Abstract
Large-scale studies on genetic risk loci for melatonin receptor 1B (MTNR1B) gene and GDM risk have not been well generalized to the Chinese population. In this study, we performed two-stage case–control study: 1.429 pregnant women: 753 GDM/676 controls in the Southern Chinese population by genotyping 5 SNPs (rs10830963, rs1387153, rs2166706, rs1447352, and rs4753426) in MTNR1B. Genotypes were determined using the Sequenom MassARRAY platform and TaqMan allelic discrimination assay. Interactions between genetic variants and age/BMI as predictors of GDM risk were evaluated under the logistic regression model. In the first stage, the SNP rs10830963 was discovered to be potentially related to GDM risk (additive model: OR = 1.27, 95%CI = 1.05–1.55, P = 0.025), which was further confirmed in the second stage with a similar effect (additive model: OR = 1.53, 95%CI = 1.19–1.98, P = 0.005). In the combined stage, the G allele of rs10830963 was potentially associated with GDM risk (additive model: OR = 1.36, 95%CI = 1.17–1.59, P < 0.001; dominant model: OR = 1.45, 95%CI = 1.15–1.83, P = 0.005). The rs10830963 interacted with age and BMI to contribute to GDM risk in the combined participants. And, the similar interactive effects for the other four SNPs also exist. These findings offer the potential to improve our understanding of the etiology of GDM, and particularly of biological mechanisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gestational diabetes mellitus (GDM), one of the most common complications of pregnancy, occurs to women who have no diabetes history. The prevalence of GDM is increasing globally, with about 14% of pregnant women affected by GDM (Xie et al. 2019). GDM is related to perinatal complications and presents an overwhelmingly increased risk of metabolic disease in both mothers and their children. Women with GDM are inclined to be attacked by depressive disorder, obesity, type 2 diabetes (T2D), and cardiovascular disease. Offspring also have a future risk of several abnormities (e.g., macrosomia) and various medical conditions such as stillbirth, fetal prematurity, obesity, and T2D across the whole life course (Zhu and Zhang 2016).
Presumable evidence suggests that pregnant women with advanced maternal age, family history of diabetes, obesity, and unhealthy lifestyle are inclined to develop GDM (Zhang et al. 2016). Based on the candidate gene strategy, previous studies have predominantly demonstrated associations of some T2D susceptibility genes with GDM risk, including transcription factor seven like two (TCF7L2), melatonin receptor 1B (MTNR1B), insulin-like growth factor two mRNA binding protein 2 (IGF2BP2), and CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1) (Zhang et al. 2013). Of these, to date, only one Genome-Wide Association Studies (GWAS) in the Asian population has been conducted and implicated rs10830962 near MTNR1B and rs7754840 in CDKAL1 as susceptibility loci of GDM (Kwak et al. 2012).
MTNR1B gene, located in chromosome region 11q14.3, encodes the G protein-coupled melatonin receptors 1B. As a kind of indole hormone, melatonin participates in the regulating progress of insulin secretion, glucose metabolism, and circadian rhythms. And, melatonin must be modulated to play the function of physiological regulation through the melatonin receptors (MTNR1A; MTNR1B) (Shen and Jin 2019). Genetic variants in MTNR1B could lead to a higher expression of MTNR1B in relative tissue cells, which combine with melatonin and induce the attenuation of insulin secretion (Lane et al. 2016). The activity and expression of MTNR1B are probably to act as markers of active glucose homeostasis with enriched production of glucose. The polymorphism in MTNR1B (rs1387153, rs10830963) was linked to the impaired function of β cell and involved in the increasing risk of various phenotypes of GDM (Tarnowski et al. 2017; Alharbi et al. 2019).
To the best of our knowledge, no studies have been previously conducted to explore the possibility that SNPs of MTNR1B predispose to GDM risk in the Southern Chinese populations. The majority of existing observational studies has inadequately adjusted for those confounding factors, including age, BMI, and parity, which has led to inconsistent results. Given the function of the MTNR1B gene, we wonder whether this association signal of SNPs could be found within the complex trait of GDM or not. Therefore, the current study was undertaken to explore the following hypotheses: some candidate SNPs are associated with GDM risk by means of an association study.
Materials and methods
Study population and study design
In this case–control study, 1,429 pregnant women were recruited from two hospitals: the Affiliated Hospital of Nantong University (AHNU) and the Nantong Maternal and Child Health Hospital (NMCHH). Of these, 912 participants, recruited from AHNU between Jul 2017 and Jan 2019, were assigned into the first stage of the study. 517 participants, collected from NMCHH from Jun 2018 to Dec 2018, were assigned into the second stage. Women with GDM were diagnosed according to the 2010 International Association of Diabetes and Pregnancy Study Group (IADPSG) criteria: fasting plasma glucose ≥ 5.1 mmol/L, and/or the 1-h plasma glucose ≥ 10.0 mmol/L, and/or the 2-h plasma glucose ≥ 8.5 mmol/L. The demographic information of all participants was collected by structured questionnaires, including age, weight, parity, family history of diabetes, etc. The obesity was categorized into three stratifications based on the Working Group on Obesity in China (WGOC) criteria recommended for the Chinese population (Zhou 2002): Underweight and Normal (BMI < 24.0 kg/m2), Overweight (24.0 kg/m2 ≤ BMI < 28.0 kg/m2), Obese (BMI ≥ 28.0 kg/m2).
All participants have signed written informed consent. The study protocol was reviewed and approved by the Ethics Committee of Nantong University. These processes were carried out following the approved guidelines. The women who were diagnosed with diabetes T1D and/or T2D before pregnancy, autoimmune and inflammatory diseases, neoplastic diseases, and chronic infections were excluded.
SNPs selection
We selected tag-SNPs in MTNR1B gene locations grounded on the Public 1000 genomes database (https://phase3browser.1000genomes.org/index.html) and NCBI database (https://www.ncbi.nlm.nih.gov/projects/SNP) with the Haploview 4.2 software. Common SNPs (Minor allele frequency, MAF ≥ 5% in Chinese Han population) were screened in MTNR1B gene regions. SNPs with low linkage disequilibrium (LD) analysis (r2 < 0.8) were retained. To promote efficiency and reproducibility, we only selected the candidate SNP rs10830963 based on the Haploview software and those previously reported studies. Furthermore, several SNPs associated with GDM/plasma glucose levels in other studies were also considered and selected as candidate SNPs (rs1387153, rs2166706, rs1447352, and rs4753426) (Staiger et al. 2008; Liao et al. 2012; Liu et al. 2016). Finally, five SNPs were genotyped in this study. One SNP rs12285500 was excluded because of the call rate < 95%.
Genotyping
The genomic DNA samples were extracted from the participants’ fasting peripheral venous blood samples collected during 24–28 weeks of gestation by a DNA Extraction Kit (Qiagen, Valencia, CA) and stored at − 30 ℃ for subsequent sequencing. Genotyping analysis in the first stage of this study was conducted through the iPLEX Sequenom MassARRAY platform (Sequenom, San Diego, California). A TaqMan allelic discrimination assay (Applied Biosystems, Inc.) was used for the second stage. The methods of the quality control of genotyping have been described in our previous study (Chu et al. 2016).
Statistical analysis
The χ2 test and Student’s t-test were used for categorical variables and continuous variables, respectively, to analyze the distribution differences of demographic characteristics, clinical variables, and genotypes between cases and controls. HWE for the distribution of each SNP was evaluated through the goodness-of-fit χ2 test by comparing the observed genotype frequencies with the expected ones. Logistic regression (adjusted the covariates such as age, weight, parity, etc.) was performed to estimate the risk of GDM in each genotype under the additive model and dominant model and calculate the OR and their 95%CIs. Stratified analysis was conducted to further control the potential influence of age and BMI. The interaction effects between gene and age or BMI were analyzed through the dominant model and additive model. The Benjamini–Yekutieli procedure of false discovery rate (FDR) was employed to adjust the significance level of the association between SNPs and GDM risk in the additive model and dominant model (Benjamini et al. 2001). All the statistical analyses were performed with Stata 13.0 (Stata, College Station, TX).
Results
Case–control study
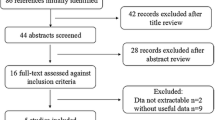
A flow diagram of sample selection is shown in (Fig. 1). In the case–control study, a sample of 1,429 pregnancy women (912 from the first stage and 517 from the second stage) were recruited and evaluated for the genotype in MTNR1B. In our study, women with GDM covered elder pregnancy women than women without GDM in two stages (P < 0.05). There was a significant difference in BMI between cases and control groups in the first stage with P value 0.004, and with P value 0.031 in the second stage. Notably, women with GDM contain more pregnant women with a family history of diabetes compared with women without GDM (P < 0.001 for the first stage; P = 0.036 for the second stage). Also, a significant difference in weight (P = 0.012) and parity (P = 0.002) between two groups only in the first stage, while not in the second stage, and the family heredity history was similar and comparable in two stages. The basic information and stratified characteristics of the participants are separately shown in (Table 1).
The genotyping success rates of all SNPs were more than 99.00%, and the genotype distribution in the control group conformed to HWE (P > 0.05) (See Supplementary Table 1). LD matrix of D´ values, as a common index of linkage disequilibrium, was calculated through the https://asia.ensembl.org/Homo_sapiens/Tools/LD. All 5 variants were in moderate to high LD in the study population (Supplementary Table 2) and a significant degree of LD likely does exist between the rs10830963 and all the other assessed SNPs (all D’ > 0.80). Therefore, the rs10830963 could serve as the tag-SNP for the five candidate SNPs.
Despite the different degrees of LD among each SNPs, the analysis of the association between each SNPs and GDM risk was also conducted and demonstrated independently. As shown in (Table 2) significant associations were detected for the five SNPs of MTNR1B and GDM risk in the additive model and dominant model in the combined populations. In the first stage, logistic regression analyses revealed that the G allele of rs10830963 was potentially associated with GDM risk (additive model: OR = 1.27, 95%CI = 1.05–1.55, P = 0.025; dominant model: OR = 1.36, 95%CI = 1.01–1.83, P = 0.067). As expected, a further examination for the second stage was conducted to test the association of the SNP rs10830963 with GDM risk and the observation was consistent in additive model except for the dominant model (OR = 1.68, 95%CI = 1.14–2.47, P = 0.045). By exerting efforts to increase the statistic power, we combined the overall participants and found that minor allele (G) of rs10830963 was significantly associated with an increased GDM risk (additive model: OR = 1.36, 95%CI = 1.17–1.59, P < 0.001; dominant model: OR = 1.45, 95%CI = 1.15–1.83, P = 0.005). Notably, similar effects were detected for SNP rs1387153 and rs2166706 in each stage definitely. Interestingly, although no significant association was detected in the first stage, SNP rs1447352 and rs4753426 could serve as protective factors for GDM in the additive model and dominant model in the combined population no matter whether being adjusted the confounding factors or not.
Subsequently, with regard to test efficiency, the associations between 5 SNPs and the susceptibility to GDM in additive model and dominant model were evaluated by stratified analysis for age and BMI. As shown in (Fig. 2), these associations for rs10830963 and rs1387153 were more evident in subjects with age ≥ 28 (for rs10830963: dominant model: OR = 1.53, 95%CI: 1.15–2.05, P = 0.004; additive model: OR = 1.44, 95%CI: 1.19–1.75, P < 0.001; for rs1387153: dominant model: OR = 1.47, 95%CI: 1.11–1.96, P = 0.008; additive model: OR = 1.44, 95%CI: 1.19–1.76, P < 0.001.). Also, this association for rs2166706 was more evident in subjects with age ≥ 28 (dominant model: OR = 1.50; 95%CI: 1.12–1.99; P = 0.006; additive model: OR = 1.44, 95%CI: 1.19–1.75, P < 0.001). Notably, the protective roles for rs1447352 were detected in subjects with age ≥ 28 (dominant model: OR = 0.67; 95%CI: 0.51–0.88; P = 0.004; additive model: OR = 0.77, 95%CI: 0.62–0.95, P < 0.013) and BMI ≥ 28 (dominant model: OR = 0.71; 95%CI: 0.51–0.98, P = 0.038; additive model: OR = 0.75, 95%CI: 0.58–0.97, P < 0.031). The protective roles for rs4753426 (OR = 0.71; 95%CI: 0.54–0.93; P = 0.012) could only be detected in the dominant model in subjects with age ≥ 28. No significant heterogeneity was observed among the stratified subgroups. Detailed information is shown in (Fig. 2).
Considering the significant differences in the distribution of age and BMI, we further explored whether the effect of SNPs on GDM was modified by age and BMI. There was no multiplicative interaction between 5 SNPs and age or BMI (all P > 0.05). The interactive analysis showed that the rs10830963 interacted with age/BMI to contribute to GDM risk (OR = 2.27, 95%CI: 1.58–3.25, P < 0.001; OR = 2.02; 95%CI: 1.46–2.78, P < 0.001) (Table 3). Similar interactive effects for the other four SNPs are shown in Supplementary table 3 and Supplementary table 4.
Discussion
Several studies have suggested that melatonin could regulate the circadian rhythm and endocrine immunity, especially insulin secretion and glucose levels (Mahanna-Gabrielli et al. 2018; Shen and Jin 2019). Variants in MTNR1B have been confirmed to be genetic components for GDM in multiple populations in the whole genome. The identified individual risk alleles were found to exert only moderate to small effects on the susceptibility to GDM.
In this study, without considering the LD, our results revealed that SNP rs10830963, rs1387153, and rs2166706 have significant associations with GDM risk in the Southern Chinese population. Outstandingly, SNP rs1447352, and rs4753426 could reduce the risk of GDM. This is consistent with several recent studies (Liao et al. 2012; Tarnowski et al. 2017). SNP rs10830963 and rs1387153 might reduce the function of insulin β-cells, which in turn affects the normal regulation of plasma glucose levels and leads to the occurrence of GDM (Caro-Gomez et al. 2018). And, the role of MTNR1B rs10830963 has been confirmed in the pathogenesis of type 2 diabetes (Gaulton et al. 2015). Furthermore, two large-scale GWAS in European and American also identified that the rs10830963 and rs1387153 pose important mechanisms of induction in the development of GDM (Rosta et al. 2017; Ding et al. 2018).
As for two SNPs, rs1447352 and rs4753426 were detected to be protective factors for GDM after adjusting potential covariates (the pooled OR = 0.82 and 0.84, respectively) in the combined stage, while not in the first stage and the second stage. Since random errors may be potential reasons for the differences in the findings among different stages. Further studies with larger sample size are warranted to test for credibility. A recent GWAS reported a significant association between rs1447352 and FPG after Bonferroni correction for multiple comparisons (Ramos et al. 2011); whereas, a contradictory conclusion was also reported (Liao et al. 2012). One study in the German population reported that rs4753426 might reduce insulin secretion and increase the level of FPG (Staiger et al. 2008). And, SNP rs4753426 was not significantly associated with GDM in white European women (Tarnowski et al. 2017). Collectively, the potential relationships between rs1447352 and rs4753426 and GDM risk have not been well documented. The possible reasons might result from differences in the genetic backgrounds of the studied population.
Further, regarding the significant degree of LD between each SNPs and rs10830963, we found that rs10830963 could be the tag-SNP of the five candidate SNPs. And as stated, in the combined stage, the OR values of additive model and dominant model of rs10830963, rs1387153 and rs2166706 were very close; these further proved the existence of tag-SNPs. Besides, the degree of LD between rs10830963 and the other four SNPs was consistent with the discovery of Liao S and Salman M (Liao et al. 2012; Salman et al. 2015). However, the real associations between SNP rs1447352, rs4753426, and rs10830963 still need to be explored in further studies. Likewise, the interaction analysis revealed that both age and BMI interacted with rs10830963 and increased GDM risk, as the same as rs1387153 and rs2166706. In the interaction analysis, we use the BMI cut-off value of ≥ 28 kg/m2 due to the stratified analysis. And, one study by Firneisz G et al. reported that the MTNR1B rs10830963 in the stratified BMI (≥ 29 kg/m2) could predict antenatal insulin therapy initiation better (Firneisz et al. 2018), which is approximately consistent with the cut-off value in our study(≥ 28 kg/m2). Additionally, a previous study found that MTNR1B rs10830963 could decrease the effect of the antenatal intervention of an individual with BMI > 30 kg/m2 and/or prior GDM in history (Grotenfelt et al. 2016). These BMI might be vital factors that influence the incidence of GDM and the effect of early medical nutrition therapy (MNT). Most importantly, since the analysis by LD matrix of D’ values showed that five SNPs were in moderate to high LD, the significant interaction associations and the haplotype analyses deserved to be further detected in the future study.
There are several strengths in this study. First, by exerting much effort to evaluate whether several SNPs in MTNR1B could be well generalized to Southern Chinese population, we systematically evaluated five SNPs in women with GDM and those without GDM, and the association for SNP rs10830963 was successfully examined in the first stage and confirmed in the second stage, and the P values remain significant after multiple testing in the combined stage. Second, the fact that participants were recruited from the two hospitals, representing relatively reasonable controls, might not result in potential selection bias. Third, the universal screening for GDM was conducted through OGTT in those women who enrolled in pregnancy complications screening, which avoided the relatively high misdiagnosis rate caused by the census performed only in pregnant women with potential risk factors for GDM (advanced maternal age, obesity, family history of diabetes and parity). Importantly, biological and environmental factors, such as maternal age, BMI, exercise, and diet, should be considered comprehensively to understand its biological plausibility and improve the identification of women at risk of GDM, which contribute to accurately evaluate risk stratification.
However, some limitations still need to be addressed. First, several SNPs in MTNR1B are located in the intron and present being silent. Considering that our study exhibiting a confirmatory manifestation in the Southern Chinese population, we selected the overlapped candidate SNP between the results of Haploview and previous studies to verify the reported association between variants of MTNR1B (especially the rs10830963) and GDM risk. Therefore, the other SNPs detected through the Haploview were not selected in this study. The LD matrix of D´ values indicates that all D’ ≥ 0.85 for the rs10830963, a significant degree of LD likely does exist between the rs10830963 and all the other assessed SNPs. In view of the comprehensiveness and stability of D’ value, we most likely assessed the same single genetic effect (with five markers) and not five independent genetic effects. Therefore, MTNR1B rs10830963 might be the truly causal SNP that drives these associations. However, the multiple comparisons test with FDR was still employed to adjust the significance level of the association in the additive model and the dominant model. Second, discrepancy among those studies is probably attributable to ethnic diversity and differences in the genotyping methods. And, the discordant results could be attributed to different diagnostic criteria. Third, little information regarding environmental exposures and serum parameters, such as diet variables and glycated hemoglobin (HbA1c) measurement, were collected and have not been adjusted for further analysis. Nevertheless, there is sufficient assurance to believe that the findings are of considerable credibility and veracity.
In summary, our results indicated that MTNR1B rs10830963 is independently associated with the susceptibility to GDM and it is the tag-SNP of the other four candidate SNPs. In the meantime, the association could be altered by maternal age and BMI. Well-designed studies are warranted to seek replication and validate our findings and extend these results.
Data availability
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available because of privacy or ethical restrictions.
References
Alharbi KK, Al-Sulaiman AM, Shedaid KMB, Al-Shangiti AM, Marie M, Al-Sheikh YA, Ali Khan I (2019) MTNR1B genetic polymorphisms as risk factors for gestational diabetes mellitus: a case-control study in a single tertiary care center. Ann Saudi Med 39:309–318
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125:279–284
Caro-Gomez MA, Naranjo-González CA, Gallego-Lopera N, Parra-Marín MV, Valencia DM, Arcos EG, Villegas-Perrasse A, Bedoya-Berrío G (2018) Association of Native American ancestry and common variants in ACE, ADIPOR2, MTNR1B, GCK, TCF7L2 and FTO genes with glycemic traits in Colombian population. Gene 677:198–210
Chu M, Rong J, Wang Y, Zhu L, Xing B, Tao Y, Zhuang X, Zhao Y, Jiang L (2016) Strong association of the polymorphisms in PBEF1 and knee OA risk: a two-stage population-based study in China. Sci Rep 6:19094
Ding M, Chavarro J, Olsen S, Lin Y, Ley SH, Bao W, Rawal S, Grunnet LG, Thuesen ACB, Mills JL, Yeung E, Hinkle SN, Zhang W, Vaag A, Liu A, Hu FB, Zhang C (2018) Genetic variants of gestational diabetes mellitus: a study of 112 SNPs among 8722 women in two independent populations. Diabetologia 61:1758–1768
Firneisz G, Rosta K, Al-Aissa Z, Hadarits O, Harreiter J, Nádasdi Á, Bancher-Todesca D, Németh L, Igaz P, Rigó J Jr, Sziller I, Kautzky-Willer A, Somogyi A (2018) The MTNR1B rs10830963 variant in interaction with pre-pregnancy BMI is a pharmacogenetic marker for the initiation of antenatal insulin therapy in gestational diabetes mellitus. Int J Mol Sci 19:5
Gaulton KJ, Ferreira T, Lee Y, Raimondo A, Mägi R, Reschen ME, Mahajan A, Locke A, Rayner NW, Robertson N, Scott RA, Prokopenko I, Scott LJ, Green T, Sparso T, Thuillier D, Yengo L, Grallert H, Wahl S, Frånberg M, Strawbridge RJ, Kestler H, Chheda H, Eisele L, Gustafsson S, Steinthorsdottir V, Thorleifsson G, Qi L, Karssen LC, van Leeuwen EM, Willems SM, Li M, Chen H, Fuchsberger C, Kwan P, Ma C, Linderman M, Lu Y, Thomsen SK, Rundle JK, Beer NL, van de Bunt M, Chalisey A, Kang HM, Voight BF, Abecasis GR, Almgren P, Baldassarre D, Balkau B, Benediktsson R, Blüher M, Boeing H, Bonnycastle LL, Bottinger EP, Burtt NP, Carey J, Charpentier G, Chines PS, Cornelis MC, Couper DJ, Crenshaw AT, van Dam RM, Doney AS, Dorkhan M, Edkins S, Eriksson JG, Esko T, Eury E, Fadista J, Flannick J, Fontanillas P, Fox C, Franks PW, Gertow K, Gieger C, Gigante B, Gottesman O, Grant GB, Grarup N, Groves CJ, Hassinen M, Have CT, Herder C, Holmen OL, Hreidarsson AB, Humphries SE, Hunter DJ, Jackson AU, Jonsson A, Jørgensen ME, Jørgensen T, Kao WH, Kerrison ND, Kinnunen L, Klopp N, Kong A, Kovacs P, Kraft P, Kravic J, Langford C, Leander K, Liang L, Lichtner P, Lindgren CM, Lindholm E, Linneberg A, Liu CT, Lobbens S, Luan J, Lyssenko V, Männistö S, McLeod O, Meyer J, Mihailov E, Mirza G, Mühleisen TW, Müller-Nurasyid M, Navarro C, Nöthen MM, Oskolkov NN, Owen KR, Palli D, Pechlivanis S, Peltonen L, Perry JR, Platou CG, Roden M, Ruderfer D, Rybin D, van der Schouw YT, Sennblad B, Sigurðsson G, Stančáková A, Steinbach G, Storm P, Strauch K, Stringham HM, Sun Q, Thorand B, Tikkanen E, Tonjes A, Trakalo J, Tremoli E, Tuomi T, Wennauer R, Wiltshire S, Wood AR, Zeggini E, Dunham I, Birney E, Pasquali L, Ferrer J, Loos RJ, Dupuis J, Florez JC, Boerwinkle E, Pankow JS, van Duijn C, Sijbrands E, Meigs JB, Hu FB, Thorsteinsdottir U, Stefansson K, Lakka TA, Rauramaa R, Stumvoll M, Pedersen NL, Lind L, Keinanen-Kiukaanniemi SM, Korpi-Hyövälti E, Saaristo TE, Saltevo J, Kuusisto J, Laakso M, Metspalu A, Erbel R, Jöcke KH, Moebus S, Ripatti S, Salomaa V, Ingelsson E, Boehm BO, Bergman RN, Collins FS, Mohlke KL, Koistinen H, Tuomilehto J, Hveem K, Njølstad I, Deloukas P, Donnelly PJ, Frayling TM, Hattersley AT, de Faire U, Hamsten A, Illig T, Peters A, Cauchi S, Sladek R, Froguel P, Hansen T, Pedersen O, Morris AD, Palmer CN, Kathiresan S, Melander O, Nilsson PM, Groop LC, Barroso I, Langenberg C, Wareham NJ, O'Callaghan CA, Gloyn AL, Altshuler D, Boehnke M, Teslovich TM, McCarthy MI, Morris AP (2015) Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet 47:1415–1425
Grotenfelt NE, Wasenius NS, Rönö K, Laivuori H, Stach-Lempinen B, Orho-Melander M, Schulz CA, Kautiainen H, Koivusalo SB, Eriksson JG (2016) Interaction between rs10830963 polymorphism in MTNR1B and lifestyle intervention on occurrence of gestational diabetes. Diabetologia 59:1655–1658
Kwak SH, Kim SH, Cho YM, Go MJ, Cho YS, Choi SH, Moon MK, Jung HS, Shin HD, Kang HM, Cho NH, Lee IK, Kim SY, Han BG, Jang HC, Park KS (2012) A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes 61:531–541
Lane JM, Chang AM, Bjonnes AC, Aeschbach D, Anderson C, Cade BE, Cain SW, Czeisler CA, Gharib SA, Gooley JJ, Gottlieb DJ, Grant SF, Klerman EB, Lauderdale DS, Lockley SW, Munch M, Patel S, Punjabi NM, Rajaratnam SM, Rueger M, St Hilaire MA, Santhi N, Scheuermaier K, Van Reen E, Zee PC, Shea SA, Duffy JF, Buxton OM, Redline S, Scheer FA, Saxena R (2016) Impact of common diabetes risk variant in MTNR1B on sleep, circadian, and melatonin physiology. Diabetes 65:1741–1751
Liao S, Liu Y, Tan Y, Gan L, Mei J, Song W, Chi S, Dong X, Chen X, Deng S (2012) Association of genetic variants of melatonin receptor 1B with gestational plasma glucose level and risk of glucose intolerance in pregnant Chinese women. PLoS ONE 7:e40113
Liu Q, Huang Z, Li H, Bai J, Liu X, Ye H (2016) Relationship between melatonin receptor 1B (rs10830963 and rs1387153) with gestational diabetes mellitus: a case-control study and meta-analysis. Arch Gynecol Obstet 294:55–61
Mahanna-Gabrielli E, Miano TA, Augoustides JG, Kim C, Bavaria JE, Kofke WA (2018) Does the melatonin receptor 1B gene polymorphism have a role in postoperative delirium? PLoS ONE 13:e0207941
Ramos E, Chen G, Shriner D, Doumatey A, Gerry NP, Herbert A, Huang H, Zhou J, Christman MF, Adeyemo A, Rotimi C (2011) Replication of genome-wide association studies (GWAS) loci for fasting plasma glucose in African-Americans. Diabetologia 54:783–788
Rosta K, Al-Aissa Z, Hadarits O, Harreiter J, Nádasdi Á, Kelemen F, Bancher-Todesca D, Komlósi Z, Németh L, Rigó J Jr, Sziller I, Somogyi A, Kautzky-Willer A, Firneisz G (2017) Association Study with 77 SNPs Confirms the Robust Role for the rs10830963/G of MTNR1B variant and identifies two novel associations in gestational diabetes mellitus development. PLoS ONE 12:e0169781
Salman M, Dasgupta S, Cholendra A, Venugopal PN, Lakshmi GL, Xaviour D, Rao J, D’Souza CJ (2015) MTNR1B gene polymorphisms and susceptibility to Type 2 diabetes: a pilot study in South Indians. Gene 566:189–193
Shen LL, Jin Y (2019) Effects of MTNR1B genetic variants on the risk of type 2 diabetes mellitus: a meta-analysis. Mol Genet Genomic Med 7:e611
Staiger H, Machicao F, Schäfer SA, Kirchhoff K, Kantartzis K, Guthoff M, Silbernagel G, Stefan N, Häring HU, Fritsche A (2008) Polymorphisms within the novel type 2 diabetes risk locus MTNR1B determine beta-cell function. PLoS ONE 3:e3962
Tarnowski M, Malinowski D, Safranow K, Dziedziejko V, Pawlik A (2017) MTNR1A and MTNR1B gene polymorphisms in women with gestational diabetes. Gynecol Endocrinol 33:395–398
Xie K, Chen T, Zhang Y, Wen J, Cui X, You L, Zhu L, Xu B, Ji C, Guo X (2019) Association of rs10830962 polymorphism with gestational diabetes mellitus risk in a Chinese population. Sci Rep 9:5357
Zhang C, Bao W, Rong Y, Yang H, Bowers K, Yeung E, Kiely M (2013) Genetic variants and the risk of gestational diabetes mellitus: a systematic review. Hum Reprod Update 19:376–390
Zhang C, Rawal S, Chong YS (2016) Risk factors for gestational diabetes: is prevention possible? Diabetologia 59:1385–1390
Zhou BF (2002) Predictive values of body mass index and waist circumference for risk factors of certain related diseases in Chinese adults–study on optimal cut-off points of body mass index and waist circumference in Chinese adults. Biomed Environ Sci 15:83–96
Zhu Y, Zhang C (2016) Prevalence of gestational diabetes and risk of progression to Type 2 diabetes: a global perspective. Curr Diab Rep 16:7
Acknowledgements
This study was supported by the Shanghai Municipal Health Commission (201840297), and supported by Construction project of Shanghai Key Laboratory of Molecular Imaging(18DZ2260400), Shanghai Municipal Education Commission (Class II Plateau Disciplinary Construction Program of Medical Technology of SUMHS, 2018–2020), and supported by Nantong Science and Technology Plan-Frontiers and Key Technologies (MS12017013-3).
Author information
Authors and Affiliations
Contributions
L.Y.J. contributed to the interpretation of results and made critical revisions. Y.L.J. drafted the protocol and wrote the final paper. X.Y.S., X.F.G., P.Z., Y.L.L, A.Y.Z., participated in the data collection. And, Y.S. reviewed the draft and made the critical revision. All authors have reviewed the final version of the manuscript and approved it for publication.
Corresponding author
Ethics declarations
Conflict of interest
Yulong Jia declares that he has no conflict of interest. Yi Shen declares that she has no conflict of interest. Xiuying Shi declares that she has no conflict of interest. Xuefeng Gu declares that he has no conflict of interest. Peng Zhang declares that he has no conflict of interest. Yuanlin Liu declares that he has no conflict of interest. Aiyong Zhu declares that he has no conflict of interest. Liying Jiang declares that she has no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Novelty statements
-
The rs10830963 in MTNR1B could modify individual susceptibility to gestational diabetes mellitus(GDM) in the study population.
-
The rs10830963 interacted with age and BMI to contribute to GDM risk. Similar interactive effects for the other 4 SNPs also exist.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jia, Y., Shen, Y., Shi, X. et al. MTNR1B gene on susceptibility to gestational diabetes mellitus: a two-stage hospital-based study in Southern China. Mol Genet Genomics 295, 1369–1378 (2020). https://doi.org/10.1007/s00438-020-01706-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-020-01706-5