Abstract
Main conclusion
A rice gene (OsSIRP2) encoding the RING Ub E3 ligase was highly induced under salinity stress and physically interacted with a transketolase (OsTKL1). Overexpression of OsSIRP2 conferred salinity and osmotic stress tolerance in plants.
The RING E3 ligases play a vital role in post transitional modification through ubiquitination-mediated protein degradation that mediate plants responses during abiotic stresses and signal transduction. In this study, we report an Oryza sativa salt induced Really Interesting New Gene (RING) finger protein 2 gene (OsSIRP2) and elucidate its role under salinity and osmotic stress. The transcript levels of OsSIRP2 in rice leaves were induced in response to different abiotic stresses, such as salt, drought, heat, and abscisic acid (ABA) exposure. In vitro ubiquitination revealed that the OsSIRP2 protein formed poly-ubiquitin products, whereas a single amino acid substitution in OsSIRP2 (OsSIRP2C149A) in the RING domain did not form ubiquitinated substrates, supporting the hypothesis that E3 ligase activity requires the functional RING domain. Using the yeast two-hybrid (Y2H) assay, O. sativa transketolase 1 (OsTKL1) was identified as an interacting partner. OsSIRP2 was localized in the nucleus, whereas its interacting partner (OsTKL1) was localized in the cytosol and plastids in the rice protoplasts. Fluorescence signals between OsSIRP2 and OsTKL1 were observed in the cytosol. The pull-down assay confirmed the physical interaction between OsSIRP2 and OsTKL1. In vitro ubiquitination assay and in vitro protein degradation assay revealed that OsSIRP2 ubiquitinates OsTKL1 and enhances the degradation of OsTKL1 through the 26S proteasomal pathway. Heterogeneous overexpression of OsSIRP2 resulted in conferring tolerance against salinity and osmotic stress. Overall, our findings suggest that OsSIRP2 may be associated with plant responses to abiotic stresses and act as a positive regulator of salt and osmotic stress tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Unfavorable conditions, such as drought, extreme temperature (cold, frost, and heat), high salinity, and metalloid toxicity negatively affect seed germination, vegetative growth, and reproductive development of plants, which ultimately reduce crop productivity (Cramer et al. 2011; Pandey et al. 2017). Among these abiotic stresses, salinity is the most significant environmental stressor in terms of reducing crop productivity and quality (Shahbaz and Ashraf 2013). Along with osmotic stress, specific ion stress, nutritional imbalances, or a combination of these factors, salinity adversely affects the physiological pathways, like respiration, photosynthesis, nitrogen fixation, and carbohydrate metabolism during plant growth (Ashraf 2004; Chen et al. 2011). Although many genes induced by salt stress have been studied and identified in the higher plants, the biological functions of these genes are still unknown. Hence, it is necessary to study the biological functions and the mechanisms of salt regulated genes to enhance crop adaptation to the saline environment.
In the soil, salt may be present naturally or introduced via irrigated water, fertilizers, or other agents, like composts and manures, which is a serious threat to the food supply of the increasing human population (Flowers 2004). Approximately more than 20% of the total cultivated and 33% of the irrigated agricultural lands are highly saline. In addition, the saline areas are increasing at a rate of 10% annually owing to various natural and agricultural activities (Jamil et al. 2011). Rice is one of the most important staple crops, as it is consumed by approximately more than half of the world’s population (Cruz et al. 2013). Rice is considered as a highly salt sensitive crop (Conde et al. 2011). Studies showed that the young seedlings of rice were damaged more by salinity, followed by the premature senescence of leaves (Maas and Hoffmann 1977).
Ubiquitin is a stable, highly conserved, and universally expressed protein composed of 76-amino acids that are linked to the lysine residues in target proteins (Herhko et al. 2000). In higher plants, ubiquitin-mediated substrate degradation plays an important role in growth, hormonal signaling, and abiotic stresses; however, the process of ubiquitination is very complex, wherein the enzymes namely E1 (Ub activating enzyme), E2 (Ub conjugating enzyme), and E3-Ub ligases are typically required (Vierstra 2009). Among them, Ub E3 ligases play a vital role in the post translational modification of proteins via binding of ubiquitin to target proteins (Dashaies and Joazeiro 2009). In the ubiquitin modification process mediated by 26S proteasome, the ubiquitin activating enzyme E1 forms a thioester bond between ubiquitin and itself by catalyzing ATP, and binds to E2 through the formation of an isopeptide bond between the carboxy terminus of ubiquitin and the lysine residue in the substrate protein, which is facilitated by an E3 ligase that can bind to both the E2 ubiquitin complex and the substrate (Schulman and Harper 2009; Komander and Rape 2012). There are three major types of ubiquitin ligases, namely cullin based E3 s (contains SCF, APC/C, and VBC complex), Homology to E6-Associated Carboxyl-Terminus (HECT)-type, and RING- finger based E3 s (Mazzucotelli et al. 2006). RING finger protein consists of cysteine (Cys) and histidine (His) residues (in the order Cys-X2-cys-X9-39-Cys-X1-3-His-X2-3-Cys/His-X2-Cys-X4-48-Cys-X2-Cys, where X is any amino acid), which bind to two zinc ligands (Freemont et al. 1991).
RING E3 ligases play an important role in the adaptation to various stress environments via different mechanisms like signal transduction, hormone biosynthesis, or transcriptional factors (Lyzenga and Stone 2012). For example, overexpression of Salt and Drought-Induced Really Interesting New Gene (SDIR1) enhanced the abscisic acid (ABA)-related phenotypes such as salt hypersensitivity and ABA hypersensitivity, and also conferred drought tolerance in Arabidopsis. SDIR1 interacted and ubiquitinated with its substrate protein, SDIR1 interacting protein1 (SDIRP1) that was self-regulated downstream of the transcription factor gene ABA-INSENSITIVE5 to control SDIRP1 stability (Zhang et al. 2007, 2015). In addition, Arabidopsis RING finger E3 ligase AtRUDF1 exhibited positive tolerance to salt and osmotic stress during germination and seedling growth (Li et al. 2013). Similarly, overexpression of tomato RING E3 ligase SpRing enhanced the salt tolerance during germination and early seedling development in Arabidopsis thaliana (Qi et al. 2016). Moreover, Arabidopsis E3 ligases AtAIRP1 and rice RING E3 ligase OsCTR1 act as positive regulators of defense mechanisms induced in response to drought (Ryu et al. 2010; Lim et al. 2013a, b). Similarly, in response to metalloid stress, Rice RING E3 ligase OsHIR1, OsAIR1, and OsAIR2 exhibit a positive role in the adaptation to cadmium- and/or arsenic-stressed environments (Lim et al. 2013a, b; Hwang et al. 2016, 2017). Under saline environment, microtubule associated rice RING E3 ligase OsRMT1 (Lim et al. 2015) and Oryza sativa salt induced RING finger protein (OsSIRP1) act as positive and negative regulators of salt tolerance in transgenic Arabidopsis, respectively (Hwang et al. 2016).
Previously, the expression patterns of 44 O. sativa RING Finger Protein Genes have been evaluated in rice roots under salinity stress (Hwang et al. 2016). In this study, we selected one salinity stress-induced RING finger protein, named O. sativa salt-induced RING Finger Protein 2 (OsSIRP2), and characterized its biological function. In brief, we attempted to determine the subcellular localization of this gene in rice protoplasts and examined the E3 ligase activity via in vitro ubiquitination assay. Subsequently, we identified the physical interaction of one partner protein (O. sativa transketolase 1, OsTKL1) and examined whether OsSIRP2 regulates the expression of the substrate protein OsTKL1 via the 26S proteasome system. Finally, we developed the OsSIRP2-overexpressed Arabidopsis plants and monitored their phenotypic effects under saline and osmotically stressed environments.
Materials and methods
Plant materials
Rice seeds (Oryza sativa ‘Donganbyeo’) were grown in mesh-supported plastic containers with half-strength Murashige and Skoog (MS) nutrient medium. These containers, along with the seeds, were incubated in a growth chamber at 16/8-h light/dark photoperiod at 25 °C with 70% relative humidity. To study the expression pattern, 2-weeks-old seedlings were treated with different abiotic stresses including salinity (200 mM), drought, heat (45 °C), and ABA, and sampled at different time points. For salinity treatment, the 2-weeks-old seedlings were transferred to fresh nutrient solution containing 200 mM NaCl, and the root tissues were sampled at 1, 6, 12, and 24 h post-treatment. For the drought-stress treatment, the 2-weeks-old seedlings were positioned without water and sampled at different time points, such as at 0, 6, 12, and 24 h. Likewise, heat stress treatment was conducted by incubating the 2-weeks-old seedlings at 45 °C and then sampling at 0, 6, 12, and 24 h. The 2 week-old seedlings were treated with 0.1 mM ABA for the hormonal treatment, and then sampled at 0, 6, 12, and 24 h. The collected samples were frozen in liquid nitrogen and kept immediately at − 80 °C until use.
Gene expression analysis
To study the expression patterns of genes, total RNA was extracted from each harvested sample using TRIzol® according to the manufacturer’s instructions (Invitrogen Life Technologies, Carlsbad, CA, USA), and the corresponding cDNA was synthesized from 2 μL of the total RNA using a cDNA synthesis kit (Takara-Bio, Otsu, Japan). After diluting the synthesized cDNA samples (30 μL), 1 μL cDNA was used as a template for quantitative real-time polymerase chain reaction (qRT-PCR). The primers for qRT-PCR were designed using the NCBI primer BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The list of all the primers used in this study is shown in Table S1. qRT-PCR was performed using the CFX Connect™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) and TOPreal™ qPCR 2× PreMIX with SYBR Green (Enzynomics, Daejeon, South Korea) in accordance with the manufacturer’s protocol. The qRT-PCR program was run as follows: denaturation at 95 °C for 10 min, followed by 50 cycles of denaturation at 95 °C for 10 s, annealing at 58 °C for 30 s, and extension at 72 °C for 30 s, with a final extension at 95 °C for 10 s and at 72 °C for 5 s. OsActinII (Os03g50885) was used as an internal control (Park et al. 2014) and the relative expression of the genes was calculated using the 2−ΔΔCT algorithm (Livak and Schmittgen 2001). The statistical significance of the gene expression level of the treated sample was calculated using the two-tailed Student’s t test in comparison to control.
Ubiquitination assay
To generate maltose binding protein (MBP)-fused OsSIRP2, the synthesized rice cDNA was amplified using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA, USA) with the gene specific primers (Table S1), followed by cloning of the PCR product into pMAL-c5X vector containing the restriction sites NdeI and BamHI (New England Biolabs, Ipswich, MA, USA), and transformed into the E. coli strain (DH5α). To generate the point mutation in the RING domain, MBP-OsSIRP2 plasmid DNA was used as the DNA template and amplified using mutagenic primers (Table S2). Then, the dsDNA of MBP-OsSIRP2C149A was transformed into the Escherichia coli strain (DH5α) to repair the nicks created in the mutated plasmid DNA. The generated mutation in the recombinant MBP-OsSIP2 C149A DNA was then confirmed by sequencing. Then, the recombinant MBP-OsSIRP2, point mutated MBP-OsSIRP2 C149A, and non-recombinant MBP (negative control) proteins were transformed and expressed in the E. coli strain BL21 (DE3) pLysS (Promega, Madison, WI, USA), respectively. The recombinant proteins were subsequently purified using affinity chromatography in an amylose resin (New England Biolabs). Besides this, the AtUBC10 fragment was cloned into the pET-28a (+) vector (Novagen, Gibbstown, NJ, USA) with a 6 × His tag, which was purified using the Ni–NTA Purification System (Invitrogen Life Technologies).
An in vitro ubiquitination assay of OsSIRP2 protein was performed as described by Park et al. (2015) with some modifications. The following reaction components were used: 50 ng yeast (E1) (Boston Biochemicals, Cambridge, MA, USA), 200 ng of purified AtUBC10 (E2), 300 ng of purified MBP-OsSIRP2 (E3), 10 µg of bovine ubiquitin (Sigma-Aldrich, St. Louis, MO, USA), and non-recombinant MBP (negative control) along with 20× reaction buffer (1 M Tris, pH 7.5; 40 mM ATP; 100 mM MgCl2; and 40 mM DTT). Corresponding reaction mixtures were prepared and incubated for 3 h at 30 °C Then, 6× sodium dodecyl sulfate (SDS) sample buffer was mixed with each of the samples (20 µL) and boiled at 100 °C for 5 min. The boiled samples were separated using SDS-PAGE and transferred to a nitrocellulose membrane (DoGen, Seoul, Korea). Immunoblot analysis was performed using anti-ubiquitin (Sigma-Aldrich) or anti-MBP (Sigma-Aldrich) as primary antibodies and goat anti-rabbit IgG peroxidase secondary antibody (Sigma-Aldrich) to detect the poly-ubiquitinated products. The horseradish peroxidase (HRP) was conjugated to the labeled antibodies and was detected using the WESTSAVE STAR™ substrate (AB Frontier, Seoul, Korea); finally, the images were captured using the ChemiDoc™ XRS+ imaging system (Bio-Rad).
To confirm the OsSIRP2-mediated ubiquitination of OsTKL1, an OsTKL1-His-Trx fusion protein was affinity-purified using the Ni–NTA purification system (Invitrogen Life Technologies). The purified protein was incubated in the presence/absence of E1, MBP-OsSIRP2, and MBP-OsSIRP2C149A in the ubiquitination mixture for 3 h at 30 °C. The proteins were immunoblotted and the signals were detected using an anti-Trx antibody (Sigma-Aldrich).
Protoplast transfection
Rice seedlings were grown on half-strength MS nutrient solution in a growth chamber at 16/8-h light/dark photoperiod at 25/23 °C with 70% relative humidity for 14 days. Leaves of 2-weeks-old seedlings were harvested, chopped, and immersed in an enzyme solution [1% cellulase R-10 (Yakult Honsha Co. Ltd, Tokyo, Japan), 0.25% macerozyme R-10 (Yakult Honsa), 0.1% BSA, 10 mM Mes, 500 mM Mannitol, and 1 mM CaCl2] for 6 h with slow shaking (Zhang et al. 2011). The enzyme solution with rice leaf samples was then filtered through a cell container and the protoplast pellets were collected by centrifugation. The harvested pellet was then suspended in W5 solution (154 mM NaCl, 5 mM glucose, 5 mM KCl, 125 mM CaCl2, and 1.5 mM Mes) and kept on ice for 3 h. This solution was again resuspended in MaMg solution (0.4 M mannitol, 15 mM MgCl2, and 4.7 mM Mes) (Cho et al. 2014). Plasmid DNA (15 µg) of the related vectors (35S:EYFP, 35S:OsSIRP2-EYFP, and 35S-OsTKL1-EYFP) were separately mixed with the protoplast solution and transfected in 40% polyethylene glycol (PEG) solution [40% PEG, 400 mM mannitol, 100 mM Ca (NO3)2] for 30 min at room temperature. For the final resuspension, the PEG solution was diluted in W5 solution, followed by resuspension of the protoplasts and incubation overnight at room temperature.
Confocal microscopy and imaging
To detect the subcellular localization, the complete OsSIRP2 or OsTKL1 gene was amplified using Pfu Turbo DNA polymerase (Stratagene) with gene specific primers. Then, the PCR products were cloned into the multiple cloning sites of the EYFP vector. In addition, full-length OsSIRP2 and OsTKL1 genes were cloned into 35S:HA-SPYCE(M) and 35S:c-Myc-SPYNE(R) vectors for the bimolecular florescence complementation (BiFC) assay, respectively. For BiFC assay, equal amounts of both vectors were mixed and transfected into rice protoplasts for transient expression. After 16 h, the transfected protoplasts were observed by confocal microscopy. The fluorescent images were obtained using the multiphoton confocal laser scanning confocal microscope (model LSM 510 META; Carl Zeiss, Jena, Germany) at the Korean Basic Science Institute Chuncheon Center.
Yeast two-hybrid screening and interaction assay
A rice cDNA library was constructed using the pGADT7-AD vector from the salt-treated 14-days-old seedlings. The complete coding sequence of OsSIRP2 was amplified using Pfu Turbo DNA polymerase with gene specific primers and fused into the pGADT7-BD vector. Then, a library screening was performed according to the commercial producers (Make Your Own ‘Mate & Plate™’ Library System; Matchmaker™ Gold Yeast Two-Hybrid System; Yeastmaker™ Yeast Transformation System 2, Clontech; Palo Alto, CA, USA) to identify the interacting proteins with the OsSIRP2 protein. At first, positive interactions were selected on double dropout media lacking leucine and tryptophan (DDO/X/A), supplemented with 40 μg/mL X-α-gal and 42 ng/mL aureobasidin A (AbA). The blue colonies selected from (DDO/X/A) screening were patched out into higher stringency QDO/X/A, which lacked adenine, histidine, leucine, and tryptophan, supplemented with 40 μg/mL X-α-gal and 42 ng/mL AbA. Finally, the positive interactions were selected as the interacting partner of OsSIRP2, which was sequenced and confirmed using the Institute for Genomic Research (TRGR) rice annotation database.
To perform the interaction assay, we cloned the complete coding region of OsTKL1 into pGADT7-AD vector and co-transformed with pGABT7-OsSIRP2 into the Y2H Gold strain vector that was grown on DDO medium for 5 days at 30 °C. When the yeast cells grew larger than 3 mm, the co-transformed cell was cultured in DDO liquid medium and adjusted to OD600 > 1 with sterilized water, and 10 μL of the yeast cells were spotted onto DDO and QDO/X/A media.
Pull-down assay
To observe the interaction between the OsSIRP2 and OsTKL1 proteins, MBP-tagged OsSIRP2 and His-tagged OsTKL1 proteins were expressed in the E. coli strain BL21. Bacterial lysate containing MBP-OsSIRP2 was incubated with amylose resin (New England Biolabs). After incubation, the lysates containing MBP- OsSIRP2 was mixed either containing the OsTKL1-His fusion protein or without the fusion protein and incubated at 4 °C for 2 h on a rotary shaker. The reaction was halted by the addition of 5× SDS sample buffer followed by boiling at 95 °C for 5 min. The proteins were separated by 7.5% SDS-PAGE and transferred to a nitrocellulose membrane. Immunoblot signals were detected using anti-MBP and anti-His antibodies (Sigma-Aldrich).
In vitro protein degradation assay
In vitro protein degradation assay was performed according to Hwang et al. (2017). The complete OsTKL1 gene was cloned into a 35S:8cMyc vector. The constructed vector was transformed to Agrobacterium strain GV3101. Then, the Agrobacterium strain harboring the recombinant construct was transfected into 3-weeks-old Nicotiana benthamiana leaves. The transfected leaves were harvested after 3 days, incubated with protease inhibitor cocktail solution (Sigma-Aldrich), and the total protein was extracted using the plant total protein extraction kit (Sigma-Aldrich). Then, each of the expressed N. benthamiana proteins were mixed with MBP-tagged OsSIRP2 protein in 1:1 volume ratio. MG132 (26S proteasome inhibitor) was added into the corresponding samples at a final concentration of 50 μM, and the reactions were stopped by adding 5× sample buffer at different time intervals between 0 and 3 h. Finally, 15 µL of each reaction was immunoblotted using anti-cMYC and anti-MBP antibodies.
Arabidopsis transformation and stress treatment
The 35S-OsSIRP2-EYFP constructs were inserted into the Agrobacterium strain GV3101 and transformed into Arabidopsis (Col-0) using the floral-dip method (Clough 2005) with some modifications. For selection of transgenic lines, the seeds of the transformed plants (T3) were harvested and selected on half-strength MS medium with 50 mg/L kanamycin. Different phenotypic observation assays were conducted to observe the phenotypic effect between the control and transgenic plants under salt and mannitol stress. For germination assay, seeds of 35S-EYFP and 35S-OsSIRP2-EYFP were grown on the half-strength MS medium containing different salt (0, 100, 150, and 200 mM) and mannitol (0, 100, 200, and 300 mM) concentrations, and was monitored every day up to 1 week, followed by calculating the percentage of germination rate. For root length assay, seeds of 35S-EYFP and 35S-OsSIRP2-EYFP were grown on the MS medium of different concentrations as described above and the germination rate was recorded for 2 weeks. Root length was measured after 2 weeks and relevant images were captured. All phenotypic assays were conducted in triplicates for confirmation, and the data obtained were analyzed using the two-tailed Student’s t test.
Results
Expression analysis of OsSIRP2 under different abiotic stress conditions
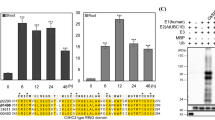
Previously, we randomly selected 44 RING finger proteins with nuclear localization sequence (NLS) and examined the expression patterns under high salinity stress (Hwang et al. 2016). Among them, one gene (Os03g58390, named OsSIRP2) was significantly induced in the 200 mM NaCl-treated rice root samples compared with the non-treated samples. To confirm this, mRNA levels of OsSIRP2 were examined in both the untreated and 200 mM salt-treated rice roots at specific time intervals (1, 6, 12, and 24 h) using qRT-PCR (Fig. 1a). We found that the expression of OsSIRP2 was quite induced in the non-treated samples; however, the expression level was significantly increased in the salt-treated samples compared with that in the non-treated samples at each time points. For example, the transcript levels in the non-treated samples were found to follow the pattern 1 h (onefold), 6 h (5.3-fold), 12 h (8.86-fold), and 24 h (9.3-fold), while in the salt-treated samples, OsSIRP2 expression was found to follow the pattern 1 h (7.06-fold), 6 h (18.97-fold), 12 h (33.39-fold), and 24 h (36.89-fold).
qRT-PCR of OsSIRP2 (LOC_Os03g58390) under abiotic stresses and ABA treated rice roots and/or leaves. a Expression pattern of OsSIRP2 in rice roots under salinity stress condition using qRT-PCR. b qRT-PCR analysis of OsSIRP2 in rice leaves response to different abiotic stressors [salt (200 mM NaCl), drought, heat (45 °C) and ABA (0.1 mM)] and qRT-PCR analysis of a specific expression marker of each abiotic stress: OsNAC10 for salt, OsSalT for drought and ABA and, OsHSP90-1 for heat (heat 45 °C). Relative levels of gene expression were presented as mean ± SD of triplicates as compared to non-treated samples in each time points, Gene expression level of OsSIRP2 was determined by qRT-PCR with OsACTINII levels as an internal control. Asterisks represent the statistically significant difference according to two-tailed Student’s t test; *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001, respectively
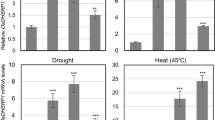
Subsequently, the transcript levels of rice leaves were examined under the three different abiotic stress treatments, including salt (200 mM NaCl), drought, heat (45 °C), and ABA (0.1 mM) at specific time intervals (0, 6, 12 and 24 h) (Fig. 1b). We found that the transcript level of OsSIRP2 was significantly increased in the rice leave tissues that were exposed to NaCl, drought, heat, and ABA treatments. For example, upon treatment with 200 mM NaCl, the transcript levels of OsSIRP2 were significantly increased after 6 h (3.9-fold) and 12 h (4.0-fold); however, the expression decreased to 2.13-fold after 24 h. Similarly, the OsSIRP2 mRNA expression level was consistently increased after 6 h (5.3-fold) and 12 h (5.6-fold) under drought stress; however, it slightly decreased, to 3.8-fold, after 24 h. In addition, the transcript levels of OsSIRP2 under heat (45 °C) stress was significantly increased to 27.09-fold after 24 h. Moreover, OsSIRP2 transcript level was highly induced up to 27 folds on ABA treatment; the mRNA expressions were highly enhanced at 6 h (7.2-fold) and then slightly decreased at 24 h. We analyzed the transcript levels of the stress-induced genes to examine the suitability of different stress treatments in rice leaves. For example, OsNAC10 for salt treatment (Jeong et al. 2010), OsHSP90-1 for heat (Hu et al. 2009), and OsSalT for drought and ABA treatments (Chen et al. 2015) were employed. Transcripts levels of OsNAC10, OsHSP90-1, and OsSaLT were significantly increased from 6 to 24 h compared to in untreated plants (0 h). Collectively, these results suggested that OsSIRP2 is induced under abiotic stresses, particularly, under salinity stress condition.
OsSIRP2 localizes in nucleus
To examine the subcellular localization, we assessed the transient expression of OsSIRP2 in the rice protoplasts using 35S:EYFP-fused OsSIRP2. We found that the 35S:EYFP-OsSIRP2 signal was distinctly located in the nucleus (Fig. 2); however, 35S-EYFP florescence was also observed in the cytosol and in other portions of the cell after DAPI staining. Previously, we found that the subcellular localization of rice RING proteins transferred their position to other organelles during abiotic stress treatment (Lim et al. 2013a, 2015). Hence, we raised the question whether the nucleus associated with OsSIRP2 could alter the position after salinity stress? Therefore, we examined the subcellular localization by treating 35S:EYFP-OsSIRP2 transfected rice protoplasts with addition of 200 mM NaCl. However, we did not observe any alternation in the localization of 35S:OsSIRP2-EYFP in the rice protoplast after salt treatment (Fig. 2, last panel).
OsSIRP2 possesses E3 ligase activity
It is suggested that the proteins with RING-HC domain possess E3 ligase activity (Stone et al. 2005). OsSIRP2, encoding a 385 amino acid protein, is predicted to have a molecular mass of 38.87 kDa, and contains the RING-HC domain at the C-terminal region. To further examine the domain structure, we retrieved and aligned putative RING-HC domains of other Poaceae orthologues, such as Sorghum bicolor (Sb01g005090), Zea mays (GRMZM2G4575544), and Brachypodium distachyon (Bradi1g05390), from the TIGR rice annotation database, which indicated that each of the RING-HC domains of the corresponding orthologues were highly conserved with the OsSIRP2 protein (Fig. 3a).
Analysis of RING domain in OsSIRP2 and ubiquitination asay. a RING domain analysis of OsSIRP2 with its ortholog genes. b Different time course reactions (0, 30, 60, 120 min) of MBP-OsSIRP2 and MBP-OsSIRP2C149A incubated in the presence of E1 (Yeast), E2 (Arabidopsis UBC 10) and E3 (OsSIRP2 and MBP-OsSIRP2C149A) and Ub. Different primary antibodies with anti-ub and anti-MBP were used to detect OsSIRP2 poly-ubiquitin chain. c In vitro ubiquitination assay of OsSIRP2 and MBPOsSIRP2C149A as point mutant by changing single amino acid. The ubiquitination assay was performed in the presence (+) and/or absence (−) of E1 (Yeast), E2 (Arabidopsis UBC 10) and E3 (OsSIRP2 and MBPOsSIRP2 C149A). Asterisk indicates the non-specific band
To identify the E3 ligase activity of OsSIRP2 with RING-HC domain, each of the MBP-tagged OsSIRP2 and the single amino acid mutated OsSIRP2C149A recombinants were constructed and incubated with reaction components (human E1, AtUBC10, and bovine ubiquitin). Immunoblot analysis showed that a high molecular mass of the poly-ubiquitin chain of MBP-OsSIRP2 was detected when it was incubated with all the reaction components (Fig. 3c, lane 4). However, no poly-ubiquitin chains were detected in the absence of E1 or AtUBC10 (Fig. 3c, lanes 2 and 3). Similarly, no poly-ubiquitin chain was detected in the presence of only MBP and OsSIRP2C149A (Fig. 4c, lanes 1 and 5). At different time intervals, in vitro ubiquitination assay revealed that the poly-ubiquitin chains in the MBP-OsSIRP2 gradually reached its highest level after 2 h of incubation (Fig. 4b, left). However, no poly-ubiquitin bands were detected in the reaction components incubated with mutated OsSIRP2C149A in both anti-Ub and anti-MBP immunoblot assays (Fig. 3b, right). These results proved that OsSIRP2 harboring a RING-HC domain acted as an E3 ubiquitin ligase.
Identification of interacting protein of OsSIRP2, their interaction, subcellular localization and gene expression analysis. a Yeast Two hybrid assay. A full length OsSIRP2 was cloned with pGBKT7, and full-length OsTKL1 was cloned into pGADT7 and each of the positive clones was transformed into Yeast Gold strain. Cells were dropped onto DDO and QDO/X/A medium. BD-murine p53 with AD-sv40 large T-antigen was used as positive control (PC) and BD-Lamin with AD-SV 40 large T-antigen as negative control (NC). b Full length sequence of OsTKL1 was fused into EYFP vector and transfected into rice protoplast, empty EYFP was taken as positive Control. c Relative expression levels of the OsTKL1 gene in rice leaves exposed to salt and drought stress as compared to controls (0 h), asterisks represent the statistically significant difference according to two-tailed Student’s t test; *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001, respectively
OsTKL1: protein interacting partner of OsSIRP2
Many studies reported that E3 ubiquitin ligases interacted with their substrates during 26S proteasome degradation (Jacobson et al. 2009; Vierstra 2009). To identify the substrate proteins of OsSIRP2, we conducted Y2H screening from salt stressed rice cDNA libraries. After Y2H screening, one interacting protein with OsSIRP2, Os05g33840 (transketolase, OsTKL1), was found based on its α-galactosidase activity (Supplemental Fig. S1). To confirm the interactions, we separately spotted OsSIRP2-pGBKT7 and OsTKL1-pGADT7 co-transformed yeast cells onto DDO and QDO/X/A medium. We detected strong physical interaction of OsSIRP2 with OsTKL1 in the QDO/X/A medium (Fig. 4a). To confirm the subcellular localization of the interacting protein, the EYFP tagged OsTKL1 was transfected in the rice protoplasts. We observed that OsTKL1 was localized in either the cytoplasm or the plastid (Fig. 4b). Moreover, the expression level of OsTKL1 was examined in salt- and drought-stressed rice leaves at different time intervals (0, 6, 12, and 24 h; Fig. 4c). Under salt stress, the mRNA expression of OsTKL1 reached the highest level (5.46-fold) after 12 h of stress treatment, however, relatively low expression was observed in the samples subjected to 24-h stress. In addition, the gene exhibited the highest expression (74.33-fold) in the 24 h drought-stressed samples. Overall, the expression pattern of OsTKL1 is also up-regulated in stress conditions, indicating the functional role of OsTKL1in abiotic stress response mechanisms.
To verify the physical interaction between the OsSIRP2 and OsTKL1 proteins, we employed the BiFC assay by generating different complementary vectors; 35S:OsSIRP2-SPYCE (M) and 35S:OsTKL1-SPYNE(R). Interestingly, the fluorescent signal of OsSIRP2/OsTKL1 complex was distinctly detected in the cytoplasm (Fig. 5a). To further examine the dimeric-complex of OsSIRP2 and OsTKL1, we performed the pull-down assay, where the MBP-OsSIRP2 protein was used as the bait and the His-tagged OsTKL1 protein as the substrate. In vitro pull-down assay clearly showed that the His-tagged OsTKL1 protein binds with OsSIRP2, whereas no other interaction was observed when pulled down with the empty-MBP protein (Fig. 5b). These results indicate that the transketolase protein OsTKL1 is one of the substrates of OsSIRP2 E3 ligase.
Interaction between OsSIRP2 and its partner protein OsTKL1. a BiFC assay of interacting protein (OsTKL1) with OsSIRP2. Full length OsTKL1 and OsSIRP2 were cloned into pSPYCE(R) and pSPYCE(M), respectively. Each constructs were transfected equally into rice protoplast and images were taken 16 h after transfection. Each of their counterpart empty vectors [Empty pSPYCE(R) and pSPYCE(M)] were used as negative controls. b In vitro pull-down assay. Full length sequences of OsSIRP2 and OsTKL1 were fused into MBP and His-tags respectively. Bacterial lysates containing MBP- OsSIRP2 was mixed with or without OsTKL1-His fusion protein and incubated at 4 °C for 2 h and MBP-OsSIRP2 was used as bait and His-OsTKL1 was used as bait protein. Immunoblot was detected by MBP and His antibody
OsSIRP2 ubiquitinates OsTKL1 and regulates protein levels via 26S proteasome
We performed further experiments to address the hypothesis that OsSIRP2 Ub E3 ligase ubiquitinates its interacting protein OsTKL1, and is subjected to proteolytic degradation in the cytoplasm via the 26S proteasome system. First, we performed the in vitro ubiquitination using each of the MBP-tagged OsSIRP2 and OsSIRP2C149A recombinant proteins. Each of the two proteins were incubated with or without the reaction components, including the His-Trx-tagged OsTKL1 protein, for 3 h. Results showed that the poly-ubiquitin chain was detected only when MBP-OsSIRP2 and His-OsTKL1 were incubated with both E1 and AtUBC10 (Fig. 6a, lane 4). However, no higher molecular band was detected in the presence of the point mutated OsSIRP2C149A and His-OsTKL1 proteins (Fig. 6a, lane 5). These results support that OsSIRP2 E3 ligase directly ubiquitinates the OsTKL1 protein using it as a substrate. Second, we constructed the OsTKL1-cMyc recombinant vector and transformed it into the tobacco leaves to test whether OsSIRP2 promotes OsTKL1 degradation via the 26S proteasome system. After the extraction of total proteins, we performed in vitro protein degradation assay and observed the protein levels of OsTKL1 in the presence and absence of MBP-OsSIRP2 protein at various time points (0, 1, and 2 h, Fig. S2). We found a clear reduction in the OsTKL1 protein level after 2 h (Fig. S2). In addition, we incubated OsSIRP2-MPB and OsTKL1-cMyc for 3 h in the presence or absence of MG132 (proteasome inhibitor), and found a dramatic reduction in the protein level of OsTKL1 when incubated without MG132; however, degradation of the substrate protein OsTKL1 did not occur in the MG132 treated reaction (Fig. 6b). These results clearly showed that the OsTKL1 protein is ubiquitinated by OsSIRP2 E3 ligase, and then leads to the proteolysis of the OsTKL1 protein via the 26S proteasome degradation pathway.
Identification of OsTKL1 protein regulation by OsSIRP2 via in vitro degradation assays. a OsSIRP2 mediates the ubiquitination of OsTKL1 protein. The full length of OsTKL1 was fussed with HiS-tags and used as substrate for ubiquitination assay, point mutation MBP-OsSIRP2C149A lane was used as negative control. Immunoblot was detected using His–antibody. b An in vitro protein degradation assay of the OsTKL1 protein. The OsTKL1-8cMyc was infiltrated into N. benthamiana leaves. The extracted OsTKL1-8cMyc was mixed with MBP-OsSIRP2 protein or empty-MBP protein and incubated at 4 °C for 3 h. The anti-MBP antibody and anti-cMyc antibody were used for immunoblotting analysis, respectively. MG132 served as an inhibitor protein to prevent the 26S proteasome pathway. Ponceau S staining of the Rubisco proteins is shown as a loading control
Overexpression of OsSIRP2 confers tolerance to salinity and osmotic stress
To evaluate the molecular function of OsSIRP2 under saline conditions, we generated the OsSIRP2-overexpressing Arabidopsis plants (Fig. 7). We performed the RT-PCR and selected three transgenic plants (#1, #2, and #4) that exhibited high OsSIRP2 gene expression (Fig. 7a). To examine the germination rates of OsSIRP2-overexpressed plants during salinity stress, 35S-EYFP and three independent transgenic plants were treated with different salt concentrations (0, 100, 150, and 200 mM NaCl). We found that both the control and transgenic plants showed no difference in the germination rate without treatment (0 mM NaCl). However, the germination rate of the transgenic plants was significantly higher than that of 35S-EYFP, when treated with 100 and 150 mM salt concentrations. Interestingly, no significant difference in the germination rate was found on treatment with 200 mM salt concentration compared to 35S-EYFP (Fig. 7b, c).
Salt-responsive phenotypes of OsSIRP2-overexpressing Arabidopsis plants under different salinity stress conditions (0, 100, 150 and 200 mM). a RT-PCR analysis of three independent 35S:OsSIRP2-EYFP T3 transgenic plants (lines #1, #2 and #4) and control plant (35S: EYFP). b, c Germination rate of control and each of transgenic lines on half-strength MS medium without NaCl and increasing NaCl concentrations. d, e Root length of three independent 35S:OsSIRP2-EYFP overexpressed plants (#1, #2, #4) and control plant (35S: EYFP). Germination percentage was evaluated after 7 days and root length was measured from 14-days-old seedling. Germination and root length data are presented as mean ± standard deviation (SD) of three independent experiments, where n = 5 for root length and n = 25 for germination rate. Asterisks represent a statistically significant difference according to two-tailed Student’s t test; *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001, respectively
Subsequently, we measured the primary root length of both 35S-EYFP and transgenic seedlings under salt stress (0, 100, 150, and 200 mM NaCl). There was no significant difference in the root length under control medium; however, OsSIRP2 overexpressing plants showed longer root length in comparison to the 35S-EYFP plants under saline condition (Fig. 8d, e). These results suggested that the OsSIRP2-overexpressing plants have a strong capacity to tolerate saline stress conditions.
Phenotypic evaluation of OsSIRP2-overexpressing plants under mannitol treatment. a Germination rates of control and three independent OsSIRP2-overexpressing plants. b, c Root length of three independent 35S:OsSIRP2-EYFP overexpressed plants (#1, #2, #4) and control plant (35S: EYFP). Plants were grown on the MS medium containing 0, 100, 200 and 300 mM mannitol for 7 days for germination and root length was measured from 14-days-old seedling and number of plants taken for each experiment are n = 5 for root length and n = 25 for germination rate. Asterisks represent a statistically significant difference according to two-tailed Student’s t test; *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001, respectively
The expression of OsSIRP2 under drought stress suggests the hypothesis on the positive functions of the gene during osmotic stress. Therefore, to examine its effect on osmotic stress (non-ionic stress), OsSIRP2, 35S-EYFP and three independent transgenic plants were treated with control and 100, 200, and 300 mM of mannitol (Fig. 8). We first evaluated the germination rates until 7 days after sowing, and found no difference in the germination rate without treatment. Interestingly, we observed rapid germination rates in the transgenic plants compared to 35S-EYFP under increasing mannitol concentrations. All transgenic plants showed 100% germination rates after 6 days in comparison to the 35S-EYFP plants under 200 mM mannitol treatment (Fig. 8a).
Subsequently, we measured the primary root length of both 35S-EYFP and transgenic seedlings at different concentrations of mannitol (0, 100, 200, 300 mM) after 14 days. The results showed no significant change in both 35S-EYFP and OsSIRP2 overexpressing plants under control condition. However, OsSIRP2 transgenic plants showed significantly higher root length compared to normal plants under mannitol stressed media (Fig. 8b, c), suggesting that OsSIRP2-overexpressing transgenic plants could confer tolerance against non-ionic/osmotic stress conditions.
Discussion
Soil salinization is becoming one of the enormous problems for crop productivity. To survive under abiotic stress conditions, including salinity, plants have evolved complex mechanisms, including signal transduction, stress perception, synthesizing stress related proteins, and transcriptional activation of stress responsive target genes, to receive external signals that mediates plant responses to the changing environment. These mechanisms facilitate plants to cope with negative environmental conditions through different physiological and biochemical manifestations. To maintain the crop productivity of saline soils, development of salt-tolerant crop varieties is the most important strategy. Therefore, our knowledge regarding the molecular functions during plant responses to salinity has to be improved substantially.
Previously, we have demonstrated the roles of several RING E3 ligases through ubiquitin-mediated protein degradation under various abiotic stress conditions (Lim et al. 2014, 2015; Park et al. 2015). In this study, we provide insights on those molecular functions of the OsSIRP2 gene encoding 385 amino acid proteins containing the RING-H2 domain. In vitro ubiquitination assay showed that the MBP-OsSIRP2 protein catalyzed the poly-ubiquitin products in the presence of E1 and E2; however, no reaction was observed by the point mutated MBP-OsSIRP2 protein. This result is consistent with previous findings, which have reported that RING-H2 domain containing proteins possess E3 ligase activity (Stone et al. 2005; Lim et al. 2014; Hwang et al. 2016). Many studies have revealed that the E3 ligase activity of the RING domain contains proteins that depend on the presence of cysteine or histidine residues involved in zinc coordination. Therefore, the generation of point mutation phenotypes could provide strong evidence on the dependence of conserved motifs of the RING domain in zinc coordination for the E3 ligase activity. For example, a single amino acid change C225A in the salt induced RING E3 ligase (OsSIRP1), OsSIRP1C225A led to the loss of protein ubiquitination (Hwang et al. 2016).
The OsSIRP2 transcripts were highly up-regulated in both the root and leaf tissues under salt stress. In addition, the OsSIRP2 transcripts were significantly increased under drought, heat and ABA treatment (Fig. 1), suggesting that the biological functions of OsSIRP2 might be related to salinity and drought in the ABA-dependent manner. However, further studies are required to determine the ABA signaling stress tolerance mechanism. The results of Y2H, BiFC, and pull-down assay clearly revealed that OsTKL1 is a protein interaction partner of OsSIRP2. Plant transketolases catalyze the reactions in the Calvin cycle of photosynthesis, and are universally required for the oxidative pentose phosphate pathway, which produces NADPH (Henkes et al. 2001). Subcellular localizations of transketolases differ within plant species, either in the plastid or the cytoplasm. For example, each transketolase is localized in the cytoplasm of Craterostigma plantagineum (Willige et al. 2009), in the plastid of Amaranthus Tricolor (Rajeendran et al. 2013), and in the chloroplast of Spinacia oleracea (Flechner et al. 1996). We identified that OsTKL1 protein is localized in either the cytoplasm or the plastid (Fig. 4b), mostly in the cytoplasm of the rice plant. Moreover, BiFC analysis demonstrated that the physical interaction between OsSIRP2 and OsTKL1 proteins occurred in the cytosol, even though the localization of OsSIRP2 is clearly associated with the nucleus. This inconsistency raises a question regarding the occurrence of the physical interactions of both proteins in the cytosol. An attractive hypothesis might be the translocation of the OsSIRP2 protein from the nucleus to the cytosol owing to salinity stress. Lim et al. (2013a, b) demonstrated that the Golgi-localized OsHCI1 protein accumulated in the nucleus owing to heat stress. However, our results showed lack of translocation of OsSIRP2 upon treatment with 200 mM NaCl (Fig. 2), suggesting different regulation mechanisms might be involved in the interaction. Therefore, considerable work still remains to be done to unravel the exact mechanism. Previous studies on the RING finger proteins showed that most of the proteins interacting with RING E3 ligase act as substrates (Lim et al. 2013a, b; Park et al. 2015; Zhang et al. 2015). Similarly, we found that OsSIRP2 ubiquitinates OsTKL1, leading to the decrease in the protein levels of OsTKL1 through the 26S proteasomal degradation pathway (Fig. 6). Overall, these results support the hypothesis that OsTKL1 is a substrate of OsSIRP2 E3 ligase.
Previously, studies demonstrated different roles of transketolases in plant growth, development, and physiological processes under normal and stressed conditions; however, the exact mechanism of transketolases in response to various stress adaptations is not yet fully understood. Henkes et al. (2001) reported that the decreased activity of transketolase in tobacco transformants demonstrated the inhibition of ribulose-1, 5-biphosphate regeneration and photosynthesis. In addition, decreased amount of transketolase was observed in the rice seedlings under salt stress, which further decreased the photosynthetic activity (Kim et al. 2005). Similar role of transketolase in animals and fungi was identified in plants, which play an important role in the stress-induced production of cytosolic NADPH (Tunc-Ozdemir et al. 2009). Such NADPH oxidases are considered as one of the major enzyme systems involved in the production of reactive oxygen species (ROS) in the apoplastic space under stress conditions (Srivastava et al. 2015). Furthermore, NADPH oxidases transport different ROS generating enzyme systems owing to their role in various signaling pathways involved in plant growth, development, and stress tolerance (Kaur et al. 2014). For example, the activity of transketolase is moderately increased under salt and oxidative stress conditions in Zea mays (Rapala-Kozik et al. 2008). In vitro studies of S. oleracea transketolase have revealed its alternative stress-protecting role through its ability to quench superoxide radicles (Kaiser 1976; Takabe et al. 1980). The production of ROS, such as superoxide, single oxygen, hydrogen peroxide, and hydroxyl radical is enhanced in response to salinity stress (Apel and Hirt 2004). There might be some inconsistency in our both findings: (1) the protein level of OsTKL1 is down-regulated by OsSIRP2 and (2) OsSIRP2-overexpressing plants confer tolerance to salinity and osmotic stress. These findings might be explained by proteostasis (Singh et al. 2015) to maintain the activity of OsTLK1 under abiotic stress, and thus confer tolerance to the OsSIRP2-overexpressing plants, although more work needs to be done to arrive at a robust conclusion. Similar findings have been reported in a previous study, wherein microtubule associated rice Ring finger protein 1 (OsRMT1) interacted with different proteins such as OsSalT, OsFKBP12, OsCPA1, and OsDH1, and degraded via the 26S proteasome pathway. However, the relative expression levels of its substrate genes were up-regulated during salt stress and OsRMT1-overexpressed Arabidopsis plants conferred positive tolerance to salinity stress (Lim et al. 2015). Similarly, interacting partner of arsenic induced E3 ligase OsAIR2 was induced upon arsenate exposure, followed by the degradation of this substrate (OsKAT1) by OsAIR2 via the 26S proteasome pathway (Hwang et al. 2017). Collectively, our findings clearly suggest that OsSIRP2 plays an important role in coping with salinity and osmotic stress through the ubiquitin-mediated target protein degradation, i.e., OsTKL1.
In conclusion, we revealed the expression patterns, subcellular localization, and the E3 ligase activity of OsSIRP2 and identified its partner protein OsTKL1. Furthermore, we performed the BiFC assay, pull-down assay, in vitro ubiquitination of OsTKL1, and in vitro protein degradation of OsTKL1 using OsSIRP2. Finally, the OsSIRP2-overexpressing plants were examined under salinity and osmotic stress, and conferred tolerance against them. Collectively, these results suggest that OsSIRP2 may act as a positive regulator in the plant response to salinity and osmotic stress via the regulation of OsTKL1 substrate protein.
Author contribution statement
SC, YCP, JHK, CSJ designed and performed the experiments. Wrote the manuscript: SC, YCP, CSJ and all authors read and approved the manuscript.
Abbreviations
- BiFC:
-
Bimolecular fluorescence complementation
- OsSIRP2 :
-
Oryza sativa salt induced Ring finger protein 2
- RING:
-
Really Interesting New Gene
- Y2H:
-
Yeast two-hybrid
References
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Ashraf M (2004) Some important physiological selection criteria for salt tolerance in plants. Flora 199:361–376
Chen W, Feng C, Guo W, Shi D, Yang C (2011) Comparative effects of osmotic-, salt- and alkali stress on growth, photosynthesis, and osmotic adjustment in cotton plants. Photosynthetica 49:417–425
Chen M, Zhao Y, Zhuo C, Lu S, Guo Z (2015) Overexpression of a NF-YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol J 13:482–491
Cho HY, Lee C, Hwang SG, Park YC, Lim HL, Jang CS (2014) Overexpression of the OsChI1 gene, encoding a putative laccase precursor, increases tolerance to drought and salinity stress in transgenic Arabidopsis. Gene 552:98–105
Clough SJ (2005) Floral dip: Agrobacterium-mediated germ line transformation. Methods Mol Biol 286:91–102
Conde A, Chaves MM, Geros H (2011) Membrane transport, sensing and signaling in plant adaptation to environmental stress. Plant Cell Physiol 52:1583–1602
Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11:163
Cruz RP, Sperotto RA, Cargnelutti D, Adamski JM, Terra TF, Fett JP (2013) Avoiding damage and achieving cold tolerance in rice plants. Food Energ Secur 2:96–119
Dashaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78:399–434
Flechner A, Dressen U, Westhoff P, Henze K, Schnarrenberger C, Martin W (1996) Molecular characterization of transketolase (EC 2.2.1.1) active in the Calvin cycle of spinach chloroplasts. Plant Mol Biol 32:475–484
Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55:307–319
Freemont PS, Hanson IM, Trowsdale J (1991) A novel cysteine-rich sequence motif. Cell 64:483–484
Henkes S, Sonnewald U, Badur R, Flachmann R, Stitt M (2001) A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 13:535–551
Herhko A, Ciechanover A, Varshavsky A (2000) The ubiquitin system. Nat Med 10:1073–1081
Hu WH, Hu GC, Han B (2009) Genome wide survey and expression during profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci 176:583–590
Hwang SG, Kim JJ, Lim SD, Park YC, Moon JC, Jang CS (2016) Molecular dissection of Oryza sativa salt-induced RING Finger Protein 1 (OsSIRP1): possible involvement in the sensitivity response to salinity stress. Physiol Plant 158:168–179
Hwang SG, Chapagain S, Han AR, Park YC, Park HM, Kim YH, Jang CS (2017) Molecular characterization of rice arsenic-induced RING Finger E3 ligase 2 (OsAIR2) and its heterogeneous overexpression in Arabidopsis thalaiana. Physiol Plant 161:372–384
Jacobson AD, Zhang NY, Xu P, Han KJ, Noone S, Peng J, Liu CW (2009) The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome. J Biol Chem 284:35485–35494
Jamil A, Riaz S, Ashraf M, Foolad MR (2011) Gene expression profiling of plants under salt stress. Crit Rev Plant Sci 30:435–458
Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197
Kaiser W (1976) The effect of hydrogen peroxide on CO2 fixation of isolated intact chloroplasts. Biochim Biophys Acta 440:476–482
Kaur G, Sharma A, Guruprasad K, Pati PK (2014) Versatile roles of plant NADPH oxidases and emerging concepts. Biotechnol Adv 32:551–563
Kim DW, Rakwal R, Agrawal GK, Jung YH, Shibato J, Jwa NS, Iwahashi Y, Iwahashi H, Kim DH, Shim IS, Usui K (2005) A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophoresis 26:4521–4539
Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81:203–229
Li J, Han Y, Zhao Q, Li C, Xie Q, Chong K, Xu Y (2013) The E3 ligase AtRDUF1 positively regulates salt stress response in Arabidopsis thaliana. PLoS One 8:e71078
Lim SD, Cho HY, Park YC, Ham DJ, Lee JK, Jang CS (2013a) The rice RING finger E3 ligase, OsHCI1, drives nuclear export of multiple substrate proteins and its heterogeneous overexpression enhances acquired thermotolerance. J Exp Bot 64:2899–2914
Lim SD, Lee CH, Jang CS (2013b) The rice RING E3 ligase, OsCTR1, inhibits trafficking to the chloroplasts of OsCP12 and OsRP1, and its overexpression confers drought tolerance in Arabidopsis. Plant Cell Environ 37:1097–1113
Lim SD, Hwang JG, Han AR, Park YC, Lee C, Ok YS, Jang CS (2014) Positive regulation of rice RING E3 ligase OsHIR1 in arsenic and cadmium uptakes. Plant Mol Biol 85:365–379
Lim SD, Hwang JG, Han AR, Park YC, Lee C, Lim CW, Kim DS, Jang CS (2015) Molecular dissection of a rice microtubule—associated RING finger protein and its potential role in salt tolerance in Arabidopsis. Plant Mol Biol 89:365–384
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
Lyzenga WJ, Stone SL (2012) Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot 63:599–616
Maas EV, Hoffmann GJ (1977) Crop salt tolerance—current assessment. J Irrig Drain Div ASCE 103:115–134
Mazzucotelli E, Belloni S, Marone D, De Leonardis AM, Guerra D, Di Fonzo N, Cattivelli L, Mastrangelo AM (2006) The E3 ubiquitin ligase gene family in plants: regulation by degradation. Curr Genomics 7:509–522
Pandey P, Irulappan V, Bagavathiannan MV, Kumar MS (2017) Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front Plant Sci 8:537
Park S, Moon JC, Park YC, Kim JH, Kim DS, Jang CS (2014) Molecular dissection of the response of a rice leucine-rich repeat receptor-like kinase (LRR-RLK) gene to abiotic stresses. J Plant Physiol 171:1645–1653
Park YC, Kim JJ, Kim DS, Jang CS (2015) Rice RING E3 ligase may negatively regulate gamma-ray response to mediate the degradation of photosynthesis-related proteins. Planta 241:1119–1129
Qi S, Lin Q, Zhu H, Gao F, Zhang W, Hua X (2016) The RING finger E3 ligase SpRing is a positive regulator of salt stress signaling in salt-tolerant wild tomato species. Plant Cell Physiol. 57(3):528–539. https://doi.org/10.1093/pcp/pcw006
Rajeendran A, Nulit R, Kalhori N, Yien CS (2013) Isolation, cloning, and subcellular localization of transketolase from Amaranthus tricolor L. J Biol 1:92–99
Rapala-Kozik M, Kowalska E, Ostrowska K (2008) Modulation of thiamine metabolism in Zea mays seedlings under conditions of abiotic stress. J Exp Bot 59:4133–4143
Ryu MY, Cho SK, Kim WK (2010) The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol 154:1983–1997
Schulman BA, Harper JW (2009) Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signaling pathways. Nat Rev Mol Cell Biol 10:319–331
Shahbaz M, Ashraf M (2013) Improving salinity tolerance in cereals. Crit Rev Plant Sci 32:237–249
Singh S, Parihar P, Singh R, Singh VP, Prasad SM (2015) Heavy metal tolerance in plants: role of transcriptomics, proteomics, metabolomics, and ionomics. Front Plant Sci 6:1143
Srivastava AK, Srivastava S, Lokhande HV, Souza SFD, Suprasanna P (2015) Salt stress reveals differential antioxidant and energetic responses in glycophyte (Brassica juncea L.) and halophyte (Sesuvium portulacastrum L.). Front. Environ Sci 3:19
Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137:13–30
Takabe T, Asami S, Akazawa T (1980) Glycolate formation catalyzed by spinach leaf transketolase utilizing the superoxide radical. Biochemistry 19:3985–3989
Tunc-Ozdemir M, Miller G, Song L, Kim J, Sodek A, Koussevitzky S, Misra AN, Mittler R, Shintani D (2009) Thiamin confers enhanced tolerance to oxidative stress in Arabidopsis. Plant Physiol 151:421–432
Vierstra RD (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10:385–397
Willige BC, Kutzer M, Tebartz F, Bartels D (2009) Subcellular localization and enzymatic properties of differentially expressed transketolase genes isolated from the desiccation tolerant resurrection plant Craterostigma plantagineum. Planta 222:659–666
Zhang YY, Yang CW, Li Y, Zheng NY, Chen H, Zhao QZ, Gao T, Guo HS, Xie Q (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19:1912–1929
Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30
Zhang H, Cui F, Wu Y, Lou L, Liu L, Tian M, Ning Y, Shu K, Tang S, Xie Q (2015) The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-interacting protein1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27:214–227
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2016R1A2B4015626).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chapagain, S., Park, Y.C., Kim, J. et al. Oryza sativa salt-induced RING E3 ligase 2 (OsSIRP2) acts as a positive regulator of transketolase in plant response to salinity and osmotic stress. Planta 247, 925–939 (2018). https://doi.org/10.1007/s00425-017-2838-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-017-2838-x