Abstract
While the importance of tight junctions in hearing is well established, the role of Claudin- 9 (CLDN9), a tight junction protein, in human hearing and deafness has not been explored. Through whole-genome sequencing, we identified a one base pair deletion (c.86delT) in CLDN9 in a consanguineous family from Turkey with autosomal recessive nonsyndromic hearing loss. Three affected members of the family had sensorineural hearing loss (SNHL) ranging from moderate to profound in severity. The variant is predicted to cause a frameshift and produce a truncated protein (p.Leu29ArgfsTer4) in this single-exon gene. It is absent in public databases as well as in over 1000 Turkish individuals, and co-segregates with SNHL in the family. Our in vitro studies demonstrate that the mutant protein does not localize to cell membrane as demonstrated for the wild-type protein. Mice-lacking Cldn9 have been shown to develop SNHL. We conclude that CLDN9 is essential for proper audition in humans and its disruption leads to SNHL in humans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hearing loss (HL) is the most common sensory deficit, affecting approximately 1 in 650 newborns (Mehl and Thomson 2002). Genetic factors account for at least half of congenital and prelingual-onset HL (Morton and Nance 2006). Hearing loss may be classified as syndromic, which presents additional clinical features, or nonsyndromic, in which there are no other clinical findings. Nonsyndromic HL comprises about 70–80% of genetic deafness (Nance 2003). To date, 115 nonsyndromic HL genes have been identified (Van Camp and Smith 2019). However, comprehensive genetic screening that includes all known deafness genes still leaves over 50% of cases unsolved (Shearer and Smith 2015).
Tight junctions are composed of transmembrane and membrane-associated proteins that serve as selective barriers to modulate paracellular transport. The permeability of tight junctions is determined by the presence of claudins, which are tetraspan integral proteins with two extracellular loops (Van Itallie and Anderson 2004). Mutations in CLDN14 (MIM 605608) encoding claudin-14 were shown to cause autosomal recessive nonsyndromic HL in humans (Wilcox et al. 2001). While claudin-9 was shown to be essential for hearing in mice (Nakano et al. 2009), CLDN9 mutations are not known to cause HL in humans.
Here, we present CLDN9 (MIM 615799) as a previously unrecognized gene associated with autosomal recessive nonsyndromic HL in humans. Via whole-genome sequencing (WGS), we identified a frameshift variant in CLDN9 in a family with nonsyndromic HL. We subsequently showed that the mutation disrupts subcellular organization of CLDN9, suggesting that this protein plays a crucial role in human hearing.
Materials and methods
Human subjects
This study was approved by the Ethics Committee of Ankara University (Turkey) and the Institutional Review Board at the University of Miami (USA). Informed consent was obtained from each participant or, in the case of a minor, from the parents.
We evaluated a Turkish family with parental consanguinity (Fig. 1a). The diagnosis of sensorineural HL (SNHL) was established following standard audiometric testing in a soundproofed room in accordance with current clinical standards. We calculated the pure-tone average over 0.5, 1, 2, and 4 kHz to determine severity of HL, applied to the better hearing ear: mild = 20–40 dB; moderate = 41–70 dB; severe = 71–95 dB; profound > 95 dB (Mazzoli et al. 2003). The clinical evaluation included a thorough physical examination and otoscopy in all cases. DNA was extracted from blood adhering to the standard procedures.
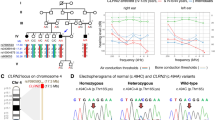
Characteristics of the family and the CLDN9 mutation. a Turkish family with SNHL, HL (black symbols) and genotypes at CLDN9 c.86delT. Double lines indicate consanguinity. b Hearing thresholds obtained from pure-tone audiograms of the family. Affected individuals show moderate to profound HL. c Electropherograms showing the CLDN9 variant. WT wild type, Hom homozygous mutant, Het heterozygous mutant. d Frameshift occurs in extracellular loop 1 (ECL1)
Sequencing and bioinformatics
We performed WGS in the proband (II:2) using the BGISEQ-500 paired-end 100 bp (PE100) (Huang et al. 2017; Bademci et al. 2018). Burrows–Wheeler Aligner was used to align reads to the human reference genome (GRCh37/hg19) (Li and Durbin 2010). We used Genome Analysis Toolkit for variant calling, BreakDancer for detecting structural variants, and CNVnator for copy number variants (McKenna et al. 2010; Chen et al. 2009; Abyzov et al. 2011).
We used Genome Aggregation Database (gnomAD), The Single Nucleotide Polymorphism (dbSNP) database, and our internal database that contains > 8500 exomes from different ethnicities, including > 1000 Turkish individuals, and > 200 genomes (Karczewski et al. 2019; Sherry et al. 2001). We used cutoffs of 0.005 and 0.0005 for recessive and dominant variants, respectively, for minor allele frequency thresholds. We also filtered variants using these combination criteria: CADD score > 20, GERP score > 2, and both PolyPhen-2 and SIFT as “damaging” (Rentzsch et al. 2018; Davydov et al. 2010; Adzhubei et al. 2010; Kumar et al. 2009). We utilized the Enlis Genome Research software to identify regions of homozygosity from WGS data (Enlis, Berkeley, CA). During the search for novel deafness genes, we focused on autozygous regions that are longer than 1 Mb. ACMG guidelines and the ClinGen Hearing Loss Gene Curation Expert Panel were followed for interpreting sequence variants (Richards et al. 2015; DiStefano et al. 2019).
We used Sanger sequencing to confirm and evaluate co-segregation of the candidate variant (Supplementary Table S1).
In vitro studies
To assess functionality, CLDN9 was cloned and expressed using a NT-GFP Fusion TOPO TA Expression Kit (cat. no. K4810-01, Invitrogen, Carlsbad, CA). Briefly, primers (Supplementary Table S1) were used to amplify the single coding exon of the gene from the human blood sample II:2 CLDN9delT (CLDN9 NM_020982.3:c.86delT) and a wild-type control. To isolate the plasmid DNA, a QIAprep Spin Miniprep Kit (cat. no. 27106, Qiagen, Hilden, Germany) was carried out according to manufacturer’s instructions. Purified DNA underwent Sanger sequencing again to choose correct orientation of the plasmid and ensure that no mutations were introduced.
Wild- type and mutant DNA samples were transfected into HEK293 cells using Lipofectamine LTX (cat no. 1533810, Invitrogen, Carlsbad, CA). After 48 h of incubation, cells were fixed in 4% PFA in PBS for 40 min at RT, permeabilized in 0.5% Triton X-100 in PBS for 10 min, and co-stained with Hoechst 33342 (cat. no. H3570, Invitrogen, Carlsbad, CA) and phalloidin CF633 conjugate (cat. no. 00046, Biotium, Fremont, CA). Cells were imaged at 40× magnification with a Zeiss LSM710 confocal microscope (Zeiss, Oberkochen, Germany).
Statistical analysis
Two-tailed t tests were calculated to determine statistical significance by measuring peaks of GFP signal intensity with the ZEN software (Zeiss, Oberkochen, Germany) (Supplementary Figure S1). The sample sizes (n), averages, and SEM are listed in each figure legend.
Results
A frameshift variant in CLDN9 is associated with nonsyndromic sensorineural hearing loss in a Turkish family
In a consanguineous family of Turkish origin, three affected individuals had SNHL (Fig. 1a). All three affected members of the family were diagnosed after age 10 years, while age of onset was not clearly delineated. Audiograms showed normal hearing in the father (I:1) and bilateral symmetric profound SNHL in the mother (I:2), moderate SNHL in the proband (II:2), and severe SNHL in an affected elder sister (II:1) (Fig. 1b). Hearing thresholds show a normal hearing level at 500 Hz and a steep decline after 1000 Hz in both sisters. In the 46-year-old mother, both 500 and 1000 Hz hearing levels show significant HL, suggesting that HL has progressed in the mother. Audiograms taken 3 years later for the sisters (II:1 and II:2) show the same hearing levels and suggest that there was not a rapid progression in HL. High-resolution computed tomography scan of the temporal bone did not show inner ear anomalies in the proband. Gross motor development was normal with no history of balance problems, vertigo, dizziness, or nystagmus. Tandem walking was normal, and the Romberg test was negative. There were no other findings affecting systems other than hearing.
Average read depth for WGS was 47.51× with at least 4× coverage for 98.87% of the genome. After filtering and excluding variants in all known deafness genes, only one variant remained mapping to an autozygous region: CLDN9 NM_020982.3:c.86delT (p.Leu29ArgfsTer4). The list of autozygous regions is provided in Supplementary Table S2. The CLDN9 variant is located in a 5.5 Mb autozygous run on chromosome 16, which is the longest of six runs over 1 Mb. The variant was not previously identified in dbSNP or gnomAD and has a CADD score of 26.5 which suggests a deleterious effect (Supplementary Table S3).
Sanger sequencing of all four family members showed co-segregation of the variant with the phenotype as an autosomal recessive trait in the family (Fig. 1a, c). The variant is located within codon Leu29, which is at the beginning of the first extracellular loop of CLDN9 (Fig. 1d). We predicted that this mutation causes a frameshift that results in a premature stop codon and a truncated protein. As CLDN9 is a single-exon gene, it is unlikely that a premature stop codon triggers nonsense-mediated decay.
Detected variant impairs CLDN9 subcellular localization
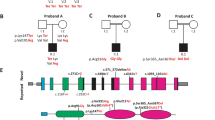
To study the cellular function, we specifically used a cloning kit that included a GFP tag at the N-terminal (Nt). The c.86delT variant occurs downstream of the Nt. We hypothesized that wild-type CLDN9 would be located in the plasma membrane, where it function as integral membrane proteins, and that the mutant CLDN9 would result in a truncated protein. As hypothesized, in the wild- type, the GFP is primarily localized in the plasma membrane (Fig. 2a). Strikingly, the GFP is localized to the cytosol only in the mutant protein (Fig. 2b). Moreover, the GFP signal is decreased in the mutant compared to the wild- type.
Representation of wild- type and mutant CLDN9 protein in HEK293 cells. a, c Wild-type CLDN9 transfected HEK293 cells show the protein expressed in both the plasma membrane and protoplasm. b, d Mutant CLDN9-transfected HEK293 cells are showing the mutant protein is limited to the cytosol. GFP-tagged wild- type or mutant CLDN9 (green), nucleus (blue), and actin (red) (scale bar: 10 μm). e Quantification of mutation on subcellular localization of CLDN9-transfected HEK293 cells. The plasma membrane-like organelle and protoplasm were measured for wild- type (n = 15) and mutant (n = 8) CLDN9. Bars represent the mean + SEM of average CLDN9-GFP fluorescence intensity in arbitrary units (AU), *p = 6.8 × 10−6, ns not significant
We measured the intensity of the GFP signal in the plasma membrane-like structure and protoplasm for both the CLDN9 control (Fig. 2c) and mutant (Fig. 2d) (Supplementary Figure S1). The CLDN9 mutant had a decreased signal in the plasma membrane compared to the wild- type (Fig. 2e). Instead, the CLDN9 is primarily found within the cytosol. However, there was no difference in intensity within the protoplasm between the two groups. This suggests that the variant disrupts the migration of CLDN9 to the plasma membrane.
Discussion
We present CLDN9 as a novel gene for autosomal recessive nonsyndromic HL. Evidence comes from a consanguineous family with a truncating mutation. To support our conclusion, our in vitro studies show that the truncated CLDN9 is confined to the cytosol, whereas in the wild- type, it is located in the plasma membrane. CLDN9 is a membrane protein and thus cannot function properly within the cytosol. This highlights that the c.86delT variant disrupts the function of CLDN9 and results in SNHL as a consequence.
Nakano et al. (2009) previously reported a claudin-9-deficient mouse strain, nfm329, which was generated using ENU-induced mutagenesis. After evaluating startle responses and measuring auditory brainstem responses (ABR), the team concluded that nmf329/nmf329 mice had severe HL at P16, which is indicative of early onset, given that the onset of hearing in wild-type mice is at P15. Wild- type and nmf329/+ mice had no difference in hearing thresholds. Similarly, the heterozygous father in our study has normal hearing. No other abnormalities were detected in the nmf329/nmf329 mice, following balance tests and histopathological experiments. Likewise, in our family, the affected individuals had no additional clinical findings in the physical examination. This draws parallels between the CLDN9 mutation homozygous individuals and the nmf329/nmf329 mice, in which genetic variations both result in recessively inherited nonsyndromic HL.
Audiograms obtained from the family have an unusual configuration. Low frequencies remain normal, while frequencies over 1000 Hz steeply decline. Studies on the nmf329 mouse line conducted by Nakano et al. (2009) further corroborate our findings. In the nmf329/nmf329 cochlea at P28, the organ of Corti was collapsed at the basal turn, lacking paracellular spaces and one of the three rows of outer hair cells (OHCs). However, the organ of Corti appeared intact at the apical turn with three rows of OHCs in nmf329 homozygotes. Similar to the results of the ABR experiment, wild- type and nmf329/+ mice were morphologically normal along the entire length of the cochlea. To better understand the progression of hair cell degeneration, Nakano et al. performed histological experiments at different stages of development. In nmf329/nmf329 mice at P8, both inner hair cells (IHCs) and OHCs were present and intact, but most of the OHCs at the basal turn had degenerated by P14. Interestingly, the effect was less pronounced at the cochlear apex. It is important to note the missing paracellular spaces in nmf329 homozygotes. Since CLDN9 is involved in paracellular permeability, this deformity is likely to impair functionality. Cochleae taken from nmf329/nmf329 mice at P80 show that the rapid degeneration that occurred in early development abated with a few OHCs remaining at the basal turn. This slow progression of HL is also observed in our family: the proband (younger sister) has moderate HL, the older sister has severe HL, and the mother is profoundly deaf. It suggests that the degeneration of hair cells is accelerated during adolescence and into adulthood and then decelerates later in life. We, therefore, conclude that CLDN9 is essential for the hair cells in the base of cochlea from early on and that c.86delT variant results in steeply sloping high frequency moderate to profound SNHL.
The findings we present here are clinically significant, because the importance of CLDN9 in human hearing was unexplored. Until this study, CLND9 was not known to be associated with HL in humans. Identifying this variant is one step closer to mapping the complete genetic landscape of deafness in humans.
References
Abyzov A, Urban AE, Snyder M, Gerstein M (2011) CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21:974–984. https://doi.org/10.1101/gr.114876.110
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. https://doi.org/10.1038/nmeth0410-248
Bademci G, Abad C, Incesulu A, Rad A, Alper O, Kolb SM, Cengiz FB, Diaz-Horta O, Silan F, Mihci E, Ocak E, Najafi M, Maroofian R, Yilmaz E, Nur BG, Duman D, Guo S, Sant DW, Wang G, Monje PV, Haaf T, Blanton SH, Vona B, Walz K, Tekin M (2018) MPZL2 is a novel gene associated with autosomal recessive nonsyndromic moderate hearing loss. Hum Genet 137:479–486. https://doi.org/10.1007/s00439-018-1901-4
Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, Shi X, Fulton RS, Ley TJ, Wilson RK, Ding L, Mardis ER (2009) BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 6:677–681. https://doi.org/10.1038/nmeth.1363
Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S (2010) Identifying a high fraction of the human genome to be under selective constraint using GERP ++. PLoS Comput Biol 6:e1001025. https://doi.org/10.1371/journal.pcbi.1001025
DiStefano MT, Hemphill SE, Oza AM, Siegert RK, Grant AR, Hughes MY, Cushman BJ, Azaiez H, Booth KT, Chapin A, Duzkale H, Matsunaga T, Shen J, Zhang W, Kenna M, Schimmenti LA, Tekin M, Rehm HL, Tayoun ANA, Amr SS, ClinGen Hearing Loss Clinical Domain Working Group (2019) ClinGen expert clinical validity curation of 164 hearing loss gene-disease pairs. Genet Med. https://doi.org/10.1038/s41436-019-0487-0
Huang J, Liang X, Xuan Y, Geng C, Li Y, Lu H, Qu S, Mei X, Chen H, Yu T, Sun N, Rao J, Wang J, Zhang W, Chen Y, Liao S, Jiang H, Liu X, Yang Z, Mu F, Gao S (2017) A reference human genome dataset of the BGISEQ-500 sequencer. GigaScience 6:1–9. https://doi.org/10.1093/gigascience/gix024
Karczewski KJ, Francioli LC, Tiao G et al (2019) Variation across 141456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. https://doi.org/10.1101/531210
Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073–1081. https://doi.org/10.1038/nprot.2009.86
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26:589–595. https://doi.org/10.1093/bioinformatics/btp698
Mazzoli M, Van Camp G, Newton V, Giarbini N, Declau F, Parving A (2003) Recommendations for the description of genetic and audiological data for families with nonsyndromic hereditary hearing impairment. Audiol Med 1:148–150. https://doi.org/10.1080/16513860301713
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Mehl AL, Thomson V (2002) The Colorado newborn hearing screening project, 1992–1999: on the threshold of effective population-based universal newborn hearing screening. Pediatrics 109:e7. https://doi.org/10.1542/peds.109.1.e7
Morton CC, Nance WE (2006) Newborn hearing screening—a silent revolution. N Engl J Med 354:2151–2164. https://doi.org/10.1056/NEJMra050700
Nakano Y, Kim SH, Kim HM, Sanneman JD, Zhang Y, Smith RJ, Marcus DC, Wangemann P, Nessler RA, Bánfi B (2009) A claudin-9-based ion permeability barrier is essential for hearing. PLoS Genet 5:e1000610. https://doi.org/10.1371/journal.pgen.1000610
Nance WE (2003) The genetics of deafness. Ment Retard Dev Disabil Res Rev 9:109–119. https://doi.org/10.1002/mrdd.10067
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M (2018) CADD: predicting the deleteriousness of variants throughout the human genome. Nucl Acids Res 47:D886–D894. https://doi.org/10.1093/nar/gky1016
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424. https://doi.org/10.1038/gim.2015.30
Shearer AE, Smith RJ (2015) Massively parallel sequencing for genetic diagnosis of hearing loss: the new standard of care. Otolaryngol Head Neck Surg 153:175–182. https://doi.org/10.1177/0194599815591156
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29:308–311. https://doi.org/10.1093/nar/29.1.308
Van Camp G, Smith RJH (2019) Hereditary hearing loss homepage. https://hereditaryhearingloss.org. Accessed 31 March 2019
Van Itallie CM, Anderson JM (2004) The molecular physiology of tight junction pores. Physiol (Bethesda) 19:331–338. https://doi.org/10.1152/physiol.00027.2004
Wilcox ER, Burton QL, Naz S, Riazuddin S, Smith TN, Ploplis B, Belyantseva I, Ben-Yosef T, Liburd NA, Morell RJ, Kachar B, Wu DK, Griffith AJ, Riazuddin S, Friedman TB (2001) Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell 104:165–172. https://doi.org/10.1016/S0092-8674(01)00200-8
Acknowledgements
We thank the study family for their participation in this study.
Funding
The study was funded by the National Institutes of Health grants R01DC009645 and R01DC012836 to MT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Ethics Committee of Ankara University (012413) and the Institutional Review Board at the University of Miami (20081138) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sineni, C.J., Yildirim-Baylan, M., Guo, S. et al. A truncating CLDN9 variant is associated with autosomal recessive nonsyndromic hearing loss. Hum Genet 138, 1071–1075 (2019). https://doi.org/10.1007/s00439-019-02037-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-019-02037-1