. 3 was more strongly correlated to sample plot variations than b3 (Table 2).
Within-sample plot heteroskedasticity in the residuals was substantially reduced by a power variance function that was expressed as a function of the predicted height [Eq. 6, with ϕ = 0.4529, estimated from data] (Fig. 3). A large deviation in the residuals was seen only for a few trees of Norway spruce (TS1) and other conifers (TS2), which were caused by extreme outlier observations. Histograms of the residuals showed the Gaussian distribution patterns, indicating that significant skewness was absent in the residuals.
The effects of HDOM, BA, and dq on the H–D relationship for each species or species group were simulated (Fig. 4). As expected, HDOM provided the largest contribution to the H–D model, followed by dq and BA. The simulation showed that tree heights increased with increasing effect of stand development and site quality described by HDOM and competition effect described by dq. However, tree height decreased with increasing BA when all other conditions were assumed the same.
Effects of dominant height (HDOM), stand basal area (BA), and ratio of DBH to quadratic mean DBH (dq) on the height–diameter relationships for different species and species groups. Curves were produced using the parameter estimates in Table 2. Mean values of the data were used for predictors except the variable of interest in the figure, which varied from approximately minimum to maximum in the measured data [TS = 1: Norway spruce; 2: other conifers; 3: European beech; 4: other broadleaved]
The calibrated H–D model described the H–D relationships adequately well in the validation dataset when random effects were predicted from height measurements of the randomly selected trees that varied from one to four, depending on the numbers of trees of a particular species or species group available on the plot. The prediction statistics R2 and RMSE for validation data were much promising, i.e., R2 varied from 0.93 to 0.97 with the largest R2 for Norway spruce and smallest R2 for other broadleaved species (TS4). The prediction statistics and graphs of the prediction errors of the calibrated H–D model applied to few major species or species groups in validation data are presented in Appendix-T3 and Appendix-F1, respectively. Testing the model against the data from four major tree species (Norway spruce, European beech, Scots pine, European larch) and two major species groups (fir species, oak species) confirmed the model’s high accuracy. We also analyzed prediction statistics and graphs of the prediction errors for several tree species falling under TS2 and TS4 in the validation dataset, and found high precisions (i.e., R2 > 0.91 and absence of substantial trends in the prediction errors).
Most of the prediction biases were found falling within ± 20% range for more than 96% sample plots in each species or species group, indicating that the calibrated H–D model for the majority of sample plots in both training and validation datasets performed adequately well (Fig. 5). However, a large bias (bias > 20%) still remained to be described for less than 4% sample plots in both datasets. A slightly larger negative skewness in the prediction bias was present in both datasets, indicating that our model slightly over-predicted the height for some extremely small trees on those sample plots where taller trees existed in abundance.
Prediction bias of a calibrated model was calculated using Eq. 10 in both training and validation datasets. The mixed-effects model was calibrated with the random effects predicted using height measurements of the randomly selected trees that varied from one to four, depending on the number of trees of a particular species or species group available on the plot [TS = 1: Norway spruce; 2: other conifers; 3: European beech; 4: other broadleaved]
We examined the sample plot-specific H–D curves produced by calibrated mixed-effects model and overlaid them on the measured height–DBH pairs of both datasets (Fig. 6). Except for few sample plots, where there were outlier measurements, the height curves produced with a calibrated model showed a complete coverage to the measured data for each species or species group. For mixed species sample plots, H–D curves were clearly differentiated, i.e., curves passed through the middle of the measured data for each species or species group.
Discussion
Based on the evaluation of four versatile growth functions, the Chapman-Richards function was selected as a base function to include covariate predictors and random effect parameters. Selection of the base functions should be based on the principles of functions’ logical behavior, high accuracy, and suitability in the practical application (Borders 1989; Castedo-Dorado et al. 2005; Goelz and Burk 1992; Liu et al. 2017), and all functions we used to fit our data would meet these requirements. The Chapman-Richards function is flexible not only for modelling H–D relationships (Lei and Parresol 2001; Fang and Bailey 1998; Huang et al. 2000; Sharma and Parton 2007; Sharma and Zhang 2004; Vargas-Larreta et al. 2009; Zhang 1997), but also for modelling other individual tree attributes (Burkhart and Tomé 2012; Carmean and Lenthall 1989; Goelz and Burk 1992; Huang and Titus 1994; Lei and Parresol 2001; Tewari and Kishan Kumar 2002). Compared to various predictors evaluated (Table 1), stand basal area (BA), ratio of DBH to quadratic mean DBH (dq) and dominant height (HDOM) more significantly contributed to the model. Among them, HDOM appeared the most influencing one (Fig. 4). This is commonly used in the H–D model, because it describes the combined effects of site quality and stand development stages on the H–D relationships (Calama and Montero 2004; Castedo-Dorado et al. 2006; Crecente-Campo et al. 2010; Eerikainen 2003; Sharma and Parton 2007; Vargas-Larreta et al. 2009). The HDOM indicates site quality in terms of growth and yield capacity of a stand. Therefore, HDOM is not used only in the H–D models, but also in other individual tree models, such as crown models (Fu et al. 2013, 2017a, 2017b; Sharma et al. 2016a, 2017a; Soares and Tomé 2001), height-to-diameter ratio models (Sharma et al. 2016c); height to crown base models (Fu et al. 2017c; Sharma et al. 2017d), crown-to-bole diameter ratio models (Sharma et al. 2017b), and crown height increment model (Short III and Burkhart 1992). However, HDOM cannot be considered as an effective measure as a site index in describing site quality, because the same HDOM can be achieved by younger stands on better site quality and older stands on poorer site quality.
With advancing stand stages through time, tree height also increases for a given stand density and competition level. The dq and BA describe the effects of stand density and competition on the H–D relationships, also contributed significantly to fitting improvement of the H–D model; however, contribution of each of them was less than that of HDOM (Fig. 4). Our data indicate that competition among individual trees increases with increasing dq, and this takes into account a level of the competition, as there is a close relationship between dq and number of trees per hectare. More dense stands tend to result in taller trees considering the same diameter, provided that all other conditions are the same (Vargas-Larreta et al. 2009). However, with increasing BA, tree height decreases and its contribution to the H–D model becomes smaller, and this may be due to the abundance of thicker trees on the majority of sample plots, where stand is relatively more sparse and tree heights are shorter. Since stand density is the most obvious influencing factor on the H–D relationships, particularly for trees growing in the mixed stands (Huang and Titus 1994; Vargas-Larreta et al. 2009).
A description of a large part of the variations in the H–D relationships without substantial trends in the residuals (Table 2; Fig. 3) indicates that chosen base model (Eq. 1) and covariate predictors (Eq. 2a), species grouping and dummy variables formed (Eq. 2b), and variance function (Eq. 6) introduced were all suited to our data. The H–D model behaves significantly differently in a particular species or species group (Fig. 4). This is due to a large effect of species-specific difference that was modeled. Only few observations could not be properly described, because they are extreme outliers (Figs. 3, 6). Each of the two species groups (TS2, TS4) contains several tree species in both datasets (Appendix-T1, T2), and therefore model may not be as accurate as that for other two tree species (TS1, TS3). This may be due to the height differences that are larger in TS2 and TS4 than in TS1 and TS3. It would be worthwhile to develop the species-specific H–D models; however, it was not possible due to the limited observations of some species (Appendix-T1). A common H–D model applicable to several tree species can be more appropriate than using separate species-specific models, as former model increases the work efficiency. The calibrated H–D model (localized model) is able to describe most of the variations in the H–D relationships for each species or species group in the validation dataset. The sample plot-specific H–D curves produced with a localized model for a particular species or species group cover most of the height measurements in each dataset (Fig. 6), indicating that our model may be adequate for precise predictions of tree heights. The calibrated curves would be very similar to the curves that would result from separately fitting of the sample plot-specific data, especially when number of trees per sample plot is adequate.
Sample plot-specific curves overlaid on the measured data (gray dots). Sample plot mean of the data were used for all predictors except ratio of DBH to quadratic mean DBH, which varied from approximately minimum to maximum range by 2 m interval. Model was calibrated with the random effects predicted using height measurements of the randomly selected trees that varied from one to four, depending on the number of trees of a particular species or species group available on the plot [TS = 1: Norway spruce; 2: other conifers; 3: European beech; 4: other broadleaved]
Even though the mixed-effects H–D model without calibration (only fixed part of the mixed model) could be used to predict height, prediction accuracy would be significantly low. Therefore, only calibrated mixed-effects H–D models are recommended for more accurate predictions (Calama and Montero 2004; Crecente-Campo et al. 2010; Robinson and Wykoff 2004; Sharma and Parton 2007; Sharma and Breidenbach 2015). However, prediction accuracies of these models depend on the application conditions of stands, such as vertical stand structure and chosen number of sample trees and representativeness of the measurements of heights that are used in calibration (Adame et al. 2008; Crecente-Campo et al. 2010; Mehtätalo et al. 2015). For a stand with homogenous structures, using height measurement of a single tree per sample plot in calibration may provide a high prediction accuracy (Trincado et al. 2007). The prediction errors could be significantly reduced for multi-layered stands, no matter whether they are monospecific or mixed stands, when height measurements of at least four trees of the same species or species group were used in calibration (Crecente-Campo et al. 2010; Sharma and Breidenbach 2015; Sharma et al. 2016b). The calibration based on a small sample size can be the most efficient while applying the model (Calama and Montero 2004; Sharma and Parton 2007). Except on less than 4% sample plots, only small prediction bias was present when calibration was done using height measurements from the randomly selected trees that varied from one to four, depending on the number of trees of a particular species or species group available on the plot (Fig. 5). Larger prediction biases for some sample plots are due to the multi-layered canopy of trees, where height differences are relatively larger. The prediction accuracies for these sample plots could be increased with increasing number of sample trees used in calibration. The higher the number of sample trees used for calibration of the mixed-effects H–D model is, the higher the prediction accuracy would be (Calama and Montero 2004; Castedo-Dorado et al. 2006; Crecente-Campo et al. 2010; Sharma and Breidenbach 2015; Sharma et al. 2016b; Temesgen et al. 2008). Existing H–D modelling studies (Calama and Montero 2004; Crecente-Campo et al. 2010; Sharma and Breidenbach 2015; Sharma et al. 2016b) and many other tree attributes modelling studies (Fu et al. 2013, 2017a, 2017c; Sharma et al. 2017a, d) show that four or five trees per sample plot are optimum for model calibration. Using more than four trees per sample plot in calibration may lead to a higher inventory cost with very little accuracy gain. Using one to four trees per sample plot may compromise between inventory cost and prediction accuracy.
Developing a simple and accurate H–D model makes it possible for model users to predict tree heights by relying on measurements of DBH and other covariate predictors that can be derived from forest inventory databases. Existing H–D models (Adamec 2015; Adamec and Drápela 2015; Sharma et al. 2016b) are based on small datasets acquired from few forest stands that are confined to small localities in the Czech Republic, and therefore these models cannot be used for forest stands of other localities. The inventory crew have been compelled to measure the heights of all trees on the NFI plots, permanent research plots, and temporary sample plots due to lack of a composite H–D model that can be applicable to several tree species. Our model will be useful for the inventory crew, who may measure the heights of only a few trees per plot and predict the heights of the remaining trees using this model. This may save time and cost required for subsequent cycles of the Czech NFI and permanent research plot inventory. For stand conditions more or less similar to the basis of this study, our model may be applicable to the forests across other European countries. However, it needs to be tested before application, as various factors that may vary from country to country, even within the same country from region to region, significantly affect the H–D relationships. Our model is parsimonious, as it has only three covariate predictors, and therefore it can be efficient for practical application. The inclusion of many predictors into the model increases forest inventory costs.
Even though tree and stand variables are assumed to be measured without systematic errors, tree height measurements may be subject to errors. These errors may be substantial (Omule 1980). In all H–D models cited in this article and others, including the model developed in our study, it is assumed that (1) tree height is a random variable and (2) other covariate predictors are fixed and measured without errors. It is known that violation of the second assumption leads to biased parameter estimates and variance, which causes invalidation of the hypothesis tests (Fu et al. 2017b; Fuller 1987; Rencher and Schaalje 2008). When predictors in the H–D model (Eq. 3) are considered to have significant errors, a complex modelling—an error-in-variable modelling approach (Kangas 1998; Sharma et al. 2011) needs to be applied.
Conclusion
Based on the modelling methods and results presented in this article, we conclude that a single generalized mixed-effects H–D model (a composite model) developed from research sample plot data using the combined approach of the mixed-effects modelling and dummy variable modelling can be used for precise height predictions for the same tree species and species groups in the NFI data. The generalized mixed-effects H–D model, which includes three covariate predictors (dominant height, ratio of individual tree DBH to quadratic mean DBH, stand basal area), and sample plot-level random effects estimated using height measurements of one to four trees depending on the number of trees of a particular species or species group available on the plot, significantly improved the prediction accuracy. The methods and model recommended in this article are based on the in-depth analyses of various graphs (residuals, prediction errors and biases, simulated height curves), goodness-of-fit statistics and prediction statistics. Unlike in the past, measuring heights of all trees on each sample plot will not be necessary in any forest inventory in the future. Instead, only heights of selected trees per sample plot may be measured and missing height measurements for the remaining trees will be imputed using our model. This will reduce the forest inventory cost. The presented model can be used for characterization of the vertical stand structures, estimation of volume and biomass, and simulation of stand dynamics. Our model will serves as an important tool in forest management decision making.
References
Adame P, del Rio M, Canellas I (2008) A mixed nonlinear height–diameter model for Pyrenean oak (Quercus pyrenaica Willd.). For Ecol Manag 256:88–98
Adamec Z (2015) Comparison of linear mixed effects model and generalized model of the tree height–diameter relationship. J For Sci 61:439–447
Adamec Z, Drápela K (2015) Generalized additive models as an alternative approach to the modelling of the tree height–diameter relationship. J For Sci 61:235–243
Akaike H (1972) A new look at statistical model identification. IEEE Trans Autom Control 19(6):716–723
Bates DM, Watts DG (1988) Nonlinear regression analysis and its applications. Wiley, New York
Bollandsås OM, Næsset E (2009) Weibull models for single-tree increment of Norway spruce, Scots pine, birch and other broadleaves in Norway. Scand J For Res 24:54–66
Borders BE (1989) Systems of equations in forest stand modeling. For Sci 35:548–556
Burkhart H, Tomé M (2012) Modeling forest trees and stands. Springer, 471 p
Burnham KP, Anderson DR (2002) Model selection and inference: a practical information-theoretic approach. Springer-Verlag, New York
Calama R, Montero G (2004) Interregional nonlinear height–diameter model with random coefficients for stone pine in Spain. Can J For Res 34:150–163
Carmean WH, Lenthall DJ (1989) Height-growth and site-index curves for jack pine in north central Ontario. Can J For Res 19:215–224
Castedo-Dorado F, Anta MB, Parresol BR, Gonzalez JGA (2005) A stochastic height–diameter model for maritime pine ecoregions in Galicia (northwestern Spain). Ann For Sci 62:455–465
Castedo-Dorado F, Dieguez-Aranda U, Anta MB, Rodriguez MS, von Gadow K (2006) A generalized height–diameter model including random components for radiata pine plantations in northwestern Spain. For Ecol Manag 229:202–213
Chapman DG (1961) Statistical problems in dynamics of exploited fisheries populations. Neyman J (ed) Proceedings of 4th Berkeley Symposium on mathematical statistics and probability, Berkeley, Vol. 4, pp 153–168
Clutter JL, Fortson JC, Pienaar LV, Brister GH, Bailey RL (1983) Timber management: a quantitative approach. Wiley, New York, 333 p
Crecente-Campo F, Tomé M, Soares P, Dieguez-Aranda U (2010) A generalized nonlinear mixed-effects height–diameter model for Eucalyptus globulus L. in northwestern Spain. For Ecol Manag 259:943–952
Duan G, Gao Z, Wang Q, Fu L (2018) Comparison of different height–diameter modelling techniques for prediction of site productivity in natural uneven-aged pure stands. Forests 9:63
Eerikainen K (2003) Predicting the height–diameter pattern of planted Pinus kesiya stands in Zambia and Zimbabwe. For Ecol Manag 175:355–366
Eerikäinen K (2009) A multivariate linear mixed-effects model for the generalization of sample tree heights and crown ratios in the Finnish National Forest Inventory. For Sci 55:480–493
Fang Z, Bailey RL (1998) Height–diameter models for tropical forests on Hainan Island in southern China. For Ecol Manag 110:315–327
FMI (2003) Inventarizace lesů, Metodika venkovního sběru dat [Forest inventory, field data collection methodology]. Brandýs nad Labem, 136 p
FMI (2007) National Forest Inventory in the Czech Republic 2001–2004; Introduction, methods, results. Brandýs nad Labem, 224 p
Fu L, Sun H, Sharma RP, Lei Y, Zhang H, Tang S (2013) Nonlinear mixed-effects crown width models for individual trees of Chinese fir (Cunninghamia lanceolata) in south-central China. For Ecol Manag 302:210–220
Fu L, Sharma RP, Hao K, Tang S (2017a) A generalized interregional nonlinear mixed-effects crown width model for Prince Rupprecht larch in northern China. For Ecol Manag 389:364–373
Fu L, Sharma RP, Wang G, Tang S (2017b) Modelling a system of nonlinear additive crown width models applying seemingly unrelated regression for Prince Rupprecht larch in northern China. For Ecol Manag 386:71–80
Fu L, Zhang H, Sharma RP, Pang L, Wang G (2017c) A generalized nonlinear mixed-effects height to crown base model for Mongolian oak in northeast China. For Ecol Manag 384:34–43
Fu L, Li X, Sharma RP (2018) Comparing height–age and height–diameter modelling approaches for estimating site productivity of natural uneven-aged forests. Forestry 91(4):419–433
Fuller WA (1987) Measurement error models. Wiley, New York, 445 p
Goelz JCG, Burk TE (1992) Development of a well-behaved site index equation-Jack pine in north central Ontario. Can J For Res 22:776–784
Gregoire TG, Schabenberger O, Barrett JP (1995) Linear modelling of irregularly spaced, unbalanced, longitudinal data from permanent-plot measurements. Can J For Res 25:137–156
Hasenauer H, Monserud RA (1996) A crown ratio model for Austrian forests. For Ecol Manag 84:49–60
Huang S, Titus SJ (1994) An age-independent individual tree height prediction model for boreal spruce-aspen stands in Alberta. Can J For Res 24:1295–1301
Huang S, Price D, Titus J S (2000) Development of ecoregion-based height–diameter models for white spruce in boreal forests. For Ecol Manag 129:125–141
Huang S, Meng SX, Yang Y (2009) Using nonlinear mixed model technique to determine the optimal tree height prediciton model for black spruce. Modern Appl Sci 3:3–18
Huuskonen S, Miina J (2007) Stand-level growth models for young Scots pine stands in Finland. For Ecol Manag 241:49–61
Kangas AS (1998) Effect of errors-in-variables on coefficients of a growth model and on prediction of growth. For Ecol Manag 102:203–212
Kučera M (2016) Výstupy NIL2—Zastoupení dřevin a věková struktura Lesa [Outputs of NIL2—representation of tree species and the age structure of forest]. In: Vašíček J, Skála V (eds): XIX. Sněm lesníků. Národní inventarizace lesů 2, Hradec Králové, 37–53
Lawrence M, McRoberts RE, Tomppo E, Gschwantner T, Gabler K (2010) Comparisons of national forest inventories. In: National forest inventories: pathways for common reporting. Springer Netherlands, Dordrecht, pp 19–32
Lei Y, Parresol BR (2001) Remarks on height–diameter modelling. In: Research Note SRS-10. USDA Forest Service, Southern Research Station, Asheville
Li Y, Deng X, Huang Z, Xiang W, Yan W, Lei P, Zhou X, Peng C (2015) Development and evaluation of models for the relationship between tree height and diameter at breast height for Chinese-Fir plantations in subtropical China. PLoS One 10, e0125118
Littell RC, Milliken GA, Stroup WW, Wolfinger RD, Schabenberger O (2006) SAS for mixed models, 2nd edn. SAS Institute, Cary, 814
Liu M, Feng Z, Zhang Z, Ma C, Wang M, Lian B-l, Sun R, Zhang L (2017) Development and evaluation of height diameter at breast models for native Chinese Metasequoia. PLoS One 12:e0182170
MacPhee C, Kershaw JA, Weiskittel AR, Golding J, Lavigne MB (2018) Comparison of approaches for estimating individual tree height–diameter relationships in the Acadian forest region. For Int J For Res 91:132–146
Mehtatalo L (2005) Height–diameter models for Scots pine and birch in Finland. Silva Fennica 39:55–66
Mehtätalo L, de-Miguel S, Gregoire TG (2015) Modeling height–diameter curves for prediction. Can J For Res 45:826–837
Monserud RA, Sterba H (1996) A basal area increment model for individual trees growing in even- and uneven-aged forest stands in Austria. For Ecol Manag 80:57–80
Montgomery DC, Peck EA, Vining GG (2001) ‘Introduction to linear regression analysis. Wiley, New York, 641 p
Nakagawa S, Schielzeth H (2013) A general and simple method for obtaining R 2 from generalized linear mixed-effects models. Methods Ecol Evol 4:133–142
Nanos N, Calama R, Montero G, Gil L (2004) Geostatistical prediction of height/diameter models. For Ecol Manag 195:221–235
Omule SAY (1980) Personal bias in forest measurements. The Forestry Chronicle 56:222–224
Paulo JA, Tomé J, Tomé M (2011) Nonlinear fixed and random generalized height–diameter models for Portuguese cork oak stands. Ann For Sci 68:295–309
Peng C (2001) Developing and validating nonlinear height–diameter models for major tree species of Ontario’s boreal forest. North J Appl For 18:87–94
Pinheiro JC, Bates DM (2000) Mixed-effects models in S and S-PLUS. Springer, New York
Rencher AC, Schaalje GB (2008) Linear models in statistics, 2nd edn. Wiley, New York
Richards FJ (1959) A flexible growth function for empirical use. J Exp Bot 10:290–300
Robinson AP, Wykoff WR (2004) Imputing missing height measures using a mixed-effects modeling strategy. Can J For Res 34:2492–2500
SAS Institute Inc (2012) SAS/ETS1 9.1.3 User’s Guide. SAS Institute Inc., Cary
Saud P, Lynch TB, KC A, Guldin JM (2016) Using quadratic mean diameter and relative spacing index to enhance height–diameter and crown ratio models fitted to longitudinal data. Forestry 89:215–229
Schmidt M, Kiviste A, von Gadow K (2011) A spatially explicit height–diameter model for Scots pine in Estonia. Eur J Forest Res 130:303–315
Schnute J (1981) A Versatile Growth Model with Statistically Stable Parameters. Can J Fish Aquat Sci 38:1128–1140
Sharma RP, Breidenbach J (2015) Modeling height–diameter relationships for Norway spruce, Scots pine, and downy birch using Norwegian national forest inventory data. For Sci Technol 11:44–53
Sharma RP, Brunner A (2017) Modeling individual tree height growth of Norway spruce and Scots pine from national forest inventory data in Norway. Scand J For Res 32:501–514
Sharma M, Parton J (2007) Height–diameter equations for boreal tree species in Ontario using a mixed-effects modeling approach. For Ecol Manag 249:187–198
Sharma M, Zhang SY (2004) Height–diameter models using stand characteristics for Pinus banksiana and Picea mariana. Scand J For Res 19:442–451
Sharma RP, Brunner A, Eid T, Øyen B-H (2011) Modelling dominant height growth from national forest inventory individual tree data with short time series and large age errors. For Ecol Manag 262:2162–2175
Sharma RP, Brunner A, Eid T (2012) Site index prediction from site and climate variables for Norway spruce and Scots pine in Norway. Scand J For Res 27:619–636
Sharma RP, Vacek Z, Vacek S (2016a) Individual tree crown width models for Norway spruce and European beech in Czech Republic. For Ecol Manag 366:208–220
Sharma RP, Vacek Z, Vacek S (2016b) Nonlinear mixed effect height–diameter model for mixed species forests in the central part of the Czech Republic. J For Sci 62:470–484
Sharma RP, Vacek Zk, Vacek S (2016c) Modeling individual tree height to diameter ratio for Norway spruce and European beech in Czech Republic. Trees 30:1969–1982
Sharma RP, Bíllek L, Vacek Z, Vacek S (2017a) Modelling crown widtH–Diameter relationship for Scots pine in the central Europe. Trees 31:1875–1889
Sharma RP, Vacek Z, Vacek S (2017b) Modelling tree crown-to-bole diameter ratio for Norway spruce and European beech. Silva Fennica 51(5):article id 1740
Sharma RP, Vacek Z, Vacek S, Jansa V (2017c) Modelling individual tree diameter growth for Norway spruce in Czech Republic using generalized algebraic difference approach. J For Sci 63:227–238
Sharma RP, Vacek Z, Vacek S, Podrázský V, Jansa V (2017d) Modelling individual tree height to crown base of Norway spruce (Picea abies (L.) Karst.) and European beech (Fagus sylvatica L.). PLoS One 12:e0186394
Short EA III, Burkhart H (1992) Prediction crown-height increment for thinned and unthinned loblolly pine plantations. For Sci 38:594–610
Šmelko ŠS, Merganič J (2008) Some methodological aspects of the national forest inventory and monitoring in Slovakia. J For Sci 54:476–483
Soares P, Tomé M (2001) A tree crown ratio prediction equation for eucalypt plantations. Ann For Sci 58:193–202
Temesgen H, Gadow KV (2004) Generalized height–diameter models: an application for major tree species in complex stands of interior British Columbia. Eur J Forest Res 123:45–51
Temesgen H, Monleon VJ, Hann DW (2008) Analysis and comparison of nonlinear tree height prediction strategies for Douglas-fir forests. Can J For Res 38:553–565
Temesgen H, Zhang CH, Zhao XH (2014) Modelling tree height–diameter relationships in multi-species and multi-layered forests: a large observational study from Northeast China. For Ecol Manag 316:78–89
Tewari VP, Kishan Kumar VS (2002) Development of top height model and site index curves for Azadirachta indica A. juss. For Ecol Manag 165:67–73
Trincado G, VanderSchaaf CL, Burkhart HE (2007) Regional mixed-effects height–diameter models for loblolly pine (Pinus taeda L.) plantations. Eur J For Res 126:253–262
Uzoh FCC, Oliver WW (2008) Individual tree diameter increment model for managed even-aged stands of ponderosa pine throughout the western United States using a multilevel linear mixed effects model. For Ecol Manag 256:438–445
Vacek S, Bílek L, Schwarz O, Hejcmanová P, Mikeska M (2013) Effect of air pollution on the health status of Spruce stands. Mt Res Dev 33:40–50
Vacek S, Vacek Z, Bilek L, Simon J, Remeš J, Hůnová I, Král J, Putalova T, Mikeska M (2016) Structure, regeneration and growth of Scots pine (Pinus sylvestris L.) stands with respect to changing climate and environmental pollution. Silva Fennica 50(4):1564
Vanclay JK (1994) Modelling forest growth and yield. Applications to mixed tropical forests. CAB International, Oxon, 312 p
Vargas-Larreta B, Castedo-Dorado F, Alvarez-Gonzalez JG, Barrio-Anta M, Cruz-Cobos F (2009) A generalized height–diameter model with random coefficients for uneven-aged stands in El Salto, Durango (Mexico). For Int J For Res 82:445–462
Vonesh EF, Chinchilli VM (1997) Linear and nonlinear models for the analysis of repeated measurements. Marcel Dekker Inc., New York, 560 p
West PW, Ratkowsky DA, Davis AW (1984) Problems of hypothesis testing of regressions with multiple measurements from individual sampling units. For Ecol Manag 7:207–224
Yang RC, Kozak A, Smith JHG (1978) The potential of Weibull-type functions as flexible growth curves. Can J For Res 8:424–431
Zeide B (1989) Accuracy of equations describing diameter growth. Can J For Res 19:1283–1286
Zhang L (1997) Cross-validation of non-linear growth functions for modelling tree height–diameter relationships. Ann Bot 79:251–257
Acknowledgements
This study was supported by Czech Ministry of Agriculture (No. QJ1520037), Faculty of Forestry and Wood Sciences, Czech University of Life Sciences Prague (IGA No. B2018, B0318, and Excellent Output 2018), and EXTEMIT-k project (No. CZ.02.1.01/0.0/0.0/15_003/0000433). We thank two anonymous reviewers for their constructive comments and insightful suggestions.
Author information
Authors and Affiliations
Contributions
Ram Sharma: conceptualized and designed the study, performed data analysis and modelling, and wrote and revised the manuscript; Zdeněk Vacek: contributed to data descriptions and produced study area maps, and worked for revision, Stanislav Vacek: provided training dataset and contributed to the manuscript improvement. Miloš Kučera: provided validation dataset and contributed to the manuscript improvement.
Corresponding author
Ethics declarations
Conflict of interest
Authors have no conflict of interest.
Additional information
Communicated by Priesack.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharma, R.P., Vacek, Z., Vacek, S. et al. Modelling individual tree height–diameter relationships for multi-layered and multi-species forests in central Europe. Trees 33, 103–119 (2019). https://doi.org/10.1007/s00468-018-1762-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-018-1762-4