Abstract
Main conclusion
A comprehensive understanding of nitrogen signaling cascades involving heterotrimeric G-proteins and their putative receptors can assist in the production of nitrogen-efficient plants.
Abstract
Plants are immobile in nature, so they must endure abiotic stresses including nutrient stress. Plant development and agricultural productivity are frequently constrained by the restricted availability of nitrogen in the soil. Non-legume plants acquire nitrogen from the soil through root membrane-bound transporters. In depleted soil nitrogen conditions, legumes are naturally conditioned to fix atmospheric nitrogen with the aid of nodulation elicited by nitrogen-fixing bacteria. Moreover, apart from the symbiotic nitrogen fixation process, nitrogen uptake from the soil can also be a significant secondary source to satisfy the nitrogen requirements of legumes. Heterotrimeric G-proteins function as molecular switches to help plant cells relay diverse stimuli emanating from external stress conditions. They are comprised of Gα, Gβ and Gγ subunits, which cooperate with several downstream effectors to regulate multiple plant signaling events. In the present review, we concentrate on signaling mechanisms that regulate plant nitrogen nutrition. Our review highlights the potential of heterotrimeric G-proteins, together with their putative receptors, to assist the legume root nodule symbiosis (RNS) cascade, particularly during calcium spiking and nodulation. Additionally, the functions of heterotrimeric G-proteins in nitrogen acquisition by plant roots as well as in improving nitrogen use efficiency (NUE) have also been discussed. Future research oriented towards heterotrimeric G-proteins through genome editing tools can be a game changer in the enhancement of the nitrogen fixation process. This will foster the precise manipulation and production of plants to ensure global food security in an era of climate change by enhancing crop productivity and minimizing reliance on external inputs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
G-proteins also referred to as guanine nucleotide-binding proteins, are grouped structurally into two distinct classes: monomeric G-proteins and heterotrimeric G-proteins (Pandey 2020; Wang and Botella 2022; Ganotra et al. 2023). The heterotrimeric G-proteins (hereafter G-proteins) comprise three different types of subunits: Gα, Gβ and Gγ (Pandey 2019; Wang and Botella 2022). The number of G-protein subunits could vary from plant to plant. For instance, the model plant Arabidopsis thaliana with a simpler genome harbor one canonical Gα (GPA1, At2g26300), three non-canonical extra-large Gα (XLG1, At2g23460; XLG2, At4g34390 and XLG3, At1g31930), one Gβ (AGB1, At4g34460), and three Gγ genes, including two canonical Gγ (AGG1, At3g63420 and AGG2, At3g22942) and one atypical Gγ (AGG3, At5g20635) (Stateczny et al. 2016; Maruta et al. 2021a; Cantos et al. 2023). Brassica napus contains 2 Gα, 6 Gβ and 13 Gγ genes, which contribute to a total of 21 G-protein family members (Xie et al. 2022). M. truncatula is a premier model legume for studies pertaining to symbiotic interactions and nitrogen fixation. A study identified two Gα, three XLG, one Gβ and five Gγ genes in the M. truncatula genome (Mt4.0) (Tang et al. 2014). Another study revealed the presence of two Gα, three XLG, one Gβ and six Gγ genes in the P. sativum genome (Pecrix et al. 2018; Bovin et al. 2022). Plants harboring complex genomes have enlarged networks of G-proteins, such as allotetraploid G. max, which has four Gα, four Gβ and ten Gγ proteins (Choudhury et al. 2011; Bisht et al. 2011).
G-proteins are molecular on–off switches, and in mammalian and yeast systems, the switch characteristic is encoded by nucleotides: a guanosine triphosphate (GTP)-bound on-state and a guanosine diphosphate (GDP)-bound off-state (Ghusinga et al. 2022). The signal perception by serpentine transmembrane receptors, namely G-protein-coupled receptors (GPCR), results in an alternation in its conformation, which subsequently functions as a guanine nucleotide exchange factor (GEF) to catalyze the GDP to GTP exchange on the Gα protein (McIntire 2009; Pandey and Vijayakumar 2018; Pandey 2019). The Gα component separates from the Gβγ subunits when it binds to GTP, allowing each of these to engage with diverse effectors to initiate downstream signaling (McIntire 2009). A signal is terminated when Gα-GTP is inactivated by GTP hydrolysis, resulting in GDP-bound Gα being released from its effector and reassociated with the Gβγ complex (McIntire 2009). A protein known as a regulator of G-protein signaling (RGS) with GTPase activating protein (GAP) activity speeds up the intrinsic GTP hydrolysis on the Gα subunit (Siderovski and Willard 2005). The G-protein signaling pathways in plants differ from the animal paradigm due to the existence of unique receptors and effectors, altered wiring of G-protein scenarios and disparate intrinsic characteristics of specific G-protein components (Pandey and Vijayakumar 2018; Maruta et al. 2019; Ghusinga et al. 2022). Notably, it has not been established that GPCRs can activate Gα by promoting GDP to GTP exchange in plants (Pandey 2020). The genetic and biochemical data imply that the exchange of nucleotides is less crucial for G-proteins to function in plants (Maruta et al. 2019). Furthermore, numerous receptor-like kinases (RLKs) have been implicated in the phosphorylation and dephosphorylation of G-proteins to mediate their mechanistic regulation (Jia et al. 2019; Pandey 2020). XLG subunit, independent of GTP-binding, has been discovered to interact with the RGS, Gβγ dimer and defense-related RLKs with an affinity similar to that of canonical Gα subunits (Liang et al. 2016; Lou et al. 2020; Maruta et al. 2021a, b). The functional investigation of RGS in A. thaliana and G. max signifies its crucial activities in the control of important physiological processes (Chen and Jones 2004; Choudhury and Pandey 2015). According to Hackenberg et al. (2017), several plants do not possess an RGS protein homolog. Therefore, it is ambiguous whether RGS-mediated deactivation is the primary mechanism controlling the G-protein cycle (Hackenberg et al. 2017).
G-proteins stimulate several intracellular signaling cascades in response to various extracellular stimuli (Majumdar et al. 2023). Almost every aspect of plant growth and development is influenced by the interaction of G-proteins with specific effectors (Roy Choudhury et al. 2019). These crucial physiological processes include regulating stomatal movement, nodulation and phytohormone signaling (Chakravorty et al. 2011; Choudhury and Pandey 2013, 2015, 2022; Jose and Choudhury 2020; Bhardwaj et al. 2020; Bovin et al. 2022). Interestingly, the knock-down of a particular G-protein subunit gene and its characterization displayed different root morphologies in both monocotyledons and dicotyledons (Table 1), thereby suggesting that G-proteins have a critical function in the development of plant roots in addition to their involvement in innate immunity and stress responses in plants (Trusov et al. 2007; Ding et al. 2008; Urano et al. 2015; Subramaniam et al. 2016; Gao et al. 2019; Maruta et al. 2021a). In this review, we have discussed the signaling mechanisms that involve G-proteins to regulate plant nitrogen nutrition.
Legumes can acquire nitrogen by atmospheric nitrogen fixation through endosymbiotic associations and the uptake of mineral nitrogen from soil (Murray et al. 2017; Roy et al. 2020; Gu et al. 2022; Zhong et al. 2022). The process of symbiosis in legumes is tightly regulated depending on the nitrogen levels in the soil. Legumes presumably evolved in nutrient-poor circumstances where the capacity to fix atmospheric nitrogen offers a significant growth benefit. The low soil nitrogen conditions encourage symbiotic associations of legumes with biological machines such as rhizobacteria (Murray et al. 2017; Roy et al. 2020). Gram-negative rhizobia (such as Rhizobium, Sinorhizobium, Azorhizobium, Bradyrhizobium, and Mesorhizobium), that associate with legume roots are diazotrophic bacteria engaged in endosymbiotic relationships to develop nodules (Geurts and Bisseling 2002; Graham and Vance 2003; Desbrosses and Stougaard 2011; Rutten and Poole 2019; Mahmud et al. 2020). Nuclear calcium oscillations are responsible for stimulating endosymbiotic programmes in response to rhizobial signals (Granqvist et al. 2012; Charpentier and Oldroyd 2013; Charpentier 2018). Moreover, calcium spiking represents one of the earliest events that can be detected and is a highly conserved component of the mutualistic signaling mechanism (Granqvist et al. 2015). This review includes information regarding the involvement of G-proteins in calcium spiking during root nodule symbiosis (RNS).
The legumes develop root cortex-based nodules in symbiosis with rhizobia (Gauthier-Coles et al. 2019). In legumes, there are two basic morphological variants of nodules, namely indeterminate and determinate. These two forms are distinguished by the existence or absence of a persistent nodule meristem, which reliably coincides with the cortical cell layers that develop nodule primordia (Kohlen et al. 2018). For instance, Glycine max and Lotus japonicus develop round determinate nodules devoid of persistent meristem (Ferguson et al. 2010; Pan and Wang 2017). In contrast, Medicago truncatula and Pisum sativum produce indeterminate nodules with a cylindrical shape in which nodule primordia generate in the inner cortex, and mature nodules retain a persistent meristem (Pan and Wang 2017; Bovin et al. 2022). Among the repertoire of proteins present in plants, the G-proteins have significant roles to play in the emergence of leguminous root nodules in response to a symbiotic relationship with rhizobia (Choudhury and Pandey 2013, 2015; Pandey 2020; Bovin et al. 2022), which have been highlighted in this review.
A major challenge for plant survival is nutrient acquisition from soil (Bhardwaj et al. 2015; Rahman et al. 2018). In plants, the detailed mechanism of nitrogen sensing pathways linked with G-proteins remains obscure. This review summarizes the developments in the mechanistic intricacies of nitrogen signaling involving G-proteins in plants. G-proteins show the ability to influence inflorescence, root architecture, seed size, number, and germination capacity, which in turn modulates essential agronomic features such as grain yield and nitrogen use efficiency (NUE) (Zhang et al. 2015; Wu et al. 2018; Liang et al. 2018; Kaur et al. 2018; Sun et al. 2018; Cui et al. 2020; Biswal et al. 2022). In this review, insights into the participation of G-proteins in regulating NUE in plants have also been discussed. This can assist in addressing the pressing issues of increasing crop growth and yield.
Possible role of G-proteins in root nodule symbiosis
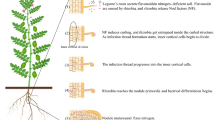
RNS is a molecular interaction between the host plant and the bacterial partner (Lazdunski et al. 2004; Mbengue et al. 2020). The outcome of a highly specific and complex signal exchange between legumes and rhizobia is the selective rhizobial colonization of legume cells within root nodules, which develop upon activation of various genes, establishing the symbiotic pathway (Desbrosses and Stougaard 2011; Das et al. 2019; Chen et al. 2021). The sensing of plant flavonoids by nitrogen-fixing rhizobia is one of the earliest steps in specificity between hosts and rhizobia (Fig. 1) (Hirsch and Fujishige 2012). Flavonoids are among the well-studied group of low molecular weight secondary metabolites (Hassan and Mathesius 2012; Dong and Song 2020) and are crucial for the sensitization of nitrogen-fixing bacteria (Liu and Murray 2016; Bag et al. 2022). Flavonoids are derived structurally from a 15-carbon skeleton composed of two benzene rings and are typically biosynthesized via the phenylpropanoid pathway (Liu and Murray 2016; Dong and Song 2020). Plants produce a range of flavonoids, with up to 10,000 found across the plant kingdom (Mathesius 2018). The specific plant flavonoids that the rhizobia in the rhizosphere interact with and recognize include isoflavonoids, daidzein, genistein, coumestrol, and naringenin, among others (Liu and Murray 2016; Bosse et al. 2021). According to Sugiyama et al. (2008), the exudates from soybean roots contain genistein and daidzein, which serve as signal molecules in the chemical communication between soybean and Bradyrhizobium japonicum. The roots of Phaseolus vulgaris when inoculated with Rhizobium leguminosarum shows the formation of nodules in the presence of genistein, daidzein, and coumestrol (Abd-Alla 2011). The flavonoid medicarpin, which is generated by both Trifolium and Medicago sp., has been shown to inhibit the growth of incompatible bacterial strains (Maxwell et al. 1989). Therefore, these studies imply that the ability of Rhizobium sp. to successfully form a symbiotic association is strongly influenced by the combination of host flavonoids present in the root exudate of legume species (Dong and Song 2020). Interestingly, in A. thaliana, G-protein signaling has been attributed to regulate flavonoid biosynthesis. The study revealed variations in the expression levels of several genes associated with the flavonoid biosynthesis in a knock-out mutant of the A. thaliana G-protein α subunit (gpa1-5) (Chakraborty et al. 2015). In future studies, the function of the G-protein complex signaling cascade can be determined in the legume flavonoid biosynthesis pathway using the G-protein subunit mutant studies. Consequently, establishing the involvement of the G-protein complex in the legume flavonoid synthesis cascade might pave avenues for enhancing nitrogen fixation by regulating the production of specific flavonoids using gene editing techniques.
Proposed model showing the role of Gα subunit in flavonoid biosynthesis and immune responses during G. max-Rhizobium symbiosis. The signal exchange between legume and rhizobia is commenced by the secretion of flavonoids from the roots of leguminous hosts. The figure depicts that Gα subunit is involved in the induction of flavonoids-responsive genes. The host flavonoids trigger the rhizobial Nod-cassette. The NodD protein, which is formed by rhizobia as a result of flavonoid perception, binds to the nod box in the promoter region of nod genes. This triggers the expression of the nod genes, which produces lipo-chitooligosaccharides (LCO), also referred to as Nod factors (NFs). These NFs have β-1,4-linked N-acetyl-d-glucosamine framework with four or five reducing and non-reducing terminal glucosamine residues (Geurts and Bisseling 2002). NFs are species-specific and undergo a range of substitutions at different positions of its non-reducing (-R1, -R2, -R3, -R4) and reducing ends (-R5, -R6) (Wang et al. 2018). Rhizobium releases NFs for symbiosis development. The intruding Rhizobium is initially perceived by the legume host as a potential pathogen, resulting in a transient defense reaction. Symbiosis receptor-like kinase (SymRK) associates directly with and suppresses the kinase activity of a positive regulator of plant immune responses, namely Brassinosteroid insensitive 1-associated receptor kinase 1 (BAK1). Further, SymRK phosphorylation of Gα prevents it from interacting with Gβγ dimer. Consequently, Gα is unable to influence host immune responses by ineffective interaction with BAK1 receptor
Despite their diversity, all rhizobia contain conserved nodABC genes for the formation of the N-acylated oligosaccharide core of the lipo-chitooligosaccharides (LCO), also called Nod factors (NFs), implying that these genes are monophyletic (Debellé et al. 2001). The flavonoid perception by rhizobia results in changes in the conformation of the NodD protein, enabling it to attach to the nod box in the promoter region of nod genes. This triggers the nod gene expression of rhizobia, which eventually culminates in the synthesis of NFs to activate various host responses (Geurts and Bisseling 2002; Hassan and Mathesius 2012; Hirsch and Fujishige 2012; Ghantasala and Roy Choudhury 2022). The legumes possess specialized lysin-motif (LysM) and leucine-rich repeat (LRR)-containing RLKs for perceiving the rhizobial NFs (Singh and Verma 2023). These complex multicomponent receptors are localized in the plasma membrane of the root epidermal cells (Ferguson et al. 2010; Roy et al. 2020). Interestingly, the interplay between the symbiotic and defense signaling mechanisms is observed in legumes during nodulation (Cao et al. 2017; Ivanova et al. 2022). It is widely accepted that rhizobia actively inhibit the host's immune response to facilitate infection and symbiotic development (Cao et al. 2017). A study in L. japonicus reported that a key symbiotic component termed as Symbiosis receptor-like kinase (SymRK) aids in the rhizobial inhibition of plant innate immunity. SymRK interacts with Brassinosteroid insensitive 1-associated receptor kinase 1 (BAK1), a positive regulator of plant innate immunity, to repress BAK1 kinase activity during rhizobial infection (Feng et al. 2021). Conversely, treatment with flagellin 22 (flg22), a conserved peptide motif in the flagellar protein of several bacteria, triggers defense responses that impede rhizobial infection and result in the delay of nodule organogenesis. This has been revealed by the adverse effect of flg22 on the spontaneous nodule production in the L. japonicus mutant, spontaneous-nodule-formation 1 (snf1). Moreover, after the symbiotic partner colonizes the host legume, the symbiotic pathway takes precedence over the defensive response (Lopez-Gomez et al. 2012). Previous studies examined the control of G-protein signaling by SymRK through protein–protein interactions and receptor-mediated in vitro phosphorylation during G. max nodulation (Choudhury and Pandey 2013, 2022). The studies reveal that SymRK phosphorylates Gα to prevent the negative regulation of nodulation (Choudhury and Pandey 2015; Pandey 2020). Therefore, it suggests that Gα might modulate host immunological responses by interacting with the SymRK-BAK1 complex, thereby affecting RNS. During RNS, the activity of G-protein subunits in the defense responses of legumes has not yet been investigated. Therefore, dissecting the specifics of all the molecular actors engaged in the RNS signaling cascade represents an exciting frontier of research. However, numerous studies illustrate the significance of G-proteins in other RNS stages, including calcium spiking and the control of nodule development in legumes (Choudhury and Pandey 2015; Mbengue et al. 2020; Pandey 2020; Bovin et al. 2022).
Involvement of G-proteins in calcium spiking
Various responses of NF signaling in the host plant include depolarization of the root hair plasma membrane, ion flux across the membrane, calcium spiking, changes in the cytoskeleton architecture, root hair curling, IT development and the production of nodule primordia (Heidstra and Bisseling 1996; Cárdenas et al. 2000; Tsyganova et al. 2018; Roy et al. 2020; Yang et al. 2022). In L. japonicus and M. truncatula, the membrane-bound Nod factor receptors (NFRs) such as LjNFR1/MtLYK3 and LjNFR5/MtNFP perceive diffusible NF released by rhizobia (Fig. 2) (Smit et al. 2007; Singh and Verma 2023). Also, LjSymRK/M. truncatula does not make infections 2 (MtDMI2) acts as a co-receptor with NFRs (Madsen et al. 2003; Antolín-Llovera et al. 2014; Singh and Verma 2023), which induces calcium spiking (or calcium oscillations) in and around the nucleus of root hair cells of host plants (Granqvist et al. 2015; Genre and Russo 2016).
Nod factor signaling cascade of events involving G-proteins occurring during legume-Rhizobium symbiosis. MtNFP/LjNFR5 and MtLYK3/LjNFR1 are receptor-like kinases (RLKs) involved in Nod factor (NF) sensing. M. truncatula Does not make infections 2 (MtDMI2)/LjSymRK functions as a co-receptor, causing intracellular signaling pathways to be activated in the legume root hair cells. The figure depicts the role of Gα in mediating early symbiotic stages, including calcium (Ca2+) influx by regulating an unknown Ca2+ channel present in the plasma membrane. Consequently, cytosolic Ca2+ spiking occurs which further generates nuclear Ca2+ spikes, where nuclear pore complexes like nucleoporin 133 (NUP133) and NUP85 play a significant role (Kanamori et al. 2006). G-protein subunits interact with G-protein-coupled receptor 1 (GCR1) and Receptor of G-protein signaling (RGS) to effect nodulation. It is hypothesized that G-protein complexes that bind with and are phosphorylated by LjNFR1 activate the release of an unidentified secondary messenger, which would promote Ca2+ spiking. The MtDMI1/LjPOLLUX and LjCASTOR cation channels get activated by this unknown secondary messenger. MtDMI1/LjPOLLUX and CNGC15s influence nodule organogenesis by regulating Ca2+ or potassium (K+) ions (Venkateshwaran et al. 2012). MtDMI3/LjCCaMK phosphorylates CYCLOPS/IPD3, which further engages with transcription factors such as DELLA, nodulation signaling pathway 1 (NSP1) and NSP2 to enhance nodule inception (NIN) expression (Cerri et al. 2017; Diédhiou and Diouf 2018; Mbengue et al. 2020). This triggers nodule organogenesis or rhizobial infection involving NIN, NF-Ys, Ets2 repressor factors required for nodulation 1 (ERN1) and early nodulin 11 (ENOD11) (Laporte et al. 2014; Laloum et al. 2014)
Recently, a study in P. sativum reveals that calcium influx activation, which is followed by root hair deformation, involves the G-protein complex (Table 2) (Bovin et al. 2022). The interaction between LysM-RLK K1 and the PsGα2 (Psat5g034360) was demonstrated in P. sativum with the aid of a Co-immunoprecipitation (Co-IP) assay (Bovin et al. 2022). According to a study, SYM10 has an apparently inactive kinase activity and associates with LysM-RLK К1 containing a YAQ motif in its kinase domain, making it functional in regulating symbiosis initiation (Nakagawa et al. 2011; Kirienko et al. 2018). This indicates the participation of G-proteins in calcium response followed by NF sensing in legumes during the initial developmental stages of symbiosis (Bovin et al. 2022). To positively control nodulation and expression of early nodulation genes in G. max, NFR1 phosphorylates RGS proteins. The phosphorylated RGS helps to initiate the signaling which results in calcium spikes while retaining the negative regulator of nodulation (Gα) in its inactive state (Choudhury and Pandey 2013, 2015; Mbengue et al. 2020). Also, in the absence of rhizobia, mastoparan 7, a G-protein agonist, mimics NF-induced early nodulin 11 (ENOD11) and ENOD12 gene expression in root hair cells of M. truncatula during symbiosis and root hair deformation in Vigna unguiculata, which is prevented by the G-protein antagonist pertussis toxin (Pingret et al. 1998; Kelly and Irving 2003; Sun et al. 2007). Mastoparan 7 has also been proven to result in calcium spiking in the roots of M. truncatula, which are not reliant on NFP and DMI2 (Sun et al. 2007). Furthermore, an inhibitor of a downstream signaling component of G-protein namely, phospholipase D (PLD), ceases the calcium oscillations (Charron et al. 2004). According to these findings, mastoparan 7 either directly or indirectly activates PLD signaling by acting downstream of NFP and the DMI2. Accordingly, a hypothesis suggested that G-protein signaling downstream of the RLK could activate the production of an unidentified secondary messenger, which in turn causes nuclear calcium oscillations (Mbengue et al. 2020). The MtDMI1/LjPOLLUX and LjCASTOR cation channels, which show the interaction with three Cyclic Nucleotide-Gated Channels, namely CNGC15a, CNGC15b and CNGC15c, get activated by an unknown secondary messenger (Charpentier et al. 2016; Mbengue et al. 2020). Overall, G-protein subunits act as downstream elements of the NF perception pathway and trigger calcium spiking, which further activates a signal cascade involving various transcription factors including CYCLOPS/IPD3, DELLA, nodulation signaling pathway 1 (NSP1), NSP2 to enhance nodule inception (NIN) expression for the development of symbiotic nodules (Lévy et al. 2004; Tirichine et al. 2006; Singh et al. 2014; Laporte et al. 2014; Laloum et al. 2014; Mbengue et al. 2020; Yuan et al. 2022; Bovin et al. 2022).
Molecular basis of root nodulation involving G-proteins, their putative receptors, and associated proteins
Indications for the participation of G-proteins in the regulation of nodule development in various leguminous species have been found in several pharmacological and biochemical tests. The development of nodules is regulated by certain G-proteins subunits (Choudhury and Pandey 2013). Overexpression of G-protein components and RNAi suppression leads to a higher and lower number of nodules, respectively, confirming their functions as positive regulators of nodule development (Choudhury and Pandey 2013). One finding is direct evidence for the significance of MtGβ1 and PsGβ1 in symbiosis development regulation in M. truncatula and P. sativum, respectively, wherein the number of indeterminate nodules generated were considerably reduced by RNAi-based inhibition of MtGβ1 (Medtr3g116500) and PsGβ1 (Psat5g006200) (Bovin et al. 2022).
A variety of membrane-bound receptors, including GPCR, RGS and RLKs, can be coupled with G-proteins in plants (Pandey 2019; Chakraborty and Raghuram 2022). In accordance with a study, the downregulation of GCR1 in L. japonicus results in a significant impact on LjNIN, a downstream component of the G-protein signaling network, which encodes for transcriptional factors crucial for nodulation (Rogato et al. 2016). According to Choudhury and Pandey (2015), a RLK protein complex i.e., NFR1–NFR5–SymRK interacts and phosphorylates G-protein subunits. Enhanced GAP activity toward the Gα subunit is demonstrated by phosphorylated RGS, implying that RLK-mediated phosphorylation of RGS enables the G-protein cycle to cease more rapidly (Choudhury and Pandey 2016). Therefore, NFR1 phosphorylation of RGS keeps Gα in an inactive state, whereas SymRK phosphorylation of Gα prevents it from interacting with Gβγ. In this scenario, the negative regulator Gα would be inactivated, and the positive regulators, i.e., Gβγ, would lead to effective nodulation (Choudhury and Pandey 2015; Pandey 2020). A recent study demonstrated that SymRK phosphorylates Gα in vitro at numerous sites, including two in the active site to prevent GTP binding (Choudhury and Pandey 2022). The two amino acids that are phosphorylated in the active region of conventional Gα are conserved in the GTP-binding pocket of XLG proteins, suggesting that XLG may also be phosphorylated and influenced by SymRK (Pandey 2020). Similar to the Gα subunit, the XLG subunit is also involved in interactions with Gβγ and RGS protein (Pandey 2020; Lou et al. 2020; Maruta et al. 2021a, b). Although XLG proteins are important regulators of defense signaling that operate in parallel with the classical Gα proteins in A. thaliana, their role in the regulation of nodulation has not been fully investigated (Maruta et al. 2015; Liang et al. 2016; Pandey 2020). This emphasizes the need for elucidating the potential role of XLG proteins in nodule formation that can be gleaned from insights into the molecular mechanism of XLG signaling transduction cascades.
Various well-known G-protein signaling downstream components, including PLC, PLD, diacylglycerol pyrophosphate, phosphatidic acid, and G-protein-related phosphoinositide 3-kinase, have been associated with the regulation of nodulation (Misra et al. 2007; Peleg-Grossman et al. 2007; Santos-Briones et al. 2009). The Co-IP experiments have confirmed the association of G-protein subunits with PLC, implying crosstalk between G-protein and PLC-mediated symbiotic signaling pathways in both M. truncatula and P. sativum (Bovin et al. 2022). In plants, Phospholipase Dα1 (PLDα1) is a crucial regulatory element of the G-protein signaling complex (Li et al. 2009; Lu et al. 2013) and it also interacts with Gα as well as Gβ proteins (Zhao and Wang 2004; Gookin and Assmann 2014). According to Roy Choudhury and Pandey (2016), PLDα1 and RGS1 are found in proximity to the G-protein complex, or perhaps PLDα1 and G-proteins form a macromolecular complex. This model positions PLDα1 and Gα downstream of RGS1, which serves as PLDα1 inhibitor rather than GAP and attributes the role of GAP to PLDα1 (Roy Choudhury and Pandey 2016). Also, pharmacological and molecular methods have been employed to uncover the role of PLDα1 in signaling during nodulation in soybean (Zhang et al. 2021). Taken together, these observations led to the suggestion of a simplified mechanism for the roles of the RGS, PLDα1, and G-proteins in RNS (Pacheco and Quinto 2022). According to this model, when NFs are perceived, the cytosolic kinase domain of NFR1 phosphorylates RGS and as a result, PLDα1 is released from its inactivated state. The G-protein complex is then rendered inactive by the active PLDα1 that act as a GAP. Consequently, the Gα subunit cannot inhibit the growth of nodules, and free Gβγ dimers serve as positive regulators of nodule development (Pacheco and Quinto 2022). Moreover, future research can also determine the role of any member of the PLD family other than PLDα1 in controlling the G-protein cycle (Pacheco and Quinto 2022). This indicates the necessity of further research to shed light on the molecular mechanisms underlying potential G-protein and PLD signaling during the root nodule development in legumes.
Soil nitrogen sensing in conjunction with G-proteins
In soils, numerous microbial communities engage in nitrogen transformations into plant-usable forms to regulate nitrogen mobilisation and fixation capacity (Robertson and Groffman 2007). In aerobic soils, most plants have nitrate as their principal source of nitrogen (Liu et al. 2015). The nitrate transporters, or channels, are grouped into five families in higher plants, namely, the nitrate transporter 1/peptide transporter family (NPF or NRT1/PTR), nitrate transporter 2 (NRT2), aluminium-activated malate transporter (ALMT), slow anion channel-associated 1 homolog 3 (SLAC1/SLAH3) and chloride channel (CLC) (Krapp et al. 2014; Léran et al. 2015; Pellizzaro et al. 2017). The first identified nitrate transporter implicated in the primary nitrate response (PNR) in A. thaliana was the moonlighting protein AtNPF6.3 (AtNRT1.1 or CHL1) (Fichtner et al. 2021; Gu et al. 2022). Members of the NPF and NRT2 families have been shown to associate with nitrate acquisition in roots (Gu et al. 2022). Moreover, plants have developed two nitrate uptake systems: a high-affinity transport system (HATS) and a low-affinity transport system (LATS) (Wang et al. 2012). In comparison with low-affinity transporters, high-affinity transporters function better at lower concentrations of nitrate while becoming saturated at greater nitrate concentrations (Muratore et al. 2021). More importantly, except for NPF6.3 (NRT1.1 or CHL1), all members of the NRT2 family are HATS, while the majority of the NPF/NRT1 family members are LATS (Ho et al. 2009; Wang et al. 2012). NPF6.3 is considered as a dual-affinity nitrate sensor and transporter in A. thaliana (Ho et al. 2009; Gu et al. 2022).
Several findings reveal the importance of G-proteins in regulating nitrate uptake and metabolism encoding genes in plants (Fig. 3) (Chakraborty et al. 2015, 2019; Liu et al. 2018; Pathak et al. 2021). Researchers are increasingly aware that GPCR is found in a multitude of cells, tissues, and organs in animals, plants, and microbes and that it shows involvement in the detection of a variety of nutrients (Moran et al. 2021; El-Defrawy and Hesham 2020; Chakraborty and Raghuram 2022). Previous findings in A. thaliana have shown genetic evidence of the GCR1-GPA1 linkage controlling the nitrate response (Chakraborty et al. 2015, 2019). In a report, gpa1-5, gcr1-5, and gpa1-5gcr1-5 mutants were studied to investigate the role of three mutants in regulating nitrate uptake and metabolism encoding genes. NRT1 was shown to be highly expressed in all three mutants as compared to the WT at low nitrate levels, but NRT1 expression was low at high nitrate levels (Chakraborty et al. 2019). By considering NRT1 as a low-affinity nitrate transporter, the low nitrate condition may have been insufficient due to mutations in GCR1 and/or GPA1, resulting in increased expression of NRT1, which was not observed at a high nitrate level (Chakraborty et al. 2019). Henceforth, according to the aforementioned study, the gcr1-5 mutant exhibits an altered dose-dependent differential nitrogen response for NRT1 gene expression. Additionally, a study has revealed the in vitro interactions of GPCR with all three subunits of G-proteins in P. sativum (Misra et al. 2007). Therefore, it develops an interest in exploring whether coupling of G-proteins with GPCR in P. sativum also effectuates significant expression level alterations of nitrate transporters. The canonical GPCRs are often implicated in agronomically significant processes in plants (Chakraborty and Raghuram 2022), but the level of association of GPCR with the Gα subunit has remained mostly unidentified (Chakraborty et al. 2019). The findings of future investigations can provide compelling scientific evidence to reassess the involvement of GPCR in plant G-protein-mediated signaling pathways.
A simplified representation of the involvement of G-protein subunits in nitrate signaling in A. thaliana. NPF6.3 (NRT1.1 or CHL1) functions as a transporter as well as a sensor of nitrate. Depending on the nitrate levels, regulation of the expression of NRT1.1 shows probable involvement of GCR1-GPA1 coupling followed by an interaction with the Arabidopsis nitrate regulated 1 (ANR1) transcription factor. This signaling subsequently regulates the nitrogen (N) responsive genes, which include nitrogen starvation/assimilation genes such as the nitrate transporter NRT2.1, isocitrate dehydrogenase (ICDH) and asparagine synthase 1 (ASN1)
Arabidopsis nitrate regulated 1 (ANR1), a MADS intervening keratin-like and C-terminal (MIKC)-type MADS-box transcription factor, was the first to be discovered in nitrate signaling pathways (Zhang and Forde 1998). In nitrate-rich localized areas, lateral root elongation is hindered in ANR1 loss-of-function mutants, which affects the root system's plasticity (Zhang and Forde 1998). Later research revealed that NRT1.1 regulates ANR1 at the transcriptional level (Remans et al. 2006). According to Chakraborty et al. (2015), NRT1.1 interacts with AtGPA1, which controls the transcription of nitrate-responsive genes through ANR1. This study employed gpa1-5, a novel GPA1 knock-out mutant, to reveal that numerous nitrogen starvation/assimilation genes, such as the nitrate transporter NRT2.1, isocitrate dehydrogenase (ICDH), and asparagine synthase (ASN1), were up-regulated in gpa1-5 (Chakraborty et al. 2015). It has been demonstrated that a Triticum aestivum Gβ gene, TaNBP1 (AK332651), regulates transcription of the nitrate transporter gene (NRT2.2) in transgenic Nicotiana benthamiana, thereby indicating a role in nitrogen uptake (Liu et al. 2018). Additionally, the role of RGA1 in nitrogen-responsive transcriptional regulation has been established by a study in Oryza sativa (Pathak et al. 2021). The genes encoding the ammonium and nitrate transporters were shown to be down-regulated in the rga1 mutant, demonstrating the relevance of RGA1 function in nitrogen uptake. Moreover, the transcription factor network analysis of the rga1 mutant revealed the importance of RGA1 in regulating the nitrogen signaling cascade with several differentially expressed genes (DEGs), including Nin-like and OsCIPK23, among others (Pathak et al. 2021). Notably, the G-protein subunit mutant studies can be beneficial for translating key research findings from commercially significant crop species to legumes for deciphering the possible participation of G-proteins in nitrogen uptake.
According to Fan et al. (2017), there are reports on nitrate transporter being used for improving crop productivity. For instance, the increased expression of OsNRT1.1B, a low-affinity nitrate transporter, in japonica rice may aid in the improved sensing of varied nitrate concentrations and increasing the capacity for nitrogen accumulation inside the grain (Hu et al. 2015). In addition, rice with overexpressed OsNRT2.3b has a better ability to absorb other nutrients, which reduces photorespiration and promotes growth and grain yield (Fan et al. 2016). Furthermore, numerous studies demonstrate that Gα subunits regulate the expression level of plant nitrate transporters (Chakraborty et al. 2015, 2019; Pathak et al. 2021). This indicates that optimizing nitrate uptake and utilization via G-protein subunits might contribute to increasing crop yield.
G-proteins and regulation of nitrogen use efficiency
NUE is a multigenic quantitative trait, including numerous nitrogen-responsive genes and mechanisms that require thorough characterization (Mandal et al. 2022). G-proteins are implicated in critical agronomic traits such as NUE, thereby directly impacting yield (Xu et al. 2016b; Stateczny et al. 2016). There are findings on the significance of the Gγ subunit in governing nitrogen assimilation and NUE. For instance, in rice, the DEP1 (Os09g0441900) gene is a plant-specific Gγ subunit that directs branching, density, and erectness of panicles and was also discovered to be a significant quantitative trait locus (QTL) for NUE (Huang et al. 2009; Sun et al. 2014, 2018; Xu et al. 2016a; Li et al. 2023). Both RGA1 and RGB1 subunits have been confirmed to interact with the DEP1 protein in vivo (Sun et al. 2014). The G protein γ-like (GGL) domain of DEP1 interacts with the rice RGB1 subunit on the plasma membrane and within the nucleus. Additionally, DEP1 binds with RGA1, and the von Willebrand factor type C (vWFC) domain at the C-terminus of DEP1 may be involved in this interaction (Sun et al. 2014). According to the study, nitrogen-mediated growth responses are likewise inhibited by reduced RGA1 or increased RGB1 activity (Sun et al. 2014). Also, plants containing the dep1-32 loss-of-function allele are unaffected by nitrogen availability, whereas plants with the dep1-1 gain-of-function allele demonstrate higher nitrogen uptake even when nitrogen is scarce, suggesting that manipulating G-protein activity could be a novel strategy for regulating NUE (Sun et al. 2014). DEP1 affects genes related to ammonium absorption and assimilation (such as OsAMT1;1, OsGS1;2 and OsNADH-GOGAT1), thereby regulating nitrogen uptake and metabolism (Huang et al. 2009). These genes showed an up-regulation in dep1 allelic plants when nitrogen levels were low. Despite the low nitrogen availability, the dep1 allelic plants exhibit higher glutamine synthase activity and accumulate more internal nitrogen than the DEP1 allelic plants (Sun et al. 2014). According to a recent study in rice, the increased stomatal conductance conferred by dep1 results in a higher photosynthetic capability under minimal nitrogen circumstances (Li et al. 2023). This research aids to comprehend the photosynthetic efficiency of the dep1 variety in low nitrogen environments by analyzing photosynthesis, stomatal function, and nitrogen uptake and assimilation (Li et al. 2023).
Furthermore, nitrogen heterogeneity in the soil is a key factor in determining root development (Araya et al. 2016). While nitrogen deficiency stops root growth, a moderate amount of nitrogen in the soil encourages the extension of lateral roots that would otherwise be stunted in high nitrogen environments (Linkohr et al. 2002; Liu et al. 2017). Altering the root system architecture is an adaptive strategy to improve NUE (Awasthi and Laxmi 2021). According to a study, the rice G-protein mutants vary from the WT in lateral root number and high nitrogen inhibition growth (Liang et al. 2018). The high nitrogen-induced suppression of root growth was abolished in the A. thaliana agb1-2 mutant. This indicates that G-protein modulates the root architecture in response to nitrogen availability (Liang et al. 2018). In T. aestivum, the nitrogen starvation response is reportedly regulated by TaNBP1 (Liu et al. 2018). In line with its function in promoting nitrogen accumulation, TaNBP1 overexpression in N. benthamiana results in enhanced phenotypic, expanded root system architecture and increased biomass for transgenic plants under nitrogen deficiency in comparison to the WT (Liu et al. 2018). Furthermore, a class C Gγ subunit of A. thaliana termed as AGG3 may also be involved in the NUE. During the early seedling stage of development, the model monocot Setaria virdis overexpressing AtAGG3 (At5g20635) showed improved root growth, enabling greater plant survival under limited nitrogen circumstances (Kaur et al. 2018). A study in B. napus revealed that under nitrogen deprivation, Gα (BnGA1) and five C-type Gγ genes (BnGG9, BnGG10, BnGG11, BnGG12 and BnGG13) were initially upregulated in roots, while Gα was initially downregulated and five C-type Gγ genes were substantially expressed at various times in leaves (Xie et al. 2022). These findings shed light on the biological processes that G-protein genes perform in response to an inadequate supply of nitrogen (Xie et al. 2022). Through genetically abrogating each of the three rice XLGs separately and synergistically using CRISPR/Cas9 genome editing in rice, a study uncovered a role for XLGs in agronomic traits. The research results reflect that non-canonical XLGs are crucial regulators of rice plant growth, grain filling and panicle phenotype (Biswal et al. 2022). Henceforth, deciphering the XLG signaling in agronomic performance can assist in establishing its association with NUE for crop improvement strategies.
Conclusions
The increasing use of nitrogen fertilizers contributes to an enormous growth in agricultural production. However, at the same time, soil quality has significantly deteriorated. Decreased soil fertility and unsustainable long-term crop yields are potential consequences of synthetic chemicals which very often suppress the symbiotic nitrogen fixation process (Akter et al. 2018; Reinprecht et al. 2020; Móring et al. 2021). With steady population growth and climate change, it has become challenging to increase crop productivity without exacerbating environmental degradation. The development of plants that could flourish independently of expensive nitrogen fertilizers would be a significant achievement in research related to nitrogen fixation. G-proteins and their potential receptors are emerging as key players in understanding root development and nodulation process. Hence, a deeper knowledge of G-protein signaling can provide insights into the control of symbiosis and root development, thereby laying the foundation for a multitude of studies in the future. Moreover, persistent efforts to incorporate the nitrogen-fixing trait into non-legume crops, particularly cereals, which constitute a significant section of the food basket, can lessen our reliance on inorganic nitrogen fertilizers. Also, uncovering the function of G-proteins in the nitrate sensing pathway and their translation into growth-enhancing reactions is crucial for enhancing NUE, crop productivity and reducing pollution caused by chemical fertilizers. The emerging research on regulating nutrient stress is supplemented by the multiple functions of G-proteins in plant root development and nitrogen signaling. Therefore, besides alleviating plant abiotic and biotic stress, G-proteins may also aid in plant adaptation to nutrient stress. This will assist in engineering more efficient crops with improved NUE by genome modifications to maximize crop yield and limit excess nitrogen being added to the environment, thereby promoting a sustainable future for modern agriculture.
Data availability
Not applicable.
References
Abd-Alla MH (2011) Nodulation and nitrogen fixation in interspecies grafts of soybean and common bean is controlled by isoflavonoid signal molecules translocated from shoot. Plant Soil Environ 57:453–458. https://doi.org/10.17221/379/2010-PSE
Akter Z, Pageni BB, Lupwayi NZ, Balasubramanian PM (2018) Biological nitrogen fixation by irrigated dry bean (Phaseolus vulgaris L.) genotypes. Can J Plant Sci 98(5):1159–1167. https://doi.org/10.1139/cjps-2017-0301
Antolín-Llovera M, Ried MK, Parniske M (2014) Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr Biol 24(4):422–427. https://doi.org/10.1016/j.cub.2013.12.053
Araya T, Kubo T, von Wirén N, Takahashi H (2016) Statistical modeling of nitrogen-dependent modulation of root system architecture in Arabidopsis thaliana. J Integr Plant Biol 58(3):254–265. https://doi.org/10.1111/jipb.12433
Awasthi P, Laxmi A (2021) Root architectural plasticity in changing nutrient availability. Rhizobiology: molecular physiology of plant roots. Springer. https://doi.org/10.1007/978-3-030-84985-6_2
Bag S, Mondal A, Majumder A, Mondal SK, Banik A (2022) Flavonoid mediated selective cross-talk between plants and beneficial soil microbiome. Phytochem Rev. https://doi.org/10.1007/s11101-022-09806-3
Bhardwaj D, Medici A, Gojon A, Lacombe B, Tuteja N (2015) A new insight into root responses to external cues: paradigm shift in nutrient sensing. Plant Signal Behav 10(12):1049791. https://doi.org/10.1080/15592324.2015.1049791
Bhardwaj D, Sahoo RK, Naqvi AR, Lakhanpaul S, Tuteja N (2020) Pea Gβ subunit of G proteins has a role in nitric oxide-induced stomatal closure in response to heat and drought stress. Protoplasma 257(6):1639–1654. https://doi.org/10.1007/s00709-020-01529-6
Bisht NC, Jez JM, Pandey S (2011) An elaborate heterotrimeric G-protein family from soybean expands the diversity of plant G-protein networks. New Phytol 190(1):35–48. https://doi.org/10.1111/j.1469-8137.2010.03581.x
Biswal AK, Wu TY, Urano D, Pelissier R, Morel JB, Jones AM, Biswal AK (2022) Novel mutant alleles reveal a role of the extra-large G protein in rice grain filling, panicle architecture, plant growth, and disease resistance. Front Plant Sci 12:2821. https://doi.org/10.3389/fpls.2021.782960
Bosse MA, da Silva MB, de Oliveira NGRM, de Araujo MA, Rodrigues C, de Azevedo JP, Dos Reis AR (2021) Physiological impact of flavonoids on nodulation and ureide metabolism in legume plants. Plant Physiol Biochem 166:512–521. https://doi.org/10.1016/j.plaphy.2021.06.007
Bovin AD, Pavlova OA, Dolgikh AV, Leppyanen IV, Dolgikh EA (2022) The role of heterotrimeric G-protein beta subunits during nodulation in Medicago truncatula Gaertn and Pisum sativum L. Front Plant Sci 12:3264. https://doi.org/10.3389/fpls.2021.808573
Cantos CF, dePamphilis CW, Assmann SM (2023) Extra-large G proteins have extra-large effects on agronomic traits and stress tolerance in maize and rice. Trends Plant Sci. https://doi.org/10.1016/j.tplants.2023.04.005
Cao Y, Halane MK, Gassmann W, Stacey G (2017) The role of plant innate immunity in the legume-Rhizobium symbiosis. Annu Rev Plant Biol 68:535–561. https://doi.org/10.1146/annurev-arplant-042916-041030
Cárdenas L, Holdaway-Clarke TL, Sánchez F, Quinto C, Feijó JA, Kunkel JG, Hepler PK (2000) Ion changes in legume root hairs responding to Nod factors. Plant Physiol 123(2):443–452. https://doi.org/10.1104/pp.123.2.443
Cerri MR, Wang Q, Stolz P, Folgmann J, Frances L, Katzer K, Li X, Heckmann AB, Wang TL, Downie JA, Klingl A (2017) The ERN1 transcription factor gene is a target of the CCaMK/CYCLOPS complex and controls rhizobial infection in Lotus japonicus. New Phytol 215(1):323–337. https://doi.org/10.1111/nph.14547
Chakraborty N, Raghuram N (2022) Life, death and resurrection of plant GPCRs. Plant Mol Biol. https://doi.org/10.1007/s11103-022-01323-3
Chakraborty N, Sharma P, Kanyuka K, Pathak RR, Choudhury D, Hooley R, Raghuram N (2015) G-protein α-subunit (GPA1) regulates stress, nitrate and phosphate response, flavonoid biosynthesis, fruit/seed development and substantially shares GCR1 regulation in A. thaliana. Plant Mol Biol 89(6):559–576. https://doi.org/10.1007/s11103-015-0374-2
Chakraborty N, Kanyuka K, Jaiswal DK, Kumar A, Arora V, Malik A, Gupta N, Hooley R, Raghuram N (2019) GCR1 and GPA1 coupling regulates nitrate, cell wall, immunity and light responses in Arabidopsis. Sci Rep 9(1):1–17. https://doi.org/10.1038/s41598-019-42084-2
Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, Assmann SM, Botella JR (2011) An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J 67(5):840–851. https://doi.org/10.1111/j.1365-313X.2011.04638.x
Charpentier M (2018) Calcium signals in the plant nucleus: origin and function. J Exp Bot 69(17):4165–4173. https://doi.org/10.1093/jxb/ery160
Charpentier M, Oldroyd GE (2013) Nuclear calcium signaling in plants. Plant Physiol 163(2):496–503. https://doi.org/10.1104/pp.113.220863
Charpentier M, Sun J, Martins TV, Radhakrishnan GV, Findlay K, Soumpourou E, Thouin J, Véry AA, Sanders D, Morris RJ, Oldroyd GE (2016) Nuclear-localized cyclic nucleotide–gated channels mediate symbiotic calcium oscillations. Science 352(6289):1102–1105. https://doi.org/10.1126/science.aae0109
Charron D, Pingret JL, Chabaud M, Journet EP, Barker DG (2004) Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol 136(3):3582–3593. https://doi.org/10.1104/pp.104.051110
Chen JG, Jones AM (2004) AtRGS1 function in Arabidopsis thaliana. Methods Enzymol 389:338–350. https://doi.org/10.1016/s0076-6879(04)89020-7
Chen WF, Wang ET, Ji ZJ, Zhang JJ (2021) Recent development and new insight of diversification and symbiosis specificity of legume rhizobia: mechanism and application. J Appl Microbiol 131(2):553–563. https://doi.org/10.1111/jam.14960
Choudhury SR, Pandey S (2013) Specific subunits of heterotrimeric G proteins play important roles during nodulation in soybean. Plant Physiol 162(1):522–533. https://doi.org/10.1104/pp.113.215400
Choudhury SR, Pandey S (2015) Phosphorylation-dependent regulation of G-protein cycle during nodule formation in soybean. Plant Cell 27(11):3260–3276. https://doi.org/10.1105/tpc.15.00517
Choudhury SR, Pandey S (2016) Interaction of heterotrimeric G-protein components with receptor-like kinases in plants: an alternative to the established signaling paradigm? Mol Plant 9(8):1093–1095. https://doi.org/10.1016/j.molp.2016.05.012
Choudhury SR, Pandey S (2022) SymRK-dependent phosphorylation of Gα protein and its role in signaling during soybean (Glycine max) nodulation. Plant J 110(1):277–291. https://doi.org/10.1111/tpj.15672
Choudhury SR, Bisht NC, Thompson R, Todorov O, Pandey S (2011) Conventional and novel Gγ protein families constitute the heterotrimeric G-protein signaling network in soybean. PLoS ONE 6(8):2336. https://doi.org/10.1371/journal.pone.0023361
Cui Y, Jiang N, Xu Z, Xu Q (2020) Heterotrimeric G protein are involved in the regulation of multiple agronomic traits and stress tolerance in rice. BMC Plant Biol 20:1–13. https://doi.org/10.1186/s12870-020-2289-6
Das DR, Horváth B, Kundu A, Kaló P, DasGupta M (2019) Functional conservation of CYCLOPS in crack entry legume Arachis hypogaea. Plant Sci 281:232–241. https://doi.org/10.1016/j.plantsci.2018.12.003
Debellé F, Moulin L, Mangin B, Dénarié J, Boivin C (2001) Nod genes and Nod signals and the evolution of the Rhizobium legume symbiosis. Acta Biochim Pol 48(2):359–365. https://doi.org/10.18388/abp.2001_3921
Desbrosses GJ, Stougaard J (2011) Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10(4):348–358. https://doi.org/10.1016/j.chom.2011.09.005
Diédhiou I, Diouf D (2018) Transcription factors network in root endosymbiosis establishment and development. World J Microbiol Biotechnol 34(3):1–14. https://doi.org/10.1007/s11274-018-2418-7
Ding L, Pandey S, Assmann SM (2008) Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J 53(2):248–263. https://doi.org/10.1111/j.1365-313X.2007.03335.x
Dong W, Song Y (2020) The significance of flavonoids in the process of biological nitrogen fixation. Int J Mol Sci 21(16):5926. https://doi.org/10.3390/ijms21165926
El-Defrawy MM, Hesham AEL (2020) G-protein-coupled receptors in fungi. Fungal biotechnology and bioengineering. Springer, Cham, pp 37–126. https://doi.org/10.1007/978-3-030-41870-0_3
Fan X, Tang Z, Tan Y, Zhang Y, Luo B, Yang M, Lian X, Shen Q, Miller AJ, Xu G (2016) Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc Natl Acad Sci 113(26):7118–7123. https://doi.org/10.1073/pnas.1525184113
Fan X, Naz M, Fan X, Xuan W, Miller AJ, Xu G (2017) Plant nitrate transporters: from gene function to application. J Exp Bot 68(10):2463–2475. https://doi.org/10.1093/jxb/erx011
Feng Y, Wu P, Liu C, Peng L, Wang T, Wang C, Tan Q, Li B, Ou Y, Zhu H, Yuan S (2021) Suppression of LjBAK1-mediated immunity by SymRK promotes rhizobial infection in Lotus japonicus. Mol Plant 14(11):1935–1950. https://doi.org/10.1016/j.molp.2021.07.016
Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, Gresshoff PM (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52(1):61–76. https://doi.org/10.1111/j.1744-7909.2010.00899.x
Fichtner F, Dissanayake IM, Lacombe B, Barbier F (2021) Sugar and nitrate sensing: a multi-billion-year story. Trends Plant Sci 26(4):352–374. https://doi.org/10.1016/j.tplants.2020.11.006
Ganotra J, Sharma B, Biswal B, Bhardwaj D, Tuteja N (2023) Emerging role of small GTPases and their interactome in plants to combat abiotic and biotic stress. Protoplasma 260(4):1007–1029 https://doi.org/10.1007/s00709-022-01830-6
Gao Y, Gu H, Leburu M, Li X, Wang Y, Sheng J, Fang H, Gu M, Liang G (2019) The heterotrimeric G protein β subunit RGB1 is required for seedling formation in rice. Rice 12(1):1–14. https://doi.org/10.1186/s12284-019-0313-y
Gauthier-Coles C, White RG, Mathesius U (2019) Nodulating legumes are distinguished by a sensitivity to cytokinin in the root cortex leading to pseudonodule development. Front Plant Sci 9:1901. https://doi.org/10.3389/fpls.2018.01901
Genre A, Russo G (2016) Does a common pathway transduce symbiotic signals in plant–microbe interactions? Front Plant Sci 7:96. https://doi.org/10.3389/fpls.2016.00096
Geurts R, Bisseling T (2002) Rhizobium Nod factor perception and signalling. Plant Cell 14:S239–S249. https://doi.org/10.1105/tpc.002451
Ghantasala S, Roy Choudhury S (2022) Nod factor perception: an integrative view of molecular communication during legume symbiosis. Plant Mol Biol. https://doi.org/10.1007/s11103-022-01307-3
Ghusinga KR, Elston TC, Jones AM (2022) Towards resolution of a paradox in plant G-protein signaling. Plant Physiol 188(2):807–815. https://doi.org/10.1093/plphys/kiab534
Gookin TE, Assmann SM (2014) Significant reduction of BiFC non-specific assembly facilitates in planta assessment of heterotrimeric G-protein interactors. Plant J 80(3):553–567. https://doi.org/10.1111/tpj.12639
Graham PH, Vance CP (2003) Legumes: importance and constraints to greater use. Plant Physiol 131(3):872–877. https://doi.org/10.1104/pp.017004
Granqvist E, Wysham D, Hazledine S, Kozlowski W, Sun J, Charpentier M, Martins TV, Haleux P, Tsaneva-Atanasova K, Downie JA, Oldroyd GE (2012) Buffering capacity explains signal variation in symbiotic calcium oscillations. Plant Physiol 160(4):2300–2310. https://doi.org/10.1104/pp.112.205682
Granqvist E, Sun J, Op den Camp R, Pujic P, Hill L, Normand P, Morris RJ, Downie JA, Geurts R, Oldroyd GE (2015) Bacterial-induced calcium oscillations are common to nitrogen-fixing associations of nodulating legumes and non-legumes. New Phytol 207(3):551–558. https://doi.org/10.1111/nph.13464
Gu B, Chen Y, Xie F, Murray JD, Miller AJ (2022) Inorganic nitrogen transport and assimilation in pea (Pisum sativum). Genes 13(1):158. https://doi.org/10.3390/genes13010158
Hackenberg D, McKain MR, Lee SG, Roy Choudhury S, McCann T, Schreier S, Harkess A, Pires JC, Wong GK, Jez JM, Kellogg EA (2017) Gα and regulator of G-protein signaling (RGS) protein pairs maintain functional compatibility and conserved interaction interfaces throughout evolution despite frequent loss of RGS proteins in plants. New Phytol 216(2):562–575. https://doi.org/10.1111/nph.14180
Hassan S, Mathesius U (2012) The role of flavonoids in root–rhizosphere signalling: opportunities and challenges for improving plant–microbe interactions. J Exp Bot 63(9):3429–3444. https://doi.org/10.1093/jxb/err430
Heidstra R, Bisseling TON (1996) Nod factor-induced host responses and mechanisms of Nod factor perception. New Phytol 133(1):25–43. https://doi.org/10.1111/j.1469-8137.1996.tb04339.x
Hirsch AM, Fujishige NA (2012) Molecular signals and receptors: communication between nitrogen-fixing bacteria and their plant hosts. Biocommun Plants. https://doi.org/10.1007/978-3-642-23524-5_14
Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138(6):1184–1194. https://doi.org/10.1016/j.cell.2009.07.004
Hu B, Wang W, Ou S, Tang J, Li H, Che R, Zhang Z, Chai X, Wang H, Wang Y, Liang C (2015) Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47(7):834–838. https://doi.org/10.1038/ng.3337
Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41(4):494–497. https://doi.org/10.1038/ng.352
Ivanova KA, Chernova EN, Kulaeva OA, Tsyganova AV, Kusakin PG, Russkikh IV, Tikhonovich IA, Tsyganov VE (2022) The regulation of pea (Pisum sativum L.) symbiotic nodule infection and defense responses by glutathione, homoglutathione, and their ratio. Front Plant Sci. https://doi.org/10.3389/fpls.2022.843565
Izawa Y, Takayanagi Y, Inaba N, Abe Y, Minami M, Fujisawa Y, Kato H, Ohki S, Kitano H, Iwasaki Y (2010) Function and expression pattern of the α subunit of the heterotrimeric G protein in rice. Plant Cell Physiol 51(2):271–281. https://doi.org/10.1093/pcp/pcp186
Jia H, Song G, Werth EG, Walley JW, Hicks LM, Jones AM (2019) Receptor-like kinase phosphorylation of Arabidopsis Heterotrimeric G-protein Gα-subunit AtGPA1. Proteomics 19(24):1900265. https://doi.org/10.1002/pmic.201900265
Jose J, Choudhury SR (2020) Heterotrimeric G-proteins mediated hormonal responses in plants. Cell Signal 76:109799. https://doi.org/10.1016/j.cellsig.2020.109799
Kanamori N, Madsen LH, Radutoi S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, Jensen TH (2006) A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci 103(2):359–364. https://doi.org/10.1073/pnas.0508883103
Kaur J, Roy Choudhury S, Vijayakumar A, Hovis L, Rhodes Z, Polzin R, Blumenthal D, Pandey S (2018) Arabidopsis type III Gγ protein AGG3 is a positive regulator of yield and stress responses in the model monocot Setaria viridis. Front Plant Sci 9:109. https://doi.org/10.3389/fpls.2018.00109
Kelly MN, Irving HR (2003) Nod factors activate both heterotrimeric and monomeric G-proteins in Vigna unguiculata (L.) Walp. Planta 216(4):674–685. https://doi.org/10.1007/s00425-002-0900-8
Kirienko AN, Porozov YB, Malkov NV, Akhtemova GA, Le Signor C, Thompson R, Saffray C, Dalmais M, Bendahmane A, Tikhonovich IA, Dolgikh EA (2018) Role of a receptor-like kinase K1 in pea Rhizobium symbiosis development. Planta 248(5):1101–1120. https://doi.org/10.1007/s00425-018-2944-4
Kohlen W, Ng JLP, Deinum EE, Mathesius U (2018) Auxin transport, metabolism, and signalling during nodule initiation: indeterminate and determinate nodules. J Exp Bot 69(2):229–244. https://doi.org/10.1093/jxb/erx308
Krapp A, David LC, Chardin C, Girin T, Marmagne A, Leprince AS, Chaillou S, Ferrario-Méry S, Meyer C, Daniel-Vedele F (2014) Nitrate transport and signalling in Arabidopsis. J Exp Bot 65(3):789–798. https://doi.org/10.1093/jxb/eru001
Laloum T, Baudin M, Frances L, Lepage A, Billault-Penneteau B, Cerri MR, Ariel F, Jardinaud MF, Gamas P, de Carvalho-Niebel F, Niebel A (2014) Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J 79(5):757–768. https://doi.org/10.1111/tpj.12587
Laporte P, Lepage A, Fournier J, Catrice O, Moreau S, Jardinaud MF, Mun JH, Larrainzar E, Cook DR, Gamas P, Niebel A (2014) The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J Exp Bot 65(2):481–494. https://doi.org/10.1093/jxb/ert392
Lazdunski AM, Ventre I, Sturgis JN (2004) Regulatory circuits and communication in Gram-negative bacteria. Nat Rev Microbiol 2(7):581–592. https://doi.org/10.1038/nrmicro924
Léran S, Edel KH, Pervent M, Hashimoto K, Corratgé-Faillie C, Offenborn JN, Tillard P, Gojon A, Kudla J, Lacombe B (2015) Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci Signal 8(375):ra43. https://doi.org/10.1126/scisignal.aaa4829
Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, Dénarié J (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303(5662):1361–1364. https://doi.org/10.1126/science.1093038
Li M, Hong Y, Wang X (2009) Phospholipase D-and phosphatidic acid-mediated signaling in plants. Biochim Biophys Acta 1791(9):927–935. https://doi.org/10.1016/j.bbalip.2009.02.017
Li Y, Lv X, Rui M, Hu J, Volkov VS, Zeng D, Wang Y (2023) Rice dep1 variety maintains larger stomatal conductance to enhance photosynthesis under low nitrogen conditions. Crop Des. https://doi.org/10.1016/j.cropd.2023.100025
Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S, Zhang Y (2016) Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. Elife 5:e13568. https://doi.org/10.7554/eLife.13568
Liang Y, Zhao X, Jones AM, Gao Y (2018) G proteins sculp root architecture in response to nitrogen in rice and Arabidopsis. Plant Sci 274:129–136. https://doi.org/10.1016/j.plantsci.2018.05.019
Linkohr BI, Williamson LC, Fitter AH, Leyser HO (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29(6):751–760. https://doi.org/10.1046/j.1365-313X.2002.01251.x
Liu CW, Murray JD (2016) The role of flavonoids in nodulation host-range specificity: an update. Plants 5(3):33. https://doi.org/10.3390/plants5030033
Liu Q, Chen X, Wu K, Fu X (2015) Nitrogen signaling and use efficiency in plants: what’s new? Curr Opin Plant Biol 27:192–198. https://doi.org/10.1016/j.pbi.2015.08.002
Liu KH, Niu Y, Konishi M, Wu Y, Du H, Sun Chung H, Li L, Boudsocq M, McCormack M, Maekawa S, Ishida T (2017) Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 545(7654):311–316. https://doi.org/10.1038/nature22077
Liu Z, Zhao Y, Wang X, Yang M, Guo C, Xiao K (2018) TaNBP1, a guanine nucleotide-binding subunit gene of wheat, is essential in the regulation of N starvation adaptation via modulating N acquisition and ROS homeostasis. BMC Plant Biol 18:1–14. https://doi.org/10.1186/s12870-018-1374-6
Lopez-Gomez M, Sandal N, Stougaard J, Boller T (2012) Interplay of flg22-induced defence responses and nodulation in Lotus japonicus. J Exp Bot 63(1):393–401. https://doi.org/10.1093/jxb/err291
Lou F, Abramyan TM, Jia H, Tropsha A, Jones AM (2020) An atypical heterotrimeric Gα protein has substantially reduced nucleotide binding but retains nucleotide-independent interactions with its cognate RGS protein and Gβγ dimer. J Biomol Struct Dyn 38(17):5204–5218. https://doi.org/10.1080/07391102.2019.1704879
Lu S, Bahn SC, Qu G, Qin H, Hong Y, Xu Q, Zhou Y, Hong Y, Wang X (2013) Increased expression of phospholipase Dα1 in guard cells decreases water loss with improved seed production under drought in Brassica napus. Plant Biotechnol J 11(3):380–389. https://doi.org/10.1111/pbi.12028
Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425(6958):637–640. https://doi.org/10.1038/nature02045
Mahmud K, Makaju S, Ibrahim R, Missaoui A (2020) Current progress in nitrogen fixing plants and microbiome research. Plants 9(1):97. https://doi.org/10.3390/plants9010097
Majumdar P, Torres Rodríguez MD, Pandey S (2023) Role of heterotrimeric G-proteins in improving abiotic stress tolerance of crop plants. J Plant Growth Regul. https://doi.org/10.1007/s00344-023-10965-6
Mandal VK, Jangam AP, Chakraborty N, Raghuram N (2022) Nitrate-responsive transcriptome analysis reveals additional genes/processes and associated traits viz height, tillering, heading date, stomatal density and yield in japonica rice. Planta 255(2):42. https://doi.org/10.1007/s00425-021-03816-9
Maruta N, Trusov Y, Brenya E, Parekh U, Botella JR (2015) Membrane-localized extra-large G proteins and Gβγ of the heterotrimeric G proteins form functional complexes engaged in plant immunity in Arabidopsis. Plant Physiol 167(3):1004–1016. https://doi.org/10.1104/pp.114.255703
Maruta N, Trusov Y, Chakravorty D, Urano D, Assmann SM, Botella JR (2019) Nucleotide exchange–dependent and nucleotide exchange–independent functions of plant heterotrimeric GTP-binding proteins. Sci Signal. https://doi.org/10.1126/scisignal.aav9526
Maruta N, Trusov Y, Jones AM, Botella JR (2021a) Heterotrimeric G proteins in plants: canonical and atypical Gα subunits. Int J Mol Sci 22(21):11841. https://doi.org/10.3390/ijms222111841
Maruta N, Trusov Y, Urano D, Chakravorty D, Assmann SM, Jones AM, Botella JR (2021b) GTP binding by Arabidopsis extra-large G protein 2 is not essential for its functions. Plant Physiol 186(2):1240–1253. https://doi.org/10.1093/plphys/kiab119
Mathesius U (2018) Flavonoid functions in plants and their interactions with other organisms. Plants 7(2):30. https://doi.org/10.3390/plants7020030
Maxwell CA, Hartwig UA, Joseph CM, Phillips DA (1989) A chalcone and two related flavonoids released from alfalfa roots induce nod genes of Rhizobium meliloti. Plant Physiol 91(3):842–847. https://doi.org/10.1104/pp.91.3.842
Mbengue MD, Hervé C, Debellé F (2020) Nod factor signaling in symbiotic nodulation. Adv Bot Res 94:1–39. https://doi.org/10.1016/bs.abr.2019.10.002
McIntire WE (2009) Structural determinants involved in the formation and activation of G protein βγ dimers. Neurosignals 17(1):82–99. https://doi.org/10.1159/000186692
Misra S, Wu Y, Venkataraman G, Sopory SK, Tuteja N (2007) Heterotrimeric G-protein complex and G-protein-coupled receptor from a legume (Pisum sativum): role in salinity and heat stress and cross-talk with phospholipase C. Plant J 51:656–669. https://doi.org/10.1111/j.1365-313X.2007.03169.x
Moran AW, Daly K, Al-Rammahi MA, Shirazi-Beechey SP (2021) Nutrient sensing of gut luminal environment. Proc Nutr Soc 80(1):29–36. https://doi.org/10.1017/S0029665120007120
Móring A, Hooda S, Raghuram N, Adhya TK, Ahmad A, Bandyopadhyay SK, Barsby T, Beig G, Bentley AR, Bhatia A, Dragosits U (2021) Nitrogen challenges and opportunities for agricultural and environmental science in India. Front Sustain Food Syst 5:505347. https://doi.org/10.3389/fsufs.2021.505347
Muratore C, Espen L, Prinsi B (2021) Nitrogen uptake in plants: the plasma membrane root transport systems from a physiological and proteomic perspective. Plants 10(4):681. https://doi.org/10.3390/plants10040681
Murray JD, Liu CW, Chen Y, Miller AJ (2017) Nitrogen sensing in legumes. J Exp Bot 68(8):1919–1926. https://doi.org/10.1093/jxb/erw405
Nakagawa T, Kaku H, Shimoda Y, Sugiyama A, Shimamura M, Takanashi K, Yazaki K, Aoki T, Shibuya N, Kouchi H (2011) From defense to symbiosis: limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume–Rhizobium symbiosis. Plant J 65(2):169–180. https://doi.org/10.1111/j.1365-313X.2010.04411.x
Pacheco R, Quinto C (2022) Phospholipase Ds in plants: their role in pathogenic and symbiotic interactions. Plant Physiol Biochem 173:76–86. https://doi.org/10.1016/j.plaphy.2022.01.025
Pan H, Wang D (2017) Nodule cysteine-rich peptides maintain a working balance during nitrogen-fixing symbiosis. Nat Plants 3(5):1–6. https://doi.org/10.1038/nplants.2017.48
Pandey S (2019) Heterotrimeric G-protein signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. https://doi.org/10.1146/annurev-arplant-050718-100231
Pandey S (2020) Plant receptor-like kinase signaling through heterotrimeric G-proteins. J Exp Bot 71(5):1742–1751. https://doi.org/10.1093/jxb/eraa016
Pandey S, Vijayakumar A (2018) Emerging themes in heterotrimeric G-protein signaling in plants. Plant Sci 270:292–300. https://doi.org/10.1016/j.plantsci.2018.03.001
Pathak RR, Mandal VK, Jangam AP, Sharma N, Madan B, Jaiswal DK, Raghuram N (2021) Heterotrimeric G-protein α subunit (RGA1) regulates tiller development, yield, cell wall, nitrogen response and biotic stress in rice. Sci Rep 11(1):1–19. https://doi.org/10.1038/s41598-021-81824-1
Pecrix Y, Staton SE, Sallet E, Lelandais-Brière C, Moreau S, Carrère S et al (2018) Whole-genome landscape of Medicago truncatula symbiotic genes. Nat Plants 4:1017–1025. https://doi.org/10.1038/s41477-018-0286-7
Peleg-Grossman S, Volpin H, Levine A (2007) Root hair curling and Rhizobium infection in Medicago truncatula are mediated by phosphatidylinositide-regulated endocytosis and reactive oxygen species. J Exp Bot 58(7):1637–1649. https://doi.org/10.1093/jxb/erm013
Pellizzaro A, Alibert B, Planchet E, Limami AM, Paven ML (2017) Nitrate transporters: an overview in legumes. Planta 246(4):585–595. https://doi.org/10.1007/s00425-017-2724-6
Pingret JL, Journet EP, Barker DG (1998) Rhizobium Nod factor signaling: evidence for a G protein-mediated transduction mechanism. Plant Cell 10:659–671. https://doi.org/10.1105/tpc.10.5.659
Rahman MA, Lee SH, Ji HC, Kabir AH, Jones CS, Lee KW (2018) Importance of mineral nutrition for mitigating aluminum toxicity in plants on acidic soils: current status and opportunities. Int J Mol Sci 19(10):3073. https://doi.org/10.3390/ijms19103073
Reinprecht Y, Schram L, Marsolais F, Smith TH, Hill B, Pauls KP (2020) Effects of nitrogen application on nitrogen fixation in common bean production. Front Plant Sci 11:1172. https://doi.org/10.3389/fpls.2020.01172
Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A (2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci 103(50):19206–19211. https://doi.org/10.1073/pnas.0605275103
Robertson GP, Groffman PM (2007) Nitrogen transformations. Soil microbiology, ecology and biochemistry. Academic Press, London, pp 341–364. https://doi.org/10.1016/B978-0-08-047514-1.50017-2
Rogato A, Valkov VT, Alves LM, Apone F, Colucci G, Chiurazzi M (2016) Down-regulated Lotus japonicus GCR1 plants exhibit nodulation signalling pathways alteration. Plant Sci 247:71–82. https://doi.org/10.1016/j.plantsci.2016.03.007
Roy S, Liu W, Nandety RS, Crook A, Mysore KS, Pislariu CI, Frugoli J, Dickstein R, Udvardi MK (2020) Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32(1):15–41. https://doi.org/10.1105/tpc.19.00279
Roy Choudhury S, Pandey S (2016) The role of PLDα1 in providing specificity to signal-response coupling by heterotrimeric G-protein components in Arabidopsis. Plant J 86:50–61. https://doi.org/10.1111/tpj.13151
Roy Choudhury S, Marlin MA, Pandey S (2019) The role of Gβ protein in controlling cell expansion via potential interaction with lipid metabolic pathways. Plant Physiol 179(3):1159–1175. https://doi.org/10.1104/pp.18.01312
Rutten PJ, Poole PS (2019) Oxygen regulatory mechanisms of nitrogen fixation in rhizobia. Adv Microb Physiol 75:325–389. https://doi.org/10.1016/bs.ampbs.2019.08.001
Santos-Briones C, Cárdenas L, Estrada-Navarrete G, Santana O, Minero-García Y, Quinto C, Sánchez F (2009) GTPγS antagonizes the mastoparan-induced in vitro activity of PIP2-phospholipase C from symbiotic root nodules of Phaseolus vulgaris. Physiol Plant 135(3):237–245. https://doi.org/10.1111/j.1399-3054.2008.01184.x
Siderovski DP, Willard FS (2005) The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. https://doi.org/10.17615/5a9d-2173
Singh J, Verma PK (2023) Role of Nod factor receptors and its allies involved in nitrogen fixation. Planta 257(3):54. https://doi.org/10.1007/s00425-023-04090-7
Singh S, Katzer K, Lambert J, Cerri M, Parniske M (2014) CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15:139–152. https://doi.org/10.1016/j.chom.2014.01.011
Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145(1):183–191. https://doi.org/10.1104/pp.107.100495
Stateczny D, Oppenheimer J, Bommert P (2016) G protein signaling in plants: minus times minus equals plus. Curr Opin Plant Biol 34:127–135. https://doi.org/10.1016/j.pbi.2016.11.001
Subramaniam G, Trusov Y, Lopez-Encina C, Hayashi S, Batley J, Botella JR (2016) Type B heterotrimeric G protein γ-subunit regulates auxin and ABA signaling in tomato. Plant Physiol 170(2):1117–1134. https://doi.org/10.1104/pp.15.01675
Sugiyama A, Shitan N, Yazaki K (2008) Signaling from soybean roots to rhizobium: an ATP-binding cassette-type transporter mediates genistein secretion. Plant Signal Behav 3(1):38–40. https://doi.org/10.4161/psb.3.1.4819
Sun J, Miwa H, Downie JA, Oldroyd GE (2007) Mastoparan activates calcium spiking analogous to Nod factor-induced responses in Medicago truncatula root hair cells. Plant Physiol 144(2):695–702. https://doi.org/10.1104/pp.106.093294
Sun H, Qian Q, Wu K, Luo J, Wang S, Zhang C, Ma Y, Liu Q, Huang X, Yuan Q, Han R (2014) Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat Genet 46(6):652–656. https://doi.org/10.1038/ng.2958
Sun S, Wang L, Mao H, Shao L, Li X, Xiao J, Ouyang Y, Zhang Q (2018) A G-protein pathway determines grain size in rice. Nat Commun 9(1):851. https://doi.org/10.1038/s41467-018-03141-y
Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, Gentzbittel L, Childs KL, Yandell M, Gundlach H, Mayer KF (2014) An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genom 15(1):1–14. https://doi.org/10.1186/1471-2164-15-312
Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, Downie A (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441(7097):1153–1156. https://doi.org/10.1038/nature04862
Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, Anderson D, Chen JG, Jones AM, Botella JR (2007) Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. Plant Cell 19(4):1235–1250. https://doi.org/10.1105/tpc.107.050096
Tsyganova AV, Kitaeva AB, Tsyganov VE (2018) Cell differentiation in nitrogen-fixing nodules hosting symbiosomes. Funct Plant Biol 45(2):47–57. https://doi.org/10.1071/FP16377
Ullah H, Chen JG, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM (2003) The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15(2):393–409. https://doi.org/10.1105/tpc.006148
Urano D, Jackson D, Jones AM (2015) A G protein alpha null mutation confers prolificacy potential in maize. J Exp Bot 66(15):4511–4515. https://doi.org/10.1093/jxb/erv215
Urano D, Leong R, Wu TY, Jones AM (2020) Quantitative morphological phenomics of rice G protein mutants portend autoimmunity. Dev Biol 457(1):83–90. https://doi.org/10.1016/j.ydbio.2019.09.007
Venkateshwaran M, Cosme A, Han L, Banba M, Satyshur KA, Schleiff E, Parniske M, Imaizumi-Anraku H, Ané JM (2012) The recent evolution of a symbiotic ion channel in the legume family altered ion conductance and improved functionality in calcium signaling. Plant Cell 24(6):2528–2545. https://doi.org/10.1105/tpc.112.098475
Wang Y, Botella JR (2022) Heterotrimeric G protein signaling in abiotic stress. Plants 11(7):876. https://doi.org/10.3390/plants11070876
Wang YY, Hsu PK, Tsay YF (2012) Uptake, allocation and signaling of nitrate. Trends Plant Sci 17(8):458–467. https://doi.org/10.1016/j.tplants.2012.04.006
Wang D, Couderc F, Tian CF, Gu W, Liu LX, Poinsot V (2018) Conserved composition of nod factors and exopolysaccharides produced by different phylogenetic lineage Sinorhizobium strains nodulating Soybean. Front Microbiol 9:2852. https://doi.org/10.3389/fmicb.2018.02852
Wu Q, Regan M, Furukawa H, Jackson D (2018) Role of heterotrimeric Gα proteins in maize development and enhancement of agronomic traits. PLoS Genet 14(4):e1007374. https://doi.org/10.1371/journal.pgen.1007374
Xie Y, Nan Y, Atif A, Hu W, Zhang Y, Tian H, Gao Y (2022) Molecular identification of the G-protein genes and their expression profiles in response to nitrogen deprivation in Brassica napus. Int J Mol Sci 23(15):8151. https://doi.org/10.3390/ijms23158151
Xu H, Zhao M, Zhang Q, Xu Z, Xu Q (2016a) The DENSE AND ERECT PANICLE 1 (DEP1) gene offering the potential in the breeding of high-yielding rice. Breed Sci 66(5):659–667. https://doi.org/10.1270/jsbbs.16120
Xu Q, Zhao M, Wu K, Fu X, Liu Q (2016b) Emerging insights into heterotrimeric G protein signaling in plants. J Genet Genomics 43(8):495–502. https://doi.org/10.1016/j.jgg.2016.06.004
Yan Y, Zhang W, Li Y, He C, Gao L, Yu X (2018) Functions of CsGPA1 on the hypocotyl elongation and root growth of cucumbers. Sci Rep 8(1):1–12. https://doi.org/10.1038/s41598-018-33782-4
Yang J, Lan L, Jin Y, Yu N, Wang D, Wang E (2022) Mechanisms underlying legume–Rhizobium symbioses. J Integr Plant Biol 64(2):244–267. https://doi.org/10.1111/jipb.13207
Yuan P, Luo F, Gleason C, Poovaiah BW (2022) Calcium/calmodulin-mediated microbial symbiotic interactions in plants. Front Plant Sci. https://doi.org/10.3389/fpls.2022.984909
Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279(5349):407–409. https://doi.org/10.1126/science.279.5349.407
Zhang DP, Zhou Y, Yin JF, Yan XJ, Lin S, Xu WF, Baluška F, Wang YP, Xia YJ, Liang GH, Liang JS (2015) Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation. J Exp Bot 66(20):6371–6384. https://doi.org/10.1093/jxb/erv350
Zhang G, Yang J, Chen X, Zhao D, Zhou X, Zhang Y, Wang X, Zhao J (2021) Phospholipase D-and phosphatidic acid-mediated phospholipid metabolism and signaling modulate symbiotic interaction and nodulation in soybean (Glycine max). Plant J 106(1):142–158. https://doi.org/10.1111/tpj.15152
Zhao J, Wang X (2004) Arabidopsis phospholipase Dα1 interacts with the heterotrimeric G-protein α-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J Biol Chem 279(3):1794–1800. https://doi.org/10.1074/jbc.M309529200
Zhong Y, Tian J, Li X, Liao H (2022) Cooperative interactions between nitrogen fixation and phosphorus nutrition in legumes. New Phytol 237(3):734–745. https://doi.org/10.1111/nph.18593
Acknowledgements
Authors acknowledge the University Grant Commission, New Delhi, India for funding this research.
Funding
No funding was received to assist with the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
NT, DB and RKS conceptualised the idea. SS and JG contributed equally to write the manuscript. SS, JG and JS designed the figures. NT, DB, RKS and JS edited the manuscript. All the authors read and approved it for publication.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Gerhard Leubner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sharma, S., Ganotra, J., Samantaray, J. et al. An emerging role of heterotrimeric G-proteins in nodulation and nitrogen sensing. Planta 258, 101 (2023). https://doi.org/10.1007/s00425-023-04251-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04251-8