Abstract
Purpose
Variants rs10830963 (C/G) and rs1387153 (C/T) in MTNR1B have been shown with an increased risk of developing type 2 diabetes and gestational diabetes mellitus. However, the results are still controversial, and evidence was not satisfied. Hence, a case–control study and a further meta-analysis will be performed in this study.
Methods
We recruited 674 GDM patients and 690 controls from Jan 2010 and Jan 2014. The SNPs were genotyped by ABI TaqMan SNP Genotyping Assays. MTNR1B rs10830963 and rs1387153 single nucleotide polymorphisms (SNPs) were performed for association analysis. Then a systematic search of all relevant studies was conducted. A meta-analysis was performed to prove the relationship between melatonin receptor 1B (rs10830963 and rs1387153) with GDM.
Results
The case–control study presented that G allele of the rs10830963 and T allele of rs1387153 were significantly associated with increased risk of GDM. The further meta-analysis included other five studies showed that the frequency of MTNR1B rs10830963 G allele and rs1387153 T allele are higher in GDM patients.
Conclusion
The case–control study proved that the risk allele (G allele) of rs10830963 and (T allele) of rs1387153 lead to a higher risk for GDM. The further meta-analysis provides additional evidence supporting the above results. Due to the limited data currently available in different race population, further studies with large sample sizes are required.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gestational Diabetes Mellitus (GDM) is defined by WHO as any degree of glucose intolerance with onset or first recognition during pregnancy [1]. The incidence of GDM is about 1–3 % of all pregnancies in the western world [2], and 5–10 % in Asian pregnancies [3]. The maternal glucose metabolism and insulin sensitivity always changes in pregnant women. In most instances, pregnant women are able to meet the increased insulin demand but in some cases these needs are not met resulting in poor glycaemic control and consequently GDM [4]. Furthermore, GDM not just increase the risk of developing type 2 diabetes mellitus, but also increasing the risk of adverse pregnancy outcomes [5]. Although the WHO guidelines for GDM have been widely used from 1999 [6], there is still no universal recommendation for screening and diagnosis of GDM [7].
Recently, a study of identifying susceptible genes of complicated diseases through genome-wide association strategy was performed. And Melatonin receptor 1B (MTNR1B) was proven be the diabetogenic genes associated with the developing of GDM [8]. The gene MTNR1B encodes a receptor for melatonin which belongs to the G protein-coupled receptors [9]. Melatonin receptors are expressed mainly in the brain, and MTNR1B has also been found in β cells, which implies that genetic variants in the MTNR1B might affect pancreatic glucose sensing, insulin secretion, and, conceivably, glucose tolerance [10].
Variants rs10830963 (C/G) and rs1387153 (C/T) in MTNR1B have been shown with an increased risk of developing type 2 diabetes [11]. And following studies reported the association of rs10830963 (C/G) and rs1387153 (C/T) with GDM [12, 13]. A meta-analysis [7] published in 2014 also reported the similar results which included five case–control studies. However, only two studies reported the association of rs1387153 (C/T) with GDM. Hence, a case–control study and further meta-analysis is performed to supplement the data of the association of rs10830963 (C/G) and rs1387153 (C/T) with GDM of Chinese population in this study.
Materials and methods
Subjects
A total of 1364 subjects were included. All pregnant women were recruited from the First Clinical Medical College of Three Gorges University between Jan 2010 and Jan 2014. The present study was approved by the Ethics Committee of the First Clinical Medical College of Three Gorges University. And all participants gave written, informed consent.
Glucose and diagnostic criteria for GDM
Included GDM cases were identified after a glucose challenge test (GCT) between weeks 24 and 28 of gestation. Height, weight, and blood pressure were also measured using standardized procedures and calibrated equipment. The standard 100 g oral glucose tolerance test (OGTT) was performed. Fasting glucose levels were measured at 1, 2 and 3 h. In this study, the GDM cases were defined as those patients who produced two or more glucose values that met or exceeded the threshold values [14]. Women diagnosed with T1DM or T2DM before pregnancy were excluded from this study.
Genotyping analysis
MTNR1B rs10830963 and rs1387153 single nucleotide polymorphisms (SNPs) were performed for association analysis. Genomic DNA was extracted from peripheral blood leukocytes using the salting-out technique [15]. The SNPs were genotyped by ABI TaqMan SNP Genotyping Assays using LightCycler 480 System [10].
Meta-analysis
Included studies had to meet the following criteria: (1) only case–control or cohort studies were included; (2) all patients meeting the diagnostic criteria for GDM, and the controls must be non-diabetic; (3) included studies should report one ore more polymorphisms (rs10830963 or rs1387153); (4) original data for calculating odds ratios (ORs) with corresponding 95 % confidence intervals (CIs) were reported; (5) genotype distribution of control for a certain polymorphism must be in Hardy–Weinberg equilibrium.
The search strategy was created with the assistance of a librarian using a combination of terms including melatonin receptor 1B, MTNR1B, gestational diabetes mellitus, GDM, gene, polymorphism, rs10830963, rs1387153, case–control study; meta-analysis; and systematic review. No language or other limitations were imposed. Two reviewers independently screened the titles and abstracts of studies identified by the search strategy and discarded clearly irrelevant studies. The same two reviewers also independently applied the selection criteria to the studies retrieved by the literature search. They discussed to resolve any disagreement; if any uncertainty remained, they consulted further reviewer and expert to decide.
Two reviewers independently extracted the data using a standardized form regarding inclusion criteria. A consensus method was used to resolve disagreements, and a third reviewer was consulted if disagreements persisted. The detailed data of the first author, year of publication, study design, total numbers, ethnicity, genotyping method, genotype distribution, and genotype distributions were extracted.
Statistical analysis
Chi square test was used to compare Allele/genotype frequencies between two groups. Hardy–Weinberg equilibrium of the genotype frequencies was tested by Chi square test. Meta-analysis was performed with STATA 12.0. The overall association between genetic polymorphisms and GDM risk was measured by OR and its 95 % CI. For rs10830963, the allelic model (G vs. C) and genotype genetic models were examined which included: (1) co-dominant effects: GG vs. CC; (2) dominant effect: GG+GC vs. CC; 3) recessive effect: GG vs. GC+CC. For rs1387153, the allelic model (T vs. C) and genotype genetic models was examined which included: (1) co-dominant effects: TT vs. CC; (2) dominant effect: TT+TC vs. CC; (3) and recessive effect: TT vs. TC+CC. We performed the meta-analysis using a fixed-effect model if no significant heterogeneity was present. A random effects model was selected to account for heterogeneity in the design and patient selection among included studies. P value less than 0.05 was considered statistically significant.
Results
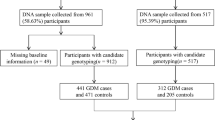
A total of 1364 subjects (674 cases and 690 controls) were included in this case–control study, 674 of them were diagnosed with GDM, the other 690 women were in control group without GDM. The distributions of the alleles and genotypes for rs10830963 and rs1387153 were described in Table 1. The genotype distribution of GDM group and control group were conformed Hardy–Weinberg equilibrium. In accordance with the genome-wide association study, the risk allele (G allele) of rs10830963 and (T allele) of rs1387153 lead to a higher risk for GDM (Table 1). G allele of the rs10830963 and T allele of rs1387153 were significantly associated with increased risk of GDM.
Meta-analysis
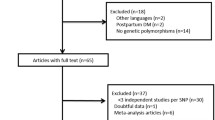
A total of 56 titles and abstracts were reviewed, and five case–control studies were included [16–20]. Figure 1 summarizes the study selection process. All 5 included trials reported the relationship between MTNR1B rs10830963 with GDM (Table 2). The distribution of genotypes and alleles in the individual studies were summarized in Table 3. The pooled analysis showed significant differences between the two groups of co-dominant effects (GG vs. CC) (OR 1.62, 95 % CI 1.34, 1.94; P = 0.000, I 2 = 29.6 %) (Fig. 2). For dominant effect (GG+GC vs. CC), the pooled analysis showed significant differences between the two groups (OR 1.29, 95 % CI 1.16, 1.45; P = 0.000, I 2 = 0.0 %) (Fig. 3). For recessive effect (GG vs. GC+CC), the pooled analysis showed significant differences between the two groups (OR 1.48, 95 % CI 1.22, 1.78; P = 0.000, I 2 = 46.4 %) (Fig. 4). Two studies reported the relationship between MTNR1B rs1387153 with GDM. For co-dominant effects: TT vs. CC, the pooled analysis showed significant differences between the two groups (OR 1.53, 95 % CI 1.26, 1.86; P = 0.000, I 2 = 0.0 %) (Fig. 5). For dominant effect (TT+TC vs. CC), the pooled analysis showed significant differences between the two groups (OR 1.23, 95 % CI 1.06, 1.42; P = 0.005, I 2 = 1.6 %) (Fig. 6). For recessive effect (TT vs. TC+CC), the pooled analysis showed significant differences between the two groups (OR 1.39, 95 % CI 1.17, 1.66; P = 0.000, I 2 = 0.0 %) (Fig. 7).
Discussion
GDM is at risk of developing type 2 diabetes mellitus, more than 50 % patients with GDM develop T2DM in 10 years after pregnancy [21]. However, the development mechanism of GDM is still not clear. Some studies have shown that gene polymorphism could be associated with the pathogenetic mechanisms and developing of GDM. MTNR1B as a member of G protein-coupled receptors has been found in β cells, which might affect pancreatic glucose sensing. The present case–control study proved that the risk allele (G allele) of rs10830963 and (T allele) of rs1387153 lead to a higher risk for GDM. G allele of the rs10830963 and T allele of rs1387153 were significantly associated with increased risk of GDM. Although the results were similar with the most published evidences, inconsistent results were still presented for MTNR1B polymorphism among studies. Hence, a comprehensive meta-analysis was needed to provide evidence with high level.
The present meta-analysis show that the frequency of MTNR1B rs10830963 G allele is higher in GDM patients than that in the healthy controls, and the frequency of MTNR1B rs1387153 T allele is also higher in GDM patients than in controls. The results demonstrated a statistically significant positive association between the risk factor rs10830963 G allele and rs1387153 T allele carriers and GDM susceptibility. The conclusion was consistent with a meta-analysis published in 2014 by Zhang et al. [7].
The development of GDM was regulated by multiple genes. A meta-analysis by Mao et al. found that eight genetic polymorphisms were significantly associated with GDM included TCF7L2 (rs7903146), IGF2BP2 (rs4402960), MTNR1B (rs10830963), CDKAL1 (rs7754840), KCNJ11 (rs5219), KCNQ1 (rs2237892 and rs2237895) and GCK (rs4607517) [22]. What’s more, the genes prompt some pathological mechanisms in process of GDM, such as impaired β cells function, insulin resistance and abnormal utilization of glucose. The genetic polymorphisms test not only can improve the levels of diagnosis, but also can provide genetic target for treating disease.
In this study, some limitation of meta-analysis should be addressed. Firstly, publication bias is a unavoidable problem in meta-analysis, because the positive results are prone to publish than negative results which leading the overestimation of effects. Secondly, the included sample size was small which may influence credibility of the results. More studies are needed in the future. Thirdly, although the baseline of included cases are comparable, no sub-group analysis of different race were performed, which may influence the stability of results.
Conclusions
The case–control study proved that the risk allele (G allele) of rs10830963 and (T allele) of rs1387153 lead to a higher risk for GDM. The further meta-analysis provides additional evidence supporting the above results. Due to the limited data currently available in different race population, further studies with large sample sizes are required.
References
World Health Organization (2002) Laboratory diagnosis and monitoring of diabetes mellitus. World Health Organization, Geneva
Hadden DR (1985) Geographic, ethnic, and racial variations in the incidence of gestational diabetes mellitus. Diabetes 34(Suppl 2):8–12
Shaat N, Groop L (2007) Genetics of gestational diabetes mellitus. Curr Med Chem 14:569–583
Macaulay S, Dunger DB, Norris SA (2014) Gestational diabetes mellitus in Africa: a systematic review. PLoS One 9(6):e97871
Nar G, Inci S, Aksan G, Unal OK, Nar R, Soylu K (2014) The relationship between epicardial fat thickness and gestational diabetes mellitus. Diabetol Metab Syndr 6(1):120
Alberti KG, Zimmet PZ (1998) Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 15:539–553
Zhang Y, Sun CM, Hu XQ, Zhao Y (2014) Relationship between melatonin receptor 1B and insulin receptor substrate 1 polymorphisms with gestational diabetes mellitus: a systematic review and meta-analysis. Sci Rep 4:6113
Cho YM, Kim TH, Lim S, Choi SH, Shin HD, Lee HK, Park KS, Jang HC (2009) Type 2 diabetes-associated genetic variants discovered in the recent genome-wide association studies are related to gestational diabetes mellitus in the Korean population. Diabetologia 52:253–261
Fredriksson R, Schioth HB (2005) The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol 67(5):1414–1425
Vejrazkova D, Lukasova P, Vankova M, Vcelak J, Bradnova O, Cirmanova V, Andelova K, Krejci H, Bendlova B (2014) MTNR1B genetic variability is associated with gestational diabetes in Czech Women. Int J Endocrinol 2014:508923
Xia Q, Chen ZX, Wang YC, Ma YS, Zhang F, Che W, Fu D, Wang XF (2012) Association between the melatonin receptor 1B gene polymorphism on the risk of type 2 diabetes, impaired glucose regulation: a meta-analysis. PLoS One 7(11):e50107
Sun XJ, Rothenberg P, Kahn CR, Backer JM, Araki E, Wilden PA, Cahill DA, Goldstein BJ, White MF (1991) Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature 352(63300):73–77
Mima A, Ohshiro Y, Kitada M, Matsumoto M, Geraldes P, Li C, Li Q, White GS, Cahill C, Rask-Madsen C, King GL (2011) Glomerular-specific protein kinase C- b-induced insulin receptor substrate-1 dysfunction and insulin resistance in rat models of diabetes and obesity. Kidney Int 79(8):883–896
Bhat M, Ramesha KN, Sarma SP, Menon S, Sowmini CV, Kumar SG (2010) Determinants of gestational diabetes mellitus: a case control study in a district tertiary care hospital in south India. Int J Diabetes Dev Ctries 30:91–96
Movva S, Alluri RV, Komandur S, Vattam K, Eppa K, Mukkavali KK, Mubigonda S, Saharia S, Shastry JC, Hasan Q (2007) Relationship between angiotensin-converting enzyme gene polymorphism with nephropathy associated with type 2 diabetes mellitus in Asian Indians. J Diabetes Complications 21:237–241
Deng Z (2011) Association of genetic variant rs10830963 of melatonin receptor 1B gene in women with gestational diabetes mellitus. Zhonghua Wei Chan Yi Xue Za Zhi 14:666–669
Wang Y, Nie M, Li W, Ping F, Hu Y, Ma L, Gao J, Liu J (2011) Association of six single nucleotide polymorphisms with gestational diabetes mellitus in a Chinese population. PLoS One 6(11):e26953
Kim JY, Cheong HS, Park BL, Baik SH, Park S, Lee SW, Kim MH, Chung JH, Choi JS, Kim MY, Yang JH, Cho DH, Shin HD, Kim SH (2011) Melatonin receptor 1 B polymorphisms associated with the risk of gestational diabetes mellitus. BMC Med Genet 12:82
Vlassi M, Gazouli M, Paltoglou G, Christopoulos P, Florentin L, Kassi G, Mastorakos G (2012) The rs10830963 variant of melatonin receptor MTNR1B is associated with increased risk for gestational diabetes mellitus in a Greek population. Hormones 11(1):70–76
Li C, Qiao B, Zhan Y, Peng W, Chen ZJ, Sun L, Zhang J, Zhao L, Gao Q (2013) Association between genetic variations in MTNR1A and MTNR1B genes and gestational diabetes mellitus in Han Chinese women. Gynecol Obstet Invest 76(4):221–227
Ben-Haroush A, Yogev Y, Hod M (2004) Epidemiology of gestational diabetes mellitus and its association with Type 2 diabetes. Diabet Med 21(2):103–113
Mao H, Li Q, Gao S (2012) Meta-analysis of the relationship between common type 2 diabetes risk gene variants with gestational diabetes mellitus. PLoS One 7(9):e45882
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Each author certifies that he or she has no commercial associations that might pose a conflict of interest related to the submitted article.
Additional information
Q. Liu and Z. Huang co-first author.
Rights and permissions
About this article
Cite this article
Liu, Q., Huang, Z., Li, H. et al. Relationship between melatonin receptor 1B (rs10830963 and rs1387153) with gestational diabetes mellitus: a case–control study and meta-analysis. Arch Gynecol Obstet 294, 55–61 (2016). https://doi.org/10.1007/s00404-015-3948-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00404-015-3948-y