Abstract
We report an in-depth characterization of two major stress proteins namely SUMO-conjugating enzyme (Sce) and peptidyl prolyl cis-trans isomerase (PPIase) in rice (Oryza sativa L.). Sce mediates addition of SUMO group to various cell proteins, through process referred to as SUMOylation. Rice nuclear genome has two putative genes encoding the Sce protein (OsSce1 and OsSce2). PCR-amplified full-length OsSce1 cDNA functionally complemented the growth defect in yeast cells lacking the equivalent Ubc9 protein (ScΔubc9). RT-PCR analysis showed that transcript levels of OsSce1 and OsSce2 in rice seedlings were regulated by temperature stress. OsSce1 protein was localized to the nucleus in onion epidermal cells as evidenced by the transient GFP expression analysis following micro-projectile gun-based shooting of an OsSce1-GFP fusion construct. PPIase proteins assist molecular chaperones in reactions associated with protein folding and protein transport across membrane. There are 23 putative genes encoding for FK506-binding proteins (FKBPs; specific class of PPIase) in rice genome. OsFKBP20 cDNA was isolated as a stress-inducible EST clone. Largest ORF of 561 bases in OsFKBP20 showed characteristic FK506-binding domain at N-terminus and a coiled-coil motif at C-terminus. RNA expression analysis indicated that OsFKBP20 transcript is heat-inducible. OsFKBP20 over-expression in yeast endowed capacity of high temperature tolerance to yeast cells. Yeast two-hybrid analysis showed that OsSce1 protein physically interacts with the OsFKBP20 protein. It is thus proposed that OsSce1 and OsFKBP20 proteins in concert mediate the stress response of rice plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Post-translational modifications of proteins can rapidly and reversibly alter the functions of pre-existing proteins, multi-protein complexes and intracellular structures (Kerscher et al. 2006). Proteins are modified by small chemical groups, sugars, lipids and by covalent attachment of other proteins like ubiquitin and SUMO (Small Ubiquitin like Modifiers; Hochstrasser 2000). SUMO, a protein of ∼101 amino acids, is structurally similar to ubiquitin. SUMOylation is a multistep process mediated by E1 (SUMO activating enzyme), E2 (SUMO-conjugating enzyme or Sce) and E3 (SUMO ligase) enzymes (Johnson 2004). Though Sce has specific structural features, which distinguish it from ubiquitin-conjugating (Ubc) enzymes, Sce proteins have been referred to as Ubc in several publications due to their close amino acid sequence similarity and were thought to help in the conjugation of ubiquitin to the target proteins. Since most of the components of the SUMOylation machinery are reported to be localized at the nuclear pore complex (Melchior et al. 2003), it has been suggested that SUMOylation of at least some proteins takes place as they enter the nucleus or is possibly involved in retention of these proteins into the nucleus (Hilgarth et al. 2004). Post-translational modification by SUMO has diverse effects on stability, localization and activity of transcriptional regulators (Gill 2005). There are several reports indicating the role of SUMOylation in stress responses. It is shown that the SUMOylation status of the proteins involved in stress is altered as they travel between the nucleus and the cytoplasm (Kurepa et al. 2003; Saracco et al. 2008). SUMO conjugation pathway is implicated in the activation of a chain of nuclear regulatory proteins whose activities may be needed at low levels in non-stressed and at high levels, in the stressed plants. The targets under negative regulation for instance include a battery of regulators that promote cell division (Sakaguchi et al. 2007). Potential targets under positive regulation include heat shock factor (HSF) like proteins: HSF1 has been reported to undergo modification at lysine298 by SUMO in HeLa cells (Hong et al. 2001). Another role proposed for SUMOylation pertains to the protection of individual proteins from unfavorable conditions by blocking their ubiquitin attachment sites (Hochstrasser 2000). Using the yeast two-hybrid system, DmUbc9 (Sce from Drosophila) was shown to interact with DmHsp23. It was also shown to interact with DmHsp27 and mammalian Hsp27 (Joanisse et al. 1998). Recently, it has been reported that SUMO interacts with parvulins (a class of molecular chaperones) in mammalian system (Mueller et al. 2006). This appears an important observation as protein-folding processes encounter negative pressures, resulting in partially folded proteins or aggregates under stress conditions and this process is minimized by molecular chaperones that promote the dissociation of protein aggregates (Vierling 1991; Boston et al. 1996; Miernyk 1999; Asadulghani et al. 2004). Peptidyl prolyl cis-trans isomerases (PPIases) are considered to assist chaperones by accelerating the slow rate-limiting isomerization steps (Dobson 2004). FK506-binding proteins (FKBP), cyclosporins (CyPs) and parvulin (Pvn) are the three major classes of PPIases, and among these, the best-studied class of PPIases is that of FKBPs by far (Buchhloz et al. 1994; Breiman and Camus 2002). It is thought that FKBPs are the master regulators of several cellular pathways like signal transduction, cell cycle and overall development (Harrar et al. 2001).
Plant Sce proteins are analyzed to a relatively lesser extent. Kurepa et al. (2003) noted that intracellular levels of SUMO conjugates were elevated after exposure to H2O2, canavanine, heat shock and ethanol stresses, which were restored upon the withdrawal of the stimuli in Arabidopsis. This group speculated that the signaling pathways initiated by accumulation of the unfolded proteins directly activate SUMOylation, which in turn activates a chain of nuclear regulatory proteins. Yeast Sce is encoded by a highly conserved, essential gene called as ubc9. In Arabidopsis, two loci-encoding related proteins have been designated as SCE1a and SCE1b (Kurepa et al. 2003). Sce1a encompasses the entire E2, whereas the SCE1b appears truncated, missing about 53 residues from the N-terminal end and is considered to be a pseudo-gene. Like Ubc9, Sce1a belongs to a family of conjugating enzymes, which have a conserved UBCC (Ub-Conjugating Enzyme Catalytic) domain that spans most of the protein. Lois et al. (2003) showed that AtUbc9 is co-localized to the nucleus along with AtSUMO although not specifically to the nuclear envelope as in other organisms. Plant FKBPs have been shown to be involved with several cellular processes such as signaling, protein trafficking, protein folding and transcription (Romano et al. 2005; Smyczynski et al. 2006). A total of 52 genes in Arabidopsis have been found to encode for immunophilins among which 23 are putative FKBPs. Of these 23 FKBPs, 16 are of low-molecular weight, while seven are high-molecular weight proteins (He et al. 2004). FKBP12 is the minimal peptide sequence homology domain in FKBPs; larger FKBPs also contain tricopeptide repeat (TPR), WW domain or calmodulin-(CaM) binding motifs for protein-protein interactions. FKBPs are shown to be localized on to diverse sub-cellular locations like cytoplasm, nucleus, rough ER, plastid stroma and mitochondrial matrix (He et al. 2004). Through various interacting protein partners, FKBP12 has been shown to regulate cell cycle (Aghdasi et al. 2001; Vespa et al. 2004). Maize FKBP66 is reportedly present in both cytosolic and nuclear compartments and interacts with calmodulin and a 36-kDa cytoplasmic protein (Hueros et al. 1998). Owens-Grillo et al. (1996) showed that FKBP52 is nuclear and is involved with targeted movement of protein complexes.
Rice, in recent years has emerged as a model species for understanding the genetics of plants and can be placed next to Arabidopsis as far as details available on its genome are concerned. Magiri et al. (2006) showed that the large FKBPs from rice (namely OsFKBP64, 65 and 75) are heat stress responsive proteins. However, detailed information on the genomic organization of rice Sce and FKBP proteins is by and large lacking. This study reports genome-wide analysis of rice Sce and FKBP encoding gene families as well as isolation and characterization of OsSce1 and OsFKBP20 genes. Association between PPIase and SUMOylation process components has not yet been marked in plant systems. Using yeast two-hybrid assay, we provide evidence that Sce and PPIase proteins may physically interact.
Materials and methods
Growth of rice seedlings, stress treatments and RNA isolation
Rice (Oryza sativa L. var. Pusa basmati 1) seeds were surface-sterilized by washing first with mild detergent, then treating with 70% ethanol (for 2 min), then washing thoroughly with water, subsequently treating with 1.2% sodium hypochlorite solution (for 30 min) and finally rinsing several times with water to remove traces of sodium hypochlorite. Seeds were sown on wet cotton, in plastic trays, which were placed in dark (for 2 days) and then in light for varied periods under culture room conditions (26 ± 2°C). Uniform-sized 7- to 10-day-old seedlings were subjected to various stress treatments. For high temperature stress (HS), seedlings were transferred to beakers, which were maintained in a water bath (for different time intervals as shown). Salt stress (SS) was imposed by placing the seedlings in beakers containing NaCl solution at indicated dosage. For low-temperature stress (LT), beakers were placed in cold room. 12% PEG8000/air-drying was used for inducing osmotic stress as indicated. Subsequent to completion of the stress intervals, tissues were harvested, frozen in liquid nitrogen and kept at −80°C. Total RNA was isolated from the tissues as per the protocol of Chomczynski and Sacchi (1987).
Isolation and characterization of OsSce1 and OsFKBP20 cDNAs
Annotated rice genome sequence has two entries for Sce, namely LOC_Os10g39120 (referred to as OsSce1 in this study) and LOC_Os03g03130 (OsSce2). The 483-bp-long coding region of OsSce1 was PCR-amplified employing specific primers using cDNA made from total RNA as template (OsSce1 Fw-5′ggaattcatgtcgggagggatcgcacg3′; Rev-5′ggggtacctcaaagcaaagcagggtac3′; restriction enzymes sites are underlined). Requisite clone was verified by nucleotide sequencing.
EST clone T4D1-98 [obtained from cDNA libraries constructed earlier by our group; EMBL accession no. AJ420710] was noted to be stress-regulated in macro-array screens (Sahi et al. 2003, 2006). The 3′ end sequencing of this clone showed that it is homologous to a peptidyl prolyl cis-trans isomerase with a highly conserved FK506-binding domain (referred to as OsFKBP20 in this study). The complete open reading frame of OsFKBP20 was PCR amplified using specific primers (OsFKBP20 Fw-5′ggaattcatggcagaggttgcagattt3′; Rev-5′ggggtaccagcatctgatgaacagtagc3′; restriction enzymes sites are underlined). PCR-amplified product was digested with EcoRI/KpnI, purified and cloned into compatible ends of pBCSK(-) (Stratagene, USA) using standard protocols (Sambrook and Russell 2001).
The nucleotide and protein sequences were analyzed using DNA analysis software DNAStar. Multiple alignments were done using MegAlign module of DNAStar. Nucleotide and deduced amino acid sequences were searched for their homology in the NCBI database using the BlastN and BlastP programs (Altschul et al. 1997). For protein domain analysis, sequences were searched against SMART (Simple Modular Architectural Research Tool) database (Schultz et al. 1998; Letunic et al. 2006) as well as at the PFAM database (http://www.sanger.ac.uk/Software/Pfam/search.shtml). Motif scan analysis of the 160-amino-acid long OsSce1 protein was carried out by ISREC-SMART database.
RNA analysis
Total RNA (10 ug) was loaded onto 1% formaldehyde-agarose gel and RNA was transferred to nylon membrane by capillary blotting as described by Sambrook and Russell (2001). PCR-amplified full-length cDNA for OsFKBP20 was radiolabeled using Megaprime Labeling Kit (Amersham, UK) and used as probe. αP32-dCTP (BRIT, India) was used as the radiolabeling molecule. Hybridization and washing of the blots was done using standard protocols (Sambrook and Russell 2001).
For semi-quantitative reverse transcriptase PCR, complementary DNAs were synthesized from 5 μg of total RNA primed with oligo (dT) primers using Promega M-MLV reverse transcriptase. RT-PCR amplification parameters were optimized to analyze individual target genes (OsSce1 primers: Fw-5′atgtcgggagggatcgcacg3′; Rev-5′cacagcacaaatggtaatgac3′, OsSce2 primers: Fw-5′ atgtcggggggaatcg3′; Rev-5′actaagcgaatgtgcagcatgg3′ and OsFKBP20 primers: Fw-5′ctcctttggagctttgatag; Rev-5′tgaagatgcaatcattaacc3′). Three RT-PCR replicates were conducted using independently isolated RNAs from all the tissues as per the following conditions: 94°C for 5 min; 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec for 25 cycles; 72°C for 1 min; and 4°C. β-actin was amplified as an internal control (Act Fw-5′ccctattgagcatggtattg3′; Rev-5′cagttgttgtaaaggaataa3′).
Protein expression, purification and western blotting
For analysis of OsSce1 protein expression, a single colony of OsSce1-transformed M15 E. coli cells was inoculated in LB (supplemented with the required antibiotic). Secondary cultures were initiated by seeding LB with O/N grown cultures. Bacterial suspension was lysed with the lysis buffer [100 mM NaH2PO4, 10 mM Tris-Cl and 8 M urea (pH 8)]. Analysis of soluble proteins was carried out using 15% denaturing SDS-gel. For western analysis, protein was transferred onto Hybond-C super membrane according to the protocol by Towbin et al. (1979). The membrane was probed with anti-D. melanogaster Ubc9 antibodies. For the analysis of OsFKBP20 protein expression, requisite ORF was excised out from pBCSK(-) and cloned into pQE30 (Qiagen, Germany). Plasmid DNA of the recombinant clone was mobilized into E. coli expression strain M15. Protein was expressed by 1 mM IPTG at 37°C for 3 h and purified by Ni-Agarose affinity column. Purified protein was employed for making antibodies in mice.
For the analysis of OsSce1 and OsFKBP20 expression in rice seedlings, fraction containing steady-state soluble proteins was isolated from rice seedlings as described by Zivy et al. (1983). An aliquot of the supernatant was used for the estimation of protein amount as described by Bradford (1976). Proteins resolved on SDS gels were electro-blotted to nitrocellulose membranes (Amersham, USA) as described by Towbin et al. (1979). Blots were probed with heterologous DmUbc9 for OsSce1 analysis and homologous OsFKBP20 antibodies for the OsFKBP20 analysis. Blots were developed by using ECL (enhanced chemiluminiscence; Amersham, USA).
Expression analysis of OsSce1 and OsFKBP20 proteins in yeast
Complete OsFKBP20 ORF was cloned into EcoRI/HindIII restriction sites in yeast centromeric plasmid pGV8 (Agarwal et al. 2003). Approximately 5 μg of plasmid DNA was linearized using EcoRV (Roche, Germany) and employed for transforming wild-type yeast strain FY3 as described by Agarwal et al. (2003). For protein isolation, 2 ml of O/N-grown primary yeast culture was spun down (6,000 rpm, 4°C). Cells were rinsed once with ice-cold sterile double distilled water. Lysis of yeast cells was carried out in Laemelli buffer containing 2% SDS with the help of glass beads (2 mm; Merck) on ice. Microcentrifuge tubes were incubated in a boiling water bath for 10 min. Subsequently, tubes were centrifuged at 10,000 rpm at 4°C. The upper phase containing the soluble protein was transferred to a fresh microcentrifuge tube. A secondary culture of yeast cells (initial OD600 0.05) was started from the O/N primary culture at 25°C (200 rpm) till OD600 reaches 0.15 (∼3 h). OD600 of the secondary culture was measured and if there was slight variation, it was normalized. Aliquots of 200 μl of the yeast cells from all the strains was made in microcentrifuge tubes, which were placed immediately in a water bath set at 50°C for different time durations. After stress, microcentrifuge tubes were immediately plunged into ice till the time of spotting. For spotting tenfold serial dilutions of the cells was prepared in sterile water and 5 μl of each was dotted on YPD-agar plates. The plates were allowed to dry in the laminar flow and finally incubated at 28°C.
For complementation of Δubc9 yeast using OsSce1 gene, YWO103 [MAT α ubc9-1 ubc9Δ::TRP1 his3-Δ 200 leu2-3, 2-112 lys2-801 trp1-1(am) ura3-52 bar1::HIS3] was used for the complementation studies (Seufert et al. 1995). The wild-type strain W303 [Mat a WT his3-Δ200,leu2-3, 2-112, lys2-801, trp1-1(am), ura3-52] was used as a control (more information about W303 can be found at http://www.yeastgenome.org/straintable.shtml#W303). GPD (glyceraldeyde-3-phosphate dehydrogenase) promoter was used for expressing OsSce1 in yeast. OsSce1 was cloned at EcoRI/SmaI sites in the MCS of pGV8 vector (Agarwal et al. 2003). The recombinant clone was confirmed by restriction analysis. Recombinant plasmid was linearized with EcoRV, while the vector was linearized with ApaI to ensure targeted insertion of these cassettes at the ura3 region in the yeast genome. Linearized plasmids were employed for transformation of yeast Δubc9 cells and transformants were selected on minimal plates (SD medium). Transformed yeast cells were checked by PCR and northern analysis for the integrity and expression of OsSce1. Yeast Δubc9 cells transformed with pGV8 were used as a negative control. For complementation assay, yeast cultures were grown at 28°C (200 rpm O/N). This culture was used to initiate 2 ml secondary culture such that the initial OD600 was kept 0.05. Secondary culture was incubated at 25°C for 3-4 h at 200 rpm so as to bring the cells to the exponential phase of growth. This culture was tenfold serially diluted and 5 μl of each of these dilutions were spotted on YPD plates. The plates were incubated at permissive (28°C) and restrictive (37°C) temperatures.
Yeast two-hybrid assay
YRG2 strain [Mat a ura3-52 his-200 ade2-101 lys2-801 trp1-901 leu2-3 112 gal4-542 gal80-538 LYS2::UASGAL1-TATAGAL1-HIS3 URA3::UASGAL4- 17mers(x3)TATAcyc1-LacZ] was used for this study. Yeast cells were grown either in rich YPAD medium, or in minimal selection medium like SD medium supplemented with required amino acids, depending upon the experiment. Yeast cells were inoculated in the requisite medium and allowed to grow at 28°C (at 200 rpm overnight).
OsSce1 ORF was sub-cloned from pQE30 vector into pBD as well as pAD vectors, while OsFKBP20 ORF was subcloned from pQE30 vector into pAD. Transformants were selected on a medium lacking both leu and trp. Yeast cultures were grown at 28°C (200 rpm O/N). These cultures were used to inoculate 2 ml secondary culture such that the initial OD600 was 0.05. Secondary culture was incubated at 25°C for 3–4 h at 200 rpm so as to bring the cells to exponential phase of growth. Five microliter of each of these cultures were also spotted on Double DO and Triple DO plates. These plates were incubated at 28°C. The ability of the cells to grow on a histidine-deficient media was used as a reporter, to assay for the protein–protein interactions (Causier and Davies 2004). 3-AT was used as a competitive inhibitor of His 3 gene product, to reduce the background due to basal His3 expression. Transformation of YRG2 with pAD-pBD was used as a vector control.
Cellular localization of OsSce1 protein
pmGFP4 (Haseloff et al. 1997) vector was used for cloning OsSce1 upstream of GFP. OsSce1 was PCR- amplified using primers containing the sites for the restriction enzymes BglII and SacI in the forward and reverse primers, respectively (Sce1GFP Fw-5′gaagatctatgtcgggagggatcgc3′; Rev-5′cgagctctcaaagcaaagcaggta3′; restriction enzymes sites are underlined). The recombinant clones were sequenced to check the fidelity of the PCR reaction. Gold particles (0.6 μm diameter: 60 mg) were incubated for 10 min in 70% ethanol and washed twice. Five microliter of gold suspension was taken and 2 μl of DNA (pmGFP4 and pmGFP4-OsSce1; 1 μg μl−1) was added to it. This mixture was vortexed for 4 min continuously. To this mix, 1 μl of spermidine (1 mM) and 5 μl of CaCl2 (2.5 M) was added. This mixture was vortexed for 3 min and centrifuged for 8–10 sec at high speed. The onion bulb scales were cut into pieces of 1.5 cm × 1.5 cm and kept on MS medium petriplates. Plasmid DNA coated onto the gold particles were bombarded on the onion bulb scales using the particle gun (PDS1000/He:Biorad, Hercules, USA) using 1,100 psi rupture discs under vaccum of 25–26 inch Hg and target distance of 10 cm. After bombardment, the explants were kept in dark at 25°C for 16–18 h. GFP expression was monitored with a Nikon Flourescence microscope (Zeiss Corp., Germany) using filter cube I (365 nm excitation, 395 nm beam splitter, 495 nm long pass emission) for mGFP4 emission. Photographs were taken using the a digital camera using the UV2A filter (330–380 nm excitation, 400 nm beam splitter and 420-nm-long pass).
Results
Analysis of Sce-encoding rice cDNAs/ genes and Sce protein
Rice genome sequence has two entries for the Sce-encoding genes namely LOC_Os10g39120 (OsSce1) and LOC_Os03g03130 (OsSce2). Various in silico characteristics of OsSce1 and 2 proteins are provided in supplementary data 1. Interestingly, OsSce1 and OsSce2 genes showed almost similar exon–intron distribution pattern and the second intron was the longest in both the cases (Fig. 1A). The 483-bp-long coding region of OsSce1 was PCR-amplified and the fidelity of the reaction was verified by nucleotide sequencing. Amino acid (aa) sequence of OsSce1 is shown in Fig. 1B. Motif-scan analysis of 160-aa-long OsSce1 by ISREC-SMART database revealed that it has highly conserved UBC domain (spanning from 8–158 aa residues; Fig. 1B). This domain is reportedly common to UBC family members. The aa sequence of OsSce1 showed high degree of conservation with Ubc9 proteins in different other eukaryotes by BLAST analysis using NCBI database [amino acid sequence in OsSce1 was 85.6%, 89.4%, 63.5%, 61.4% and 63.1% homologous to AtUbc9 (from A. thaliana), NtUbc9 (N. tabacum), HsUbc9 (H. sapiens), SpHus5 (S. pombe) and ScUbc9 (S. cerevisiae), respectively]. Phylogenetic tree analysis reflected that OsSce1 has high degree of homology with counterpart proteins from diverse organisms indicating that plant Sce proteins may have evolved from a common ancestor (Fig. 2A). PCR-amplified OsSce1 ORF was cloned into pBCSK vector. The fidelity of PCR reaction was confirmed by nucleotide sequencing using T3 primer. OsSce1 was subsequently subcloned from pBCSK into pQE30 vector, to express the OsSce1 recombinant protein in E. coli M15 cells. OsSce1 in M15 cells was expressed as ∼21-kDa-sized protein on 15% SDS-gel. Expressed protein from M15 cells was purified to near-homogeneity using Ni-NTA agarose column under denaturing conditions. Western analysis of the purified OsSce1 protein carried out employing anti-Dmubc9 antibodies showed a distinct cross-reaction (Fig. 2B). At the transcript levels, OsSce1 and OsSce2 genes showed differential stress-induced patterns. OsSce1 transcript was repressed, while OsSce2 transcript was strongly induced by 24 h of LT stress treatment. OsSce1 showed a slightly higher level of transcription during HS treatment as compared to OsSce2. Both the transcripts showed moderate induction at 60 min of HS treatment (Fig. 2C). Salt and desiccation stress also resulted in up-regulation of both these transcripts (Fig. 2C).
A The exon–intron structure of OsSce1 and OsSce2 proteins are shown. The grey boxes represent the exons while the lines represent the introns. Vertical lines in the first exons represent the start point of ORFs. The positions of the exons based on the genomic sequence (on the top; position 1 represents transcription initiation site) and ORF (at the bottom; position 1 represents start codon) and the length of the introns are mentioned. B Structure-based sequence alignment of rice OsSce1 (Os10g39120) and OsSce2 (Os03g03130) with Sce from other systems. CAE45567: Sce from Nicotiana benthamiana, CAD29823: Populus x canadensis, AF246690: Dictyostelium discoideum, ABG79538: Triticum turgidum, NP_191346: Arabidopsis thaliana, YDL069W: Saccharomyces cerevisiae. On the top of the amino acid sequence, helices are lettered while strands are numbered. The asterisk marks the active site cysteine residue
A Phylogenetic relationship of OsSce1 with other eukaryotic Sce proteins as determined by CLUSTAL using DNAStar. The length of each pair of branches represents the distance between sequence pairs, while the units at the bottom of the tree indicate the number of substitution events. CAE45567: Sce from Nicotiana benthamiana, CAD29823: Populus x canadensis, AF246690: Dictyostelium discoideum, ABG79538: Triticum turgidum, NP_191346: Arabidopsis thaliana, YDL069W: Saccharomyces cerevisiae. B Dm-UBC9 antibodies cross-reaction with OsSce1 protein (which was expressed in E. coli). C RT-PCR based expression profile of the transcripts of OsSce1 and OsSce2 genes in response to different abiotic stresses. 5 μg of total RNA was used for reverse transcription and equal volume was taken for subsequent PCRs. HS: heat shock treatment (45°C; 10′, 20′, 30′ and 1 h). LT: low-temperature stress (8 ± 2°C; 24 h and 48 h). SS: NaCl stress (150 mM; 3 h and 6 h). DS: PEG treatment (12% PEG8000; 3 h and 6 h)
Complementation analysis of ScΔubc9 mutant with the OsSce1 gene
As mentioned above, OsSce1 and ScUbc9 proteins are 63.1% homologous. The relatedness between the rice and yeast Sce proteins was also reflected from the comparison of their hydropathy plots (Fig. 3A). In both rice and yeast Sce proteins, almost similar profiles of hydrophobic and hydrophilic aa were seen throughout the length of the protein. Next, OsSce1 cDNA was subcloned from pQE30 to pGV8 vector (under the control of GPD promoter) for expression in yeast (Fig. 3B). Linearized pGV8-OsSce1 and empty pGV8 plasmid DNAs were transformed into ScΔubc9 cells. Northern analysis carried out for the wild type (lane 1), ScΔubc9 (mutant cells; lane 2), ScΔubc9+pGV8 (mutant cells carrying empty plasmid; lane 3) and ScΔubc9+pGV8-OsSce1 (mutant cells with OsSce1 encoding gene cloned in pGV8; lane 4) yeast cells using radiolabeled OsSce1 cDNA as probe showed cross-reacting band in RNA samples from ScΔubc9+pGV8-OsSce1 cells only (Fig. 3C). These cells (WT, ScΔubc9, ScΔubc9+pGV8, ScΔubc9+pGV8-OsSce1) were spotted on YPD plates (at four different dilutions): one set of plates was incubated at 28°C while the other set of plates was kept at 37°C. ScΔubc9 cells were notably impaired in growth as compared to the WT cells; this effect was seen at both the incubation temperatures analyzed in this study but the effect was more pronounced at 37°C as compared to 28°C. Importantly, it was observed that OsSce1 cDNA functionally complements the growth defect of the ScΔubc9 strain (Fig. 3D).
A Hydropathy plots of OsSce1 and ScUbc9 proteins. Hydropathy values are assigned for all amino acids and are then averaged over a user defined window. Residue hydropathy assignments are derived from water-vapor transfer free energies and the interior–exterior distribution of residue side-chains. B Linear map of pGV8-OsSce1 vector. C Northern analysis of yeast cells. RNA was isolated from wild type (lane 1), ScΔubc9 mutant (lane 2), ScΔubc9+pGV8 (lane 3) and ScΔubc9+pGV8-OsSce1 (lanes 4) cells. PCR-amplified OsSce1 was used as probe. Equal amount of RNA is depicted in the ethidium bromide stained-lower panel. D Thermotolerance plate assay. Genetic constitution of different yeast cells used in this assay are shown toward the left. The cultures were serially diluted to a final concentration of 10−3. 5 μl of different dilutions were spotted on YPD plates. After spotting, one plate was kept at 28°C and other at 37°C. UD is the undiluted concentration of yeast cells
Analysis of FKBP20-encoding rice cDNAs/ genes and FKBP20 protein
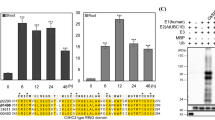
Full-length OsFKBP20 cDNA was amplified from the rice EST clone. Full-length OsFKBP20 cDNA was cloned in pBCSK(-) and was sequenced using T3 and T7 primers to confirm the authenticity of the amplicon. OsFKBP20 nucleotide sequence corresponded to the LOC_Os05g38370 locus of the TIGR rice database. An extensive search made at the TIGR rice database using OsFKBP20 aa sequence as a query showed that there are 23 putative FKBP proteins in rice genome. Based on their molecular weights, these 23 proteins can be classified as low-molecular weight (LMW) FKBPs and high-molecular weight (HMW) FKBPs (He et al. 2004). According to our analysis, there are nine LMW and 14 HMW FKBPs in rice. In silico search for their cellular localization revealed that eight of these FKBPs were organellar. A detailed description of these members in terms of their molecular weights, isoelectric points, predicted localization and domain architecture is provided in supplementary data 2. Supplementary data 3 shows phylogenetic relationship among the Arabidopsis and rice proteins. Grouping of OsFKBPs under different clades indicates that rice FKBPs have evolved independently of each other and may be involved in diverse functions in the cell. 561 bp ORF of OsFKBP20 clone encodes for a 186-amino-acid-long protein of ∼20 kDa (EMBL accession no. AJ833645). Translated protein contained FK506 binding domain toward the N-terminus, while it had a low complexity coiled-coil region (as predicted by SMART database) at the C-terminus (Fig. 4A). The coiled-coil motif contains a bipartite NLS, which suggests that the rice protein might be nuclear. Blast analysis using NCBI database showed that OsFKBP20 is homologous to peptidyl prolyl cis-trans isomerases in the database, including Arabidopsis 20 kDa FKBP (AL132975). Sequence alignment of the conceptual translated amino acids showed that OsFKBP20 belongs to highly conserved protein family across the species (supplementary data 4).
A Schematic representation of OsFKBP20 protein as revealed by domain analysis at Pfam (http://www.sanger.ac.uk/Software/Pfam/search.shtml). OsFKBP20 contains a FKBP domain at the N-terminus while the NLS is present at the C-terminus. B OsFKBP20 transcript analysis by northern blotting. 1-week-old PB1 rice seedlings were heat stressed at different temperatures for 2 h (left panel) or for different time periods at 42°C (right panel) as indicated. 10 μg of total RNA was loaded on denaturing 1% agarose gel. The blots were probed with radiolabeled OsFKBP20 cDNA. Methylene blue stained bands of rRNA depict equal RNA loading. C RT-PCR based expression profile analysis of OsFKBP20 transcript in response to different abiotic stresses. 5 μg of total RNA was used for reverse transcription and equal volume was taken for subsequent PCRs. HS: heat shock treatment (45°C; 10′, 20′, 30′ and 1 h), LT: low temperature stress (8 ± 2°C; 24 h and 48 h), SS: NaCl stress (150 mM; 3 h and 6 h). DS: PEG treatment (12% PEG8000; 3 h and 6 h). The PCR products were run on 1% agarose gels. D Stress-regulated expression of OsFKBP20 protein in rice. 30 μg of total soluble shoot proteins isolated from rice seedlings subjected to different stresses (as indicated at the top of the panel) were loaded on 15% SDS-gel. C-unstressed control, HS: heat shock treatment (45°C; 2 h), SS: NaCl stress (200 mM; 24 h), DS: air-drying (2 h), LT: low temperature stress (4°C; 48 h). The proteins were electro-blotted on to nitrocellulose membrane. Blot was probed with primary antibody (1:1,000) raised against OsFKBP20 protein. The blot was probed with HRP-conjugated anti-mouse secondary antibody (1:2,000) after which the blot was processed for enhanced chemiluminescence-based development. E Thermotolerance plate assay. Equal number of yeast cells from different strains (as shown on right side of the panel) was stressed at 50°C for 30 and 45 min (as indicated at the bottom of the respective panels). 5 μl of different dilutions including the undiluted (UD) stock of yeast cell (as indicated at the top) were dotted on the YPD-agar medium. Plates were put on recovery at 28 ± 2°C for 2 days. FY3: wild-type yeast cells; FY3-pGV8: vector control; FY3-pGV8-OsFKBP20; wild-type yeast cells expressing the rice OsFKBP20. Panel on the right indicated by W depict the western analysis of different yeast strains
On northern blots, OsFKBP20 transcript was noted to be HS-inducible (Fig. 4B). The induction was notably significant at 42°C (2 h): the induction of OsFKBP20 transcript was noticeable even at 20 min sampling interval at this temperature and was followed by further increase in transcript levels with increasing stress durations. RT-PCR results showed that OsFKBP20 transcript is induced by HS, salt and desiccation-stress treatments. Unlike OsSce2, exposure of seedlings to low-temperature stress caused down-regulation in the levels of this transcript (Fig. 4C). Western blotting analysis showed that OsFKBP20 protein is present in unstressed rice shoots. Levels of this protein were noted to be slightly up-regulated in response to HS, SS and air-drying (Fig. 4D); protein amounts appeared comparable in unstressed control and low-temperature-stressed seedlings. Yeast cells harboring OsFKBP20 cDNA were examined for expression by northern (data not shown) and western analysis. On western blot, OsFKBP20 protein was detected only in FY3-pGV8-OsFKBP20 yeast cells (Fig. 4E; right side panel). Next, FY3-pGV8-OsFKBP20 yeast cells were noted to be more efficient in tolerating 50° C HS as compared to FY3 (WT) and FY3-pGV8 yeast cells. After 30 min of HS, FY3 (WT) and FY3-pGV8 yeast cells grew at a much lower rate as compared to FY3-pGV8-OsFKBP20 yeast cells. Yeast cells expressing OsFKBP20 were observed even at 100-fold dilution after 45 min of continuous HS stress (Fig. 4E).
Interaction of OsSce1 and OsFKBP20 proteins
Testing for the auxotrophy for histidine in the presence of 3-AT was taken as a criterion to adjudge interaction of OsSce1 and OsFKBP20 proteins in a yeast two-hybrid assay. Figure 5 shows that in the double dropout medium (lacking leu and trp), yeast cells transformed with pBD-OsSce1/ pAD-OsFKBP20 as well as cells transformed with pAD/pBD, pAD+pBD-OsSce1 and pAD-OsSce1+pBD were able to grow. However, in the triple dropout medium (lacking leu, trp and his), only the cells containing pBD-OsSce1+pAD-OsFKBP20 grew. This indicates that these two proteins may interact. The experiment was repeated several times and similar results were obtained.
Yeast two-hybrid assay showing interaction of OsSce1 with OsFKBP20. An equal number of cells were spotted on yeast synthetic dropout medium lacking leu, trp (Double Drop Out) and that lacking leu, his, trp and ura (Triple Drop Out). Ura and 3-AT (1 mM) were added later to the media. After spotting, plates were placed at 28°C
Analysis of cellular location of OsSce1 using onion scale cells
OsSce1 cDNA was cloned into pmGFP4 vector to yield pmGFP4-OsSce1 plasmid (Fig. 6A). Bombardment of onion scale cells with the vector-control plasmid (pmGFP4) showed fluorescence emanating from the expression of GFP almost from the entire cytoplasm of the transformed cells (Fig. 6B; position of the three cells showing fluorescence in the right panel are indicated by arrowheads in the left panel). On the other hand, shooting onion cells with pmGFP4-OsSce1 plasmid showed the emission of fluorescence localized predominantly to the nucleus (Fig. 6d and f; position of the cell showing fluorescence emanating from the nucleus in the right panel is indicated by arrowhead in the left panel; the position of the nucleus in this cell is shown by the arrow). It is thus inferred that OsSce1 is localized predominantly to the nucleus.
A Schematic map of pmGFP4-OsSce1 construct. B Fluorescence expression profile of onion scale epidermal cells shot with pmGFP4 (a, b) and pmGFP4-OsSce1 (c–f) constructs. The signal emanating from the GFP protein was analyzed using fluorescence microscope. The arrows in brightfield indicate the fluorescence emanating from the constructs in the respective onion cells. Arrowheads in the left panel indicate the cells showing fluorescence in the right panel. Arrow shows the position of nucleus
Discussion
This study provides an in-depth analysis of Sce and FKBP proteins in rice. From the genome-wide search parameters, it appeared that two gene loci namely, LOC_Os10g39120 (OsSce1) and LOC_Os03g03130 (OsSce2) localized on chromosomes 10 and 3, respectively, encode for the Sce protein in the annotated rice genome (Fig. 1). These two rice Sce genes showed 86% homology at the nucleotide and 93% similarity at the amino acid levels (supplementary data 1). As the length of the exons in these two genes was noted to be similar, differences in their size were thus contributed by the length of introns. Interestingly, the positions of the introns were also noted to be conserved in these two genes (Fig. 1A). Further, search for segmental duplication in rice at TIGR (http://www.tigr.org/tdb/e2k1/osa1/segmental_dup/500kb/duplication_listing.html) showed that this gene pair was a result of a duplication event. Putative cis-elements in 1 kb region upstream of both the genes at PLACE database (http://www.dna.affrc.go.jp/PLACE/) showed notable differences: the region upstream of OsSce2 possessed higher abundance of stress-related elements like LTREs, DREs, ABREs (Hirayama and Shinozaki 2007; Seki et al. 2007), CCAAT (Gelinas et al. 1985), and AREs (Olive et al. 1991), as compared to the region upstream of OsSce1 (data not shown). It would thus be interesting to analyze and compare the regulatory aspects of OsSce1 and OsSce2 gene promoters in future course. Blast analysis showed that OsSce1 protein has high degree of homology with different counterpart proteins of higher eukaryotes (Figs. 1B, 2A), highlighting conservation of this protein across biological kingdoms. Saracco et al. (2008) noted that Arabidopsis Sce1 mutants were embryonic lethal, indicating that Sce protein may be performing critical role in cellular functioning. In our study, Western analysis with E. coli cells producing OsSce1 protein showed cross-reaction with anti-DmUbc9 (Sce protein from D. melanogaster) antibodies (Fig. 2B), further stressing that Sce protein was highly conserved. However, we did not find cross-reaction of anti-DmUBC9 antibodies with protein extracts made from control or stressed (high temperature, low temperature, salt stressed) rice seedlings. While the exact reason for this lack of cross-reaction is not worked out as yet, it may be that the OsSce1 protein is present in extremely low amounts in rice seedlings. Hydropathy plot showed that the profile of the OsSce1 and ScUbc9 proteins was nearly identical (Fig. 3A). OsSce1 protein was further seen to genetically complement growth defect observed in ScΔubc9 cells (Fig. 3D). Earlier, DmUbc9 (Sce from D. melanogaster) (Joanisse et al. 1998), NbUbc9 (Sce from N. benthamiana) (Castillo et al. 2004) and Hsubc9 (Sce from H. sapiens) (Yasugi and Howley 1996) proteins have been shown to complement ScΔubc9 phenotype.
Earlier studies have shown that the SUMOylation activity gets increased in response to different stresses (Zhou et al. 2004). In Arabidopsis, Saracco et al. (2008) recently indicated that the pool of SUMO conjugates is elevated in response to heat stress. RT-PCR analysis in the present study showed that OsSce transcript levels were altered in rice seedlings under different stresses (Fig. 3C). OsSce1 and OsSce2 transcript levels were moderately up-regulated by high-temperature stress: on relative basis, transcript levels of OsSce1 under control and heat shock conditions were higher than OsSce2 levels at the respective sampling intervals. Importantly, OsSce1 transcript levels declined during the first 24 h of low-temperature stress, while there was a significant increase in the levels of OsSce2 transcripts at the same sampling time. OsSce1 transcript levels declined at 6 h of 12% PEG treatment as compared to 3 h treatment, while there was no change in the levels of OsSce2 in the same time interval. These observations lead to the inference that OsSce1 and OsSce2 transcripts are differentially regulated by stress conditions in rice.
Analysis of cellular localization of OsSce1 by bombardment of OsSce1 cDNA into onion epidermal cells showed that OsSce1 protein is nuclear localized (Fig. 6d, f). This observation is in conformity with the observations made earlier that the Sce protein is primarily associated with the nuclear pore complex (NPC; Melchior et al. 2003). It has been suggested that the NPC-associated localization of Sce may be of importance for (1) altering the function of the proteins as they enter the nucleus and/or (2) cycles of modification and cleavage at the NPC may be required for the actual translocation. However, it is not possible for us to comment whether OsSce1 associates with NPC or not based on the present results. Although AtUbc9 protein is known to be co-localized to the nucleus along with AtSUMO (Lois et al. 2003) in this case also, it is not known where exactly in nucleus it is localized.
There is no published data on genomic organization of rice FKBP members. Genome-wide search for FKBPs in this study indicated that there are 23 FKBP member proteins in rice. These proteins were noted to be widely distributed in the cell. Various in silico characteristics of these members are shown in supplementary data 2 and 3. Rice chromosomes 11 and 12 are reported to have undergone extensive duplication events (The Rice Chromosomes 11 and 12 Sequencing Consortia 2005; Paterson 2006). It is interesting to note that Os11g05090 and Os12g05090 gene pair which showed significant homology (88% at the nucleotide level) is predicted to represent one such event (http://www.tigr.org/tdb/e2k1/osa1/segmental_dup/500kb/duplication_listing.html): the rice genome browser showed that some genetic regions are conserved at the loci surrounding Os11g05090 and Os12g05090 genes. We isolated cDNA encoding for OsFKBP20 in this work (supplementary data 4, Fig. 4A). RT-PCR analysis indicated that OsFKBP20 transcript is induced by high-temperature stress in rice shoots (Fig. 4C). RNA blot analysis also showed that OsFKBP20 transcript was strongly induced by high-temperature stress (Fig. 4B). The maximum expression of OsFKBP20 transcript was noted at 42°C in this experiment. To examine the possible cellular role of plant FK506-binding protein in high-temperature stress tolerance response, OsFKBP20 full-length cDNA was cloned in a yeast expression vector under the control of constitutive GPD promoter. OsFKBP20 over-expression imparted significant heat stress tolerance to WT yeast cells (Fig. 4E). Even after 45 min at 50°C, 100-fold more tolerance was scored for OsFKBP20 expressing yeast cells over WT and vector-transformed yeast cells. Our results are in accordance with the earlier findings suggesting that PPIases are implicated in stress (including high temperature) responses in varied organisms (Meza-Zepeda et al. 1998; Andreeva et al. 1999; Dwivedi et al. 2003). Yeast ess1 mutant for PPIase activity shows a temperature-sensitive phenotype in yeast (Morris et al. 1999). Most of the FKBP transcripts analyzed till now have shown heat inducibility. It is therefore possible that cis-trans isomerization is a critical step in protein folding, which is compromised under conditions of high-temperature stress and OsFKBP20 may help to mitigate the stress effects through their role in maintaining proper protein folding.
As mentioned earlier, there is evidence that SUMO and parvulins show physical interaction in mammalian cells (Mueller et al. 2006). There is no report on interaction of these proteins in plant systems. Our observation that OsSce1 and OsFKBP20 interact in a yeast two-hybrid assay (Fig. 5) is a novel finding. Lois and Lima (2005) showed that E1 enzyme and Ubc9 protein, physically bind with each other. We suggest that interactions between SUMO moiety and parvulins (OsFKBP20 in this study) involving E1 enzyme may also be assisted by Sce protein. It will be important to show and isolate physical-binding states of these components in future studies. OsFKBP20 protein has a bipartite NLS signature sequence, indicating that OsFKBP20 protein might be nucleus-localized protein (supplementary data 2, Fig. 4A). Thus, it is likely that the physical interactions discussed above involve nucleus interface.
Finally, this study suggests that Sce, may have a key role in cellular trafficking of stress proteins in plants. We provide evidence that rice Sce is localized in nucleus; it may have NPC localization as shown for Sce of other systems. It can be speculated that NPC-localized OsSce1 might be involved in the transportation of OsFKBP20 protein to the inside of the nucleus as shown for nuclear transport of other substrate proteins (Pichler and Melchoir 2002; Stade et al. 2002; Nacerddine et al. 2005). PPIase have been implicated in chromatin remodeling and cell cycle progression as well as in regulation of protein kinases to control the activity or stability of key regulatory proteins (Shaw 2007). It may be the case that PPIase, SUMO and Sce proteins are co-involved in this event. For unveiling further details, it is important that future studies address where in nucleus Sce is active and what other proteins it interacts with.
References
Agarwal M, Sahi C, Katiyar-Agarwal S, Agarwal S, Young T, Gallie DR, Sharma VM, Ganesan K, Grover A (2003) Molecular characterization of rice hsp101: complementation of yeast hsp104 mutation by disaggregation of protein granules and differential expression in indica and japonica rice types. Plant Mol Biol 51:543–553
Aghdasi B, Ye K, Resnick A, Huang A, Ha HC, Guo X, Dawson TM, Dawson VL, Snyder SH (2001) FKBP12, the 12-kDa FK506-binding protein, is a physiologic regulator of the cell cycle. Proc Natl Acad Sci USA 98:2425–2430
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Andreeva L, Heads R, Green CJ (1999) Cyclophilins and their possible role in the stress response. Int J Exp Pathol 80:305–315
Asadulghani, Nitta K, Kaneko Y, Kojima K, Fukuzawa H, Kosaka H, Nakamoto H (2004) Comparative analysis of the hspA mutant and wildtype Synechocystis sp. strain PCC6803 under salt stress: evaluation of the role of hspA in salt stress management. Arch Microbiol 182:487–497
Boston RS, Viitanen PV, Vierling E (1996) Molecular chaperones and protein folding in plants. Plant Mol Biol 32:191–222
Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Breiman A, Camus I (2002) The involvement of mammalian and plant FK506-binding proteins (FKBPs) in development. Transgenic Res 11:321–335
Buchholz WG, Harris-Haller L, DeRose RT, Hall TC (1994) Cyclophilins are encoded by a small gene family in rice. Plant Mol Biol 25:837–843
Castillo AG, Kong LJ, Hanley-Bowdoin L, Bejarano ER (2004) Interaction between a geminivirus replication protein and the plant sumoylation system. J Virol 78:2758–2769
Causier B, Davies B (2004) Analyzing protein-protein interactions with the yeast two-hybrid system. Plant Mol Biol 50:855–870
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
Dobson CM (2004) Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol 15:3–16
Dwivedi RS, Breiman A, Herman EM (2003) Differential distribution of the cognate and heat-stress-induced isoforms of high Mr cis-trans prolyl peptidyl isomerase (FKBP) in the cytoplasm and nucleoplasm. J Exp Bot 54:2679–2689
Gelinas R, Endlich B, Pfeiffer C, Yagi M, Stamatoyannopoulos G (1985) G-substitution to A-substitution in the distal CCAAT box of the gamma-globin gene in Greek hereditary persistence of fetal hemoglobin. Nature 313:323–325
Gill G (2005) Something about SUMO inhibits transcription. Curr Opin Genet Dev 15:536–541
Harrar Y, Bellini C, Faure JD (2001) FKBPs: at the crossroads of folding and transduction. Trends Plant Sci 6:426–431
Haseloff J, Siemering KR, Prasher DC, Hodges S (1997) Removal of a cryptic intron and sub-cellular localization of GFP are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci 94:2122–2127
He Z, Li L, Luan S (2004) Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol 134:1–20
Hilgarth RS, Murphy LA, O’Connor CM, Clark JA, Park-Sarge OK, Sarge KD (2004) Identification of Xenopus heat shock transcription factor-2: conserved role of sumoylation in regulating deoxyribonucleic acid-binding activity of heat shock transcription factor-2 proteins. Cell Stress Chaperones 9:214–220
Hirayama T, Shinozaki K (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12:343–351
Hochstrasser M (2000) All in the ubiquitin family. Science 289:563–564
Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD (2001) Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem 276:40263–40267
Hueros G, Rahfeld J, Salamini F, Thompson R (1998) A maize FK506-sensitive immunophilin, mzFKBP-66, is a peptidylproline cis-trans-isomerase that interacts with calmodulin and a 36-kDa cytoplasmic protein. Planta 205:121–131
Joanisse DR, Inaguma Y, Tanguay RM (1998) Cloning and developmental expression of a nuclear ubiquitin-conjugating enzyme (DmUbc9) that interacts with small heat shock proteins in Drosophila melanogaster. Biochem Biophys Res Commun 244:102–109
Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73:355–382
Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22:159–180
Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278:6862–6872
Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P (2006) SMART 5: domains in the context of genomes and networks. Nucleic Acids Res 34:D257–D260
Lois LM, Lima CD (2005) Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J 24:439–451
Lois LM, Lima CD, Chua NH (2003) Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15:1347–1359
Magiri EN, Farchi-Pistany O, Avni A, Breiman A (2006) The expression of the large rice FK506 binding proteins demonstrate tissue specificity and heat stress responsiveness. Plant Sci 170:695–704
Melchior F, Schergaut M, Pichler A (2003) SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem Sci 28:612–618
Meza-Zepeda LA, Bando MM, Palva ET, Heino P (1998) Isolation and characterization of a cDNA corresponding to a stress activated cyclophilin gene in Solanum commersoni. J Exp Bot 49:1451–1452
Miernyk JA (1999) Protein folding in the plant cell. Plant Physiol 121:695–703
Morris DP, Phatnani HP, Greenleaf AL (1999) Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3′-end formation. J Biol Chem 274:31583–31587
Mueller JW, Kessler D, Neumann D, Stratmann T, Papatheodorou P, Hartmann-Fatu C, Bayer P (2006) Characterization of novel elongated Parvulin isoforms that are ubiquitously expressed in human tissues and originate from alternative transcription initiation. BMC Mol Biol 7:9
Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, pandolfi PP, Dejean A (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation on mice. Dev Cell 9:769–779
Olive MR, Peacock WJ, Dennis ES (1991) The anaerobic responsive element contains two GC-rich sequences essential for binding a nuclear protein and hypoxic activation of the maize Adh1 promoter. Nucleic Acids Res 19:7053–7060
Owens-Grillo JK, Stancato LF, Hoffmann K, Pratt WB, Krishna P (1996) Binding of immunophilins to the 90 kDa heat shock protein (hsp90) via a tetratricopeptide repeat domain is a conserved protein interaction in plants. Biochem 35:15249–15255
Paterson AH (2006) Leafing through the genomes of our major crop plants: strategies for capturing unique information. Nat Rev Genet 7:174–184
Pichler A, Melchoir F (2002) Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3:381–387
Romano P, Gray J, Horton P, Luan S (2005) Plant immunophilins: functional versatility beyond protein maturation. New Phytol 166:753–769
Sahi C, Agarwal M, Reddy MK, Sopory SK, Grover A (2003) Isolation and expression analysis of salt stress-associated ESTs from contrasting rice cultivars using a PCR-based subtraction method. Theor Appl Genet 106:620–628
Sahi C, Singh A, Kumar K, Blumwald E, Grover A (2006) Salt stress response in rice: genetics, molecular biology, and comparative genomics. Funct Integr Genomics 6:263–284
Sakaguchi K, Koshiyama A, Iwabata K (2007) Meiosis and ubiquitin-related modifier (SUMO)-conjugating enzyme, Ubc9. FEBS J 274:3519–3531
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2008) Genetic analysis of sumoylation in Arabidopsis: heat-induced conjugation of SUMO1 and 2 is essential. Plant Physiol (in press). doi:10.1104/pp.107.102285
Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95:5857–5864
Seki M, Umezawa T, Urano K, Shinzoaki K (2007) Regulatory metabolic networks in drought stress responses, Curr Opin Plant Biol 10:296–302
Seufert W, Futcher B, Jentsch S (1995) Role of a ubiquitin-conjugating enzyme in degradation of S- and N-phase cyclins. Nature 373:78–81
Shaw PE (2007) Peptidyl-prolyl cis/trans isomerases and transcription: is there a twist in the tail? EMBO Rep 8:40–45
Smyczynski C, Roudier F, Gissot L, Vaillant E, Grandjean O, Morin H, Masson T, Bellec Y, Geelen D, Faure JD (2006) The C-terminus of the immunophilin PASTICCINO1 is required for plant development and for interaction with a NAC-like transcription factor. J Biol Chem 281:25475–25484
Stade K, Vogel F, Schwienhorst J, Meusser B, Volkwein C, Nentwig Bm Dihmen RJ, Sommer J (2002) A lack of SUMO conjugation affects cNLS-dependent nuclear protein import in yeast. J Biol Chem 277:49554–49561
The Rice Chromosomes 11, 12 Sequencing Consortia (2005) The sequence of rice chromosome 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol 3:20
Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76:4350–4354
Vespa L, Vachon G, Berger F, Perazza D, Faure JD, Herzog M (2004) The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol 134:1283–1292
Vierling E (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42:579–620
Yasugi T, Howley PM (1996) Identification of the structural and functional human homolog of the yeast conjugating enzyme UBC9. Nucleic Acids Res 24:2005–2010
Zhou W, Ryan JJ, Zhou H (2004) Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem 279:32262–32268
Zivy M, Thiellement H, deVienne D, Hofmann JP (1983) Study on nuclear and cytoplasmic genome expression in wheat by two-dimensional gel electrophoresis. Theor Appl Genet 66:1–7
Acknowledgments
AG thanks Department of Biotechnology, Government of India for the Bioscience Career Award Project grant. We thank Prof. Robert Tanguay for providing the anti-DmUBC9 antibodies and Prof. Stefan Jentsch for the yeast wild type and Δubc9 mutant strain. NN acknowledges Monsanto (India) and Rotary Club of India while CS and AS acknowledge the Council of Scientific and Industrial Research, New Delhi for the fellowship awards.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.-K. Zhu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nigam, N., Singh, A., Sahi, C. et al. SUMO-conjugating enzyme (Sce) and FK506-binding protein (FKBP) encoding rice (Oryza sativa L.) genes: genome-wide analysis, expression studies and evidence for their involvement in abiotic stress response. Mol Genet Genomics 279, 371–383 (2008). https://doi.org/10.1007/s00438-008-0318-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-008-0318-5