Abstract
Main conclusion
The nitrate transporters, belonging to NPF and NRT2 families, play critical roles in nitrate signaling, root growth and nodule development in legumes.
Nitrate plays an essential role during plant development as nutrient and also as signal molecule, in both cases working via the activity of nitrate transporters. To date, few studies on NRT2 or NPF nitrate transporters in legumes have been reported, and most of those concern Lotus japonicus and Medicago truncatula. A molecular characterization led to the identification of 4 putative LjNRT2 and 37 putative LjNPF gene sequences in L. japonicus. In M. truncatula, the NRT2 family is composed of 3 putative members. Using the new genome annotation of M. truncatula (Mt4.0), we identified, for this review, 97 putative MtNPF sequences, including 32 new sequences relative to previous studies. Functional characterization has been published for only two MtNPF genes, encoding nitrate transporters of M. truncatula. Both transporters have a role in root system development via abscisic acid signaling: MtNPF6.8 acts as a nitrate sensor during the cell elongation of the primary root, while MtNPF1.7 contributes to the cellular organization of the root tip and nodule formation. An in silico expression study of MtNPF genes confirmed that NPF genes are expressed in nodules, as previously shown for L. japonicus, suggesting a role for the corresponding proteins in nitrate transport, or signal perception in nodules. This review summarizes our knowledge of legume nitrate transporters and discusses new roles for these proteins based on recent discoveries.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
World agricultural production has increased markedly since the 1950s, as part of an intensive production model to ensure food security. However, these intensive systems have serious negative consequences for the environment. In fact, it has been estimated that 85–90 million tons of nitrogen fertilizers are added to the soil around the world every year (Good et al. 2004). However, the sustainability of intensive production is increasingly challenged. In the context of plant nitrogen nutrition, the use of green manure, as cover crops, is increasing during crop rotation: species of the Fabaceae family, more commonly referred to as legumes, are sown only to be subsequently crushed and incorporated into the soil. Thus, legumes contribute to nitrogen soil enrichment, but they also represent a key source of protein for human and animal nutrition through forage legumes such as alfalfa and soybean.
Many minerals, such as nitrogen (N), have a considerable impact on plant development, especially of the root system development, right from the early stages after seed germination (López-Bucio et al. 2003). Plants can absorb both organic and inorganic nitrogen forms. However, there is no direct evidence that organic nitrogen contributes significantly to the nitrogen nutrition of the plant (Näsholm et al. 2009). Thus, most of the nitrogen taken-up by the plant is in an inorganic form, mainly nitrate (NO3 −) and ammonium (NH4 +). For most cultivated crops, nitrate is the major source of nitrogen for plant growth (Crawford and Glass 1998). However, plants grow under fluctuating environmental conditions and are consequently exposed to frequent changes in mineral nutrient availability. To respond to the availability of nitrate in the soil, plants adapt their root system architecture (Zhang and Forde 2000) and their absorption mechanisms.
In legumes, nitrate transporters have received little attention, perhaps because legumes have the ability to establish symbioses with Rhizobium bacteria for fixing atmospheric N2. Effective symbiotic interaction is supported by a complex network of nutrient exchange between the two partners, plant and Rhizobium, and is controlled by a well-regulated carbon/nitrogen balance (Lodwig et al. 2003; Libault 2014). Indeed, photosynthate and other plant nutrients are transported to nodules to reduce optimally atmospheric N2 (Lodwig et al. 2003; Libault 2014). However, nitrate uptake remains important for legumes, especially during early seedling growth when the ability to fix atmospheric nitrogen has not developed. Moreover, this nitrate absorption by the root system appears essential for legumes since some environmental factors have been reported to interfere with nodule establishment and functioning, and consequently with plant nutrition (Marino et al. 2007). Understanding nitrate uptake and its control of plant development are important challenges as we seek to optimize legume growth by controlling nitrogen input. This review aims to provide an update on current knowledge on nitrate transporters in legumes and their roles in nitrate signal transduction and plant development.
Molecular basis of nitrate transport in legumes
Our understanding of the molecular basis of nitrogen transport in higher plants has increased markedly since the identification of the first gene encoding a nitrate transporter initially named CHL1 (Chlorate resistant 1; Tsay et al. 1993). Many transporters are now characterized, transporting inorganic and organic nitrogen, both in some model species and plants of agronomic interest. In higher plants, nitrate transporters, or channels, belong to five families of proteins: nitrate transporter1/peptide transporter family (NPF), nitrate transporter2 (NRT2), chloride channel (CLC), aluminum-activated malate transporter (ALMT) and slow anion channel-associated 1 homolog 3 (SLAC1/SLAH3). Given that only members of the NPF and NRT2 families have been shown to be involved in nitrate uptake in roots (see review by Nacry et al. 2013), the present review focuses on these two families.
NPF family
Recently, a unified nomenclature, NPF, has been proposed for the nitrate transporter1/peptide transporter (NRT1/PTR) family (Léran et al. 2014). Previous studies have shown that a wide variety of molecules, such as nitrate, abscisic acid (ABA), auxin, dipeptide or glucosinolates, is transported by members of this NPF family, with some members able to transport two different substrates (see review by Léran et al. 2014).
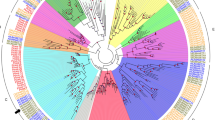
In legumes, limited data are available on NPFs, unlike with Arabidopsis thaliana in which 18 of the 53 AtNPF members have been characterized as nitrate and/or dipeptide transporters (see review by Wang et al. 2012; Hsu and Tsay 2013). All those characterized as nitrate transporters are low-affinity nitrate transporters, except AtNPF6.8 (AtNRT1.1; CHL1) which is a dual-affinity nitrate transporter (Liu et al. 1999). In Glycine max, a cDNA encoding a putative NPF has been cloned, but functional characterization has not been published (Yokoyama et al. 2001). Except for this result, our knowledge comes from studies on two model legumes, Lotus japonicus and Medicago truncatula. A molecular characterization of the NPF family in L. japonicus led to the identification of 37 putative LjNPF sequences (Criscuolo et al. 2012). Up to now, no gene encoding an NPF has been isolated from L. japonicus. Concerning M. truncatula, the Mt3.5 annotation version of the sequenced genome enabled identification of 80 genes encoding putative NPFs, belonging to eight subfamilies (Léran et al. 2014). With the new genome annotation of M. truncatula (Mt4.0; Tang et al. 2014), we are able to identify 97 putative MtNPF sequences predicted to encode proteins ranging from 388 to 647 amino acids (Supplementary Table SI). Among the 80 sequences previously identified by Léran et al. (2014), 65 were retained in this study and 15 were deleted, either because the amino acid sequence was too small (less than 215 amino acids) or because the sequence no longer existed in the Mt4.0 version genome annotation. So, through this present study, 32 new sequences of putative MtNPFs have been identified. The 97 putative MtNPF sequences are distributed in eight clades (Fig. 1) as previously described by Léran et al. (2014) for the 80 MtNPF sequences. Such a phylogenetic study can be taken as an insightful clue to start an analysis of the protein family. For example, following a phylogenetic study on nitrate and peptide transporters, including several species of angiosperms, it has been proposed that each separate group of NPF protein sequences identified within the NPF family shares common function (von Wittgenstein et al. 2014). However, although phylogenetically close, two genes may exhibit different patterns of expression. We investigated the in silico expression profiles of 44 MtNPF, available among the 97 MtNPFs genes of M. truncatula, by analysis of transcript microarray data (Fig. 2). Some genes were specifically expressed during nodule development (for example MtNPF4.7), seed development (for example MtNPF4.12), or in a particular plant tissue (for example MtNPF6.8). It can be noted that genes belonging to the same clade (same color in Fig. 2) are not necessary clustered. Two phylogenetically close proteins can also have different substrates. Indeed, AtNPF7.3 (AtNRT1.5) and OsNPF7.3 (OsPTR6), although belonging to the same clade (Léran et al. 2014), specifically transport nitrate (Lin et al. 2008) or dipeptides (Ouyang et al. 2010), respectively. So, it seems difficult to speculate on the nature of the substrate transported by these NPFs, or on their substrate affinity, without experimental characterization. Among genes encoding MtNPFs in M. truncatula, two genes have been extensively studied and characterized as being nitrate transporters. The first gene, MtNPF1.7 (LATD/NIP), was cloned by Yendrek et al. (2010). The second gene, MtNPF6.8 (MtNRT1.3), was isolated following a quantitative genetic study of post-germination stage seedlings (Morère-Le Paven et al. 2011). The potential roles of these two nitrate transporters in the plant are discussed in the following sections of this review.
Phylogenetic tree of MtNPF family of the model plant M. truncatula. Research in silico protein sequences was performed using the BLAST algorithm of the specific Medicago JCVI database (http://www.jcvi.org/medicago/) using the last version of the annotation genome (Mt4.0) (Tang et al. 2014). The phylogenetic tree was obtained through MEGA5 software. The analysis consists of 97 protein sequences identified as putative MtNPFs and grouped into eight clades numbered from 1 to 8 (Léran et al. 2014). The proteins belonging to a same clade are of the same color. The newly identified sequences, through the Mt4.0 genome annotation, are presented in light color. Black boxes indicated nitrate transporters already characterized
Expression profile cluster analysis of MtNPF genes. The expression data were clustered using Genesis software and distributed according to the tissues (A), nodule development (B) and seed development (C). The gene names are of the same color as in Fig. 1. DPI days after inoculation, DAP days after pollination. Microarray data come from Benedito et al. (2008)
NRT2 family
In legumes, a cDNA encoding a putative high-affinity nitrate transporter of G. max has been cloned (Amarasinghe et al. 1998). Subsequently, two studies were published describing the identification and molecular characterizations of the NRT2 gene family; one using L. japonicus, the other M. truncatula (Criscuolo et al. 2012; Pellizzaro et al. 2015). In L. japonicus, interrogation of the genome database led to the identification of four putative LjNRT2 named LjNRT2.1, LjNRT2.2, CM0001.20 and CM0161.180 (Criscuolo et al. 2012). Using similar approaches, Pellizzaro et al. (2015) revealed, for M. truncatula, a small NRT2 gene family composed of three MtNRT2 genes (named MtNRT2.1, MtNRT2.2 and MtNRT2.3), indicating a similar NRT2 family size in legume or non-legume species. Indeed, seven AtNRT2 genes were identified in A. thaliana (Orsel et al. 2002a, b; Okamoto et al. 2003) and four in rice (Araki and Hasegawa 2006; Cai et al. 2008; Feng et al. 2011). In these non-legume species, all characterized NRT2s have been reported to be strictly high-affinity nitrate transporters (Feng et al. 2011; Krapp et al. 2014). The function of almost all NRT2 of higher plants depends on a small protein of about 200 amino acids called NAR2 (NRT3), which interacts directly with NRT2 (see review by Krapp et al. 2014). In M. truncatula, in silico search revealed an MtNAR2 gene family composed only of two genes (named MtNAR2.1 and MtNAR2.2) (Pellizzaro et al. 2015), as in A. thaliana and O. sativa (Okamoto et al. 2003; Cai et al. 2008).
Function of nitrate transport in legumes
Role of transporters in HATS and LATS
To respond to soil nitrate availability, which can vary from μM to mM, plants adapt their absorption system. On the basis of their kinetics, two nitrate absorption systems have been described in many species: (a) a high-affinity transport system (HATS), and (b) a low-affinity transport system (LATS). The HATS is involved in nitrate uptake at low external nitrate concentrations (as low as 1 µM) while the LATS operates at higher concentration (>0.5 mM) (see review by Nacry et al. 2013). Both systems possess a constitutive component (cHATS; cLATS) and an inducible component (iHATS; iLATS) (Siddiqi et al. 1989, 1990; Faure-Rabasse et al. 2002).
Among the nine LjNPF genes studied in L. japonicus, expression of only CM0608.1210 was induced by nitrate. However, without biochemical information and genetic characterizations, the authors were not able to define the functional role in LATS (Criscuolo et al. 2012). In M. truncatula, both MtNPFs studied were expressed and characterized in Xenopus oocytes. The first studied protein, MtNPF1.7, was demonstrated to be a high-affinity nitrate transporter (Bagchi et al. 2012). This result indicates that the classification made until now (indicating that low-affinity nitrate transporters belong to the NPF family and high-affinity transporters belong to the NRT2 family) is too simplistic. MtNPF1.7 can restore chlorate sensitivity in resistant mutants of A. thaliana, but is not involved in the nitrate uptake in M. truncatula (Bagchi et al. 2012; Salehin et al. 2013). The second studied protein, MtNPF6.8, has the proprieties of a dual-affinity nitrate transporter when expressed in Xenopus oocytes (Morère-Le Paven et al. 2011). However, MtNPF6.8 does not contribute to HATS in planta (neither the constitutive, nor the inducible component) (Pellizzaro et al. 2014). Indeed, MtNPF6.8 is specifically involved in the inducible component of the LATS. The knockdown of MtNPF6.8 affected the entire inducible component of the LATS, suggesting that MtNPF6.8 is an essential actor of the inducible LATS (Pellizzaro et al. 2014). So, to date, both studied nitrate transporters from M. truncatula, MtNPF6.8 and MtNPF1.7, can transport nitrate at low concentrations when expressed in heterologous transport system, without having a HATS activity in planta (Morère-Le Paven et al. 2011; Bagchi et al. 2012; Pellizzaro et al. 2014). Interestingly, AtNPF6.3 of A. thaliana, known to be a dual-affinity nitrate transporter in oocytes, has been shown to be involved in HATS in planta (Wang et al. 1998; Liu et al. 1999; Ho et al. 2009), although this is a matter of debate (Glass and Kotur 2013).
Concerning NRT2, at least in post-germinative M. truncatula seedlings, MtNRT2.1 is expressed predominantly in roots and is induced by nitrate (Pellizzaro et al. 2015). Based on the observation that, in A. thaliana, a strong correlation exists between induction of gene expression in response to nitrate and HATS activity (Krouk et al. 2006), it was suggested that MtNRT2.1 could be a nitrate transporter with a major contribution to HATS activity in M. truncatula (Pellizzaro et al. 2015). In addition, co-expression of MtNRT2.1 and MtNAR2 suggests interaction of these two proteins for nitrate uptake (Pellizzaro et al. 2015). In L. japonicus, two of the four NRT2 genes, LjNRT2.1 and CM0161.180, showed a major increase of transcript abundance in the roots of plants grown under low nitrate concentrations (Criscuolo et al. 2012), suggesting involvement of both putative nitrate transporters in the HATS activity.
Long-distance nitrate transport
It is now accepted that the role of nitrate transporters extends beyond their function in the active uptake of nitrate from the environment (Wang et al. 2012). Indeed, nitrate transporters are expressed in all plant organs, allowing both long-distance transport of nitrate, and tailored delivery to cell types. Provision of nitrate to aerial organs first requires xylem loading by nitrate transporters (Lin et al. 2008; Li et al. 2010; Wang and Tsay 2011) and, therefore, identification of the transporters responsible may be assisted by knowledge of the tissue patterning of gene expression. MtNPF6.8 is expressed in shoots as well as in the pericycle region in roots (Pellizzaro et al. 2014). In addition, study of MtNPF6.8 expression in 28 day-old plants (Benedito et al. 2008; Fig. 2) or in 2-month-old plants (Morère-Le Paven, personal data) showed the presence of transcripts in all plant organs and mainly in the leaves. While analyses suggest that MtNPF6.8 does not participate significantly in nitrate translocation at the seedling stage (10 day-old plants), it is possible the functional contribution changes in more advanced developed stages. Concerning the NRT2 family, MtNRT2.3 is expressed in both roots and shoots, a pattern similar to that observed for OsNRT2.3 in O. sativa, or AtNRT2.4 in A. thaliana, suggesting a role in the transport of nitrate inside the plant from the root to shoot (Feng et al. 2011; Kiba et al. 2012; Pellizzaro et al. 2015). Furthermore, the tissue-wide expression pattern of MtNAR2.2 suggests that this protein may interact with MtNRT2.3 for this function (Pellizzaro et al. 2015).
Nitrate transporters and nitrate signal perception
Nitrate is not only a nutrient; it is also a molecular signal governing root development and massive reprogramming of gene expression (see review by Nacry et al. 2013). In this context, it is appropriate to discuss these signaling aspects and to highlight the molecular and physiological responses, the molecular players and the signaling pathways likely involved in nitrate signaling in legumes.
Primary nitrate response
It is well established that nitrate is a signal molecule inducing massive and rapid change in gene expression (Wang et al. 2003). Following a period of nitrate deficiency, new nitrate supply, even at low concentrations (250 µM nitrate), induces rapid (within 30 min) changes in the expression of over 1000 genes. This response is named the Primary Nitrate Response (PNR) (Wang et al. 2003; Krapp et al. 2014; Medici and Krouk 2014). In M. truncatula, the PNR has been studied (summarized on Fig. 3), especially in the nodule after 4 h or 8 h of nitrate supply (Cabeza et al. 2014) where massive regulation of gene expression, affecting 20% of all genes expressed in the treatments and/or control, was observed. The expression of some genes involved in nitrate transport (encoding low- and high-affinity nitrate transporters) or nitrate assimilation (encoding the nitrate reductase (NR) for example) is up-regulated (Fig. 3). Cabeza et al. (2014) have also identified modulation of gene expression induced by nitrate that impacts the activity of the nodules. For example, all nine of the expressed genes for leghemoglobin, the symbiotic hemoglobin involved in the scavenging of O2 and its transport for symbiotic nitrogen fixation, were down-regulated. Furthermore, strong evidences suggesting that the PNR in M. truncatula is mediated by MtNPF6.8 are provided (Fig. 3) (Pellizzaro et al. 2014). Indeed, nitrate induction of NR genes is impaired in npf6.8 mutants at 5 mM but also at 250 µM of nitrate supply. Since MtNPF6.8 does not influence the nitrate uptake at 250 µM of nitrate supply nor the global nitrogen status of the plant, it is possible that the role of MtNPF6.8 in response to nitrate is independent of its nitrate transport function. So, MtNPF6.8 could be a nitrate transceptor, having a nitrate-transporter and a nitrate-sensor function (Pellizzaro et al. 2014), as found for AtNPF6.3 (Ho et al. 2009; Ho and Tsay 2010). In addition, Pellizzaro et al. (2014) noticed that the induction of NR1 and NR2 was reduced in npf6.8 mutants by 60%, suggesting the existence of other nitrate sensors in M. truncatula (Fig. 3). Finally, the capacity of MtNPF6.8 to transport ABA, and its involvement in the PNR as nitrate-sensor, suggest a possible role of ABA in the PNR signaling response (Fig. 3). This hypothesis is supported by recent results showing that, in A. thaliana, ABI2 (ABA-insensitive 2; a protein phosphatase 2C) enhanced AtNPF6.3-dependent nitrate transport, nitrate sensing and nitrate signaling (Léran et al. 2015).
Schematic representation of primary nitrate response (PNR) in M. truncatula. Results obtained in roots indicated that MtNPF6.8 is a receptor of the PNR; MtNPF6.8 has a role in nitrate signaling inducing changes in gene expression involved in nitrate assimilation (Pellizzaro et al. 2014). Other nitrate sensors may also exist. The involvement of ABA in the signaling pathway remains a hypothesis. Results obtained in nodules after nitrate supply (4 or 8 h) show some genes involved in nitrate transport or assimilation/reduction are up-regulated while other genes, involved in the activity of the nodules, are down-regulated (Cabeza et al. 2014). PM plasma membrane
Regulation of root system architecture
Developmental plasticity enables plants to respond to nutrient availability by changing the architecture of their root system (Zhang and Forde 2000; Linkohr et al. 2002). Nitrate is one of the major nutrients known to act as a signal molecule in the regulation of root architecture through the regulation of primary root growth and by influencing the emergence and development of lateral roots (LR) (Zhang and Forde 1998, 2000; Zhang et al. 1999; Tian et al. 2008; Vidal et al. 2010; Celis-Arámburo et al. 2011). This effect is complicated due to the changes in root architecture caused by changes in root environment and their influence on plant physiology. For example, hormones coordinate the responses modulated by external environment, regulating both primary root growth and LR initiation and development. Auxin (indole-3-acetic acid, IAA) and ABA have both been proposed to play a role in mediating nitrate effects on LR development (Signora et al. 2001; De Smet et al. 2006; Walch-Liu et al. 2006; Zhang et al. 2007).
In M. truncatula, studies (summarized on Fig. 4) have shown that both characterized nitrate transporters, MtNPF1.7 and MtNPF6.8, play a role in root development (Harris and Dickstein 2010; Bagchi et al. 2012; Pellizzaro et al. 2014). MtNPF1.7 is expressed in the tips of primary and lateral roots (including the root cap, meristem and proximal portion of the elongation zone) as well as in the nodule meristem (Fig. 4) (Yendrek et al. 2010). Accordingly, MtNPF1.7 plays a role in perception of ABA (Yendrek et al. 2010) and contributes to LR formation, nodule meristem development and primary root growth organization of the root tip (Veereshlingam et al. 2004; Liang et al. 2007). MtNPF1.7 expression is controlled by several hormones, in particular by ABA, but is not regulated by nitrate (Yendrek et al. 2010). Interestingly, root development defect of mutants affected in MtNPF1.7 (noted latd) can be rescued by ABA addition (Liang et al. 2007), indicating a positive role of ABA in the establishment, or the maintenance, of root meristem function (Fig. 4) (Harris 2015). Since ABA levels in latd mutants are indistinguishable from those in wild type (Liang et al. 2007), the defect in latd mutants is likely to be in ABA transport or signaling. However, to date, MtNPF1.7 has not been shown to be an ABA transporter. Recently, Zhang et al. (2014) demonstrated that ABA restores root length in the latd mutant by reducing ROS accumulation levels and by increasing cell length demonstrating an important role of MtRbohC (encoding a NADPH oxidase) as a negative regulator of root elongation. Zhang et al. (2014) propose that one hypothesis to explain this finding could be that MtNPF1.7 and ABA act in parallel to regulate ROS levels and root elongation (Fig. 4).
Role of both characterized nitrate transporters of M. truncatula, MtNPF1.7 and MtNPF6.8, on root system development. The data concerning MtNPF6.8 and MtNPF1.7 are indicated on the left and on the right, respectively. Regions, where genes are expressed, are indicated in red. MtNPF6.8 is a nitrate sensor acting on cell elongation of the primary root. ABA acts downstream of MtNPF6.8 in the signaling pathway (Pellizzaro et al. 2014). MtNPF1.7 contributes to nodule meristem development, LR formation and primary root growth organization of the root tip (see review by Harris 2015). Zhang et al. (2014) propose that one hypothesis is that ABA and MtNPF1.7 could function in parallel to modulate root elongation via reactive oxygen species (ROS) generated by MtRbohC NADPH oxidase
Nitrate perceived by a NPF transporter and nitrate impact on root architecture have been studied in M. truncatula. In this species, nitrate has an inhibitory effect on primary root growth at low and high concentrations in two different ecotypes, A17 and R108 (Yendrek et al. 2010; Morère-Le Paven et al. 2011; Pellizzaro et al. 2014). A quantitative genetic approach allowed us to identify a major QTL involved in the control of M. truncatula radicle elongation in post-germination phase in both N-free medium and medium supplied with 5 mM nitrate. The MtNPF6.8 gene, which coincided with the peak of the QTL, appeared to be a significant candidate involved in the control of primary root growth and nitrate-sensing (Morère-Le Paven et al. 2011). Further experiments using npf6.8 mutants indicated that MtNPF6.8 has the hallmarks of a nitrate sensor (Fig. 4), which can regulate primary root growth via control of primary root cell elongation (Pellizzaro et al. 2014). The fact that MtNPF6.8 expression was stimulated by ABA, which also restored the inhibitory effect of nitrate on primary root growth in npf6.8 mutants, allowed us to hypothesize that ABA acts downstream MtNPF6.8 in this nitrate-signaling pathway. MtNPF6.8 was also demonstrated to be an ABA facilitator/transporter when expressed in Xenopus oocytes, confirming the ability of a single protein to transport both nitrate and ABA as previously shown for the AtNPF4.6 transporter of A. thaliana (Huang et al. 1999; Kanno et al. 2012). However, no direct link between a nitrate signal mediated by MtNPF6.8 acting on the primary root growth and ABA transport has been demonstrated.
In summary, in M. truncatula, two nitrate transporters, MtNPF1.7 and MtNPF6.8, are implicated in primary root growth by regulating root cell length, both in connection with ABA signaling. As noted by Harris (2015), it is surprising that 10 µM ABA rescues the wild type phenotype either by increasing (for latd mutants) or decreasing (for nitrate-treated npf6.8 mutants) root length, suggesting contradictory results (Fig. 4). However, the genotypes used in both cases were different. It is also important to note that studies on MtNPF6.8 concern nitrate effect on primary root length, since MtNPF6.8 was shown to function as a nitrate sensor. The role of MtNPF1.7 in nitrate sensing is still unknown. Finally, whether MtNPF6.8 and MtNPF1.7 contribute to the same signaling pathway in root growth control is still to be unravelled.
Nitrate transporters and their possible roles in rhizobial symbiosis
In legumes, it has been determined that nitrate is involved in the regulation of nodulation (Heath et al. 2010; Saito et al. 2014). Nitrate transporters are thus likely to have important roles to play during symbiosis establishment. Indeed, in a recent study, Damiani et al. (2016) analysed the root hair transcriptome from roots of control M. truncatula or following treatment with nod-factors (NF). NF are signaling molecules produced by rhizobia, inducing many of the plant responses during the early stages of plant-bacteria interaction. The authors paid particular attention to proteins likely to play a role in nutrient ion uptake. Medtr5g012290 (also named MtNPF6.7; Léran et al. 2014), a homolog of AtNPF6.3, was the most highly expressed NPF member in M. truncatula root hairs, while MtNPF1.7 expression was 6 times lower than that of MtNPF6.7 and MtNPF6.8 was not expressed. Upon NF treatment, the expression of MtNPF6.7 was up-regulated (Damiani et al. 2016). In the NRT2 family, MtNRT2.1 and MtNRT2.2 are also expressed in root hairs and up-regulated upon NF treatment (Damiani et al. 2016). In the present study, the microarray data analysis showed that many M. truncatula NPF genes are expressed in nodules (Fig. 2). For some genes (NPF4.7, NPF3.2, NPF7.4, NPF5.4, NPF7.6, NPF3.3, NPF1.4, NPF5.11, NPF1.2 and NPF5.6), the expression in nodules (Fig. 2B) is higher than in other plant tissues (Fig. 2A), or during seed development (Fig. 2C). Furthermore in M. truncatula, MtNRT2s and MtNAR2s are expressed in mature nodules (Criscuolo et al. 2012; Pellizzaro et al. 2015). This expression of NPF and NRT2 genes in nodules raise the question of a possible role of the corresponding transporters in nitrate transport inside the nodule. However, direct involvement of nitrate transporters in the exchange of nutrients between roots and nodules has never been reported.
One might ask, what are the roles attributed to these transporters in nodules, if they are not involved in transporting nitrate? In M. truncatula, it has been shown that MtNPF1.7 is essential in the formation and maintenance of nodule meristems and in rhizobial invasion during nodulation (Veereshlingam et al. 2004; Harris and Dickstein 2010). Using different mutants of MtNPF1.7, Bagchi et al. (2012) proposed that the ability of MtNPF1.7 to transport nitrate is correlated with the abilities of mutants to form and maintain nodules. However, these same authors also proposed that MtNPF1.7 should have another unknown activity, besides nitrate transport, in nodule development and in root architecture. MtNPF1.7 may be important for nitrate redistribution within the plant or could transport nitrate that would be useful as a precursor for the synthesis of nitric oxide (NO), a potent signaling molecule shown to be an important regulatory player in the Rhizobium-legume symbiotic interaction (Hichri et al. 2015). Otherwise, since new nodule formation is partially dependent on the suppression of lateral root emergence, MtNPF1.7, which is expressed in both of these organs, could play a role in coordinating development of lateral roots and nodules (Harris and Dickstein 2010). Alternatively, the role of MtNPF1.7 in ABA signaling in root meristem function, as shown by Liang et al. (2007), suggests that nodule formation, controlled by MtNPF1.7, may require ABA (Harris and Dickstein 2010). High expression of MtNRT2.1 and MtNRT2.3 in M. truncatula nodules indicates that nitrate transporters of the MtNRT2 family could also be involved in signaling pathways governing nodule formation, development and/or function (Pellizzaro et al. 2015). The involvement of nitrate transporters in the establishment of the symbiotic interaction has also been proposed after studying some members of the NPF and NRT2 families in another species of legume, L. japonicus (Criscuolo et al. 2012). These authors suggested the possible involvement of LjNRT2 and LjNPF in the high and/or low nitrate-signaling pathways affecting nodule formation, but also a possible role for a LjNPF, named CM0826.370, in mature nodules. All these data provide a basis for future experiments that will help to understand the possible roles of these nitrate transporters in the nodule.
Conclusion
Fundamental studies to understand mechanisms that govern nitrate transport and signaling offer prospects to better control nitrogen use efficiency in plants. Mining of extant datasets enables identification of candidate genes and consideration of their possible roles in planta. To date, few studies of legume nitrate transporters have been undertaken, and thus much remains undiscovered, in particular the role of these transporters in nodules.
Experimental characterization of nitrate transporters is usually performed under optimal or at least unchallenging conditions. However, plants are continuously subjected to ever changing environmental conditions. Their sessile lifestyle exposes them to unavoidable frequent and severe climatic changes and attacks by herbivores and microorganisms. It is well known that legume plants adapt their root architecture and their metabolism to abiotic stress through signaling pathways involving ABA and NO (Planchet et al. 2011, 2014). Given that studies on both characterized nitrate transporters, MtNPF1.7 and MtNPF6.8, connect these NPFs with root architecture and ABA, it will be valuable to fully investigate the role of legume NPFs in response to challenging environmental conditions.
Author contribution statement
AP designed the outline of the article. BA investigated the in silico gene expression profiles. AP and BA composed figures and the supplementary table. AP, EP, AL and MCML wrote the manuscript.
References
Amarasinghe BH, de Bruxelles GL, Braddon M, Onyeocha I, Forde BG, Udvardi MK (1998) Regulation of GmNRT2 expression and nitrate transport activity in roots of soybean (Glycine max). Planta 206:44–52
Araki R, Hasegawa H (2006) Expression of rice (Oryza sativa L.) genes involved in high-affinity nitrate transport during the period of nitrate induction. Breed Sci 56:295–302
Bagchi R, Salehin M, Adeyemo OS, Salazar C, Shulaev V, Sherrier DJ, Dickstein R (2012) Functional assessment of the Medicago truncatula NIP/LATD protein demonstrates that it is a high-affinity nitrate transporter. Plant Physiol 160:906–916. doi:10.1104/pp.112.196444
Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, Moreau S, Niebel A, Frickey T, Weiller G, He J, Dai X, Zhao PX, Tang Y, Udvardi MK (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55:504–513. doi:10.1111/j.1365-313X.2008.03519.x
Cabeza R, Koester B, Liese R, Lingner A, Baumgarten V, Dirks J, Salinas-Riester G, Pommerenke C, Dittert K, Schulze J (2014) An RNA sequencing transcriptome analysis reveals novel insights into molecular aspects of the nitrate impact on the nodule activity of Medicago truncatula. Plant Physiol 164:400–411. doi:10.1104/pp.113.228312
Cai C, Wang JY, Zhu YG, Shen QR, Li B, Tong YP, Li ZS (2008) Gene structure and expression of the high-affinity nitrate transport system in rice roots. J Integr Plant Biol 50:443–451. doi:10.1111/j.1744-7909.2008.00642.x
Celis-Arámburo TJ, Carrillo-Pech M, Castro-Concha LA, Miranda-Ham ML, Martínez-Estévez M, Echevarría-Machado I (2011) Exogenous nitrate induces root branching and inhibits primary root growth in Capsicum chinense Jacq. Plant Physiol Biochem 49:1456–1464. doi:10.1016/j.plaphy.2011.09.003
Crawford NM, Glass AD (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3:389–395
Criscuolo G, Valkov VT, Parlati A, Alves LM, Chiurazzi M (2012) Molecular characterization of the Lotus japonicus NRT1(PTR) and NRT2 families. Plant Cell Environ 35:1567–1581. doi:10.1111/j.1365-3040.2012.02510.x
Damiani I, Drain A, Guichard M, Balzergue S, Boscari A, Boyer JC, Brunaud V, Cottaz S, Rancurel C, Da Rocha M, Fizames C, Fort S, Gaillard I, Maillol V, Danchin EGJ, Rouached H, Samain E, Su YH, Thouin J, Touraine B, Puppo A, Frachisse JM, Pauly N, Sentenac H (2016) Nod Factor effects on root hair-specific transcriptome of Medicago truncatula: focus on plasma membrane transport systems and reactive oxygen species networks. Front Plant Sci 7:1–22. doi:10.3389/fpls.2016.00794
De Smet I, Zhang H, Inzé D, Beeckman T (2006) A novel role for abscisic acid emerges from underground. Trends Plant Sci 11:434–439. doi:10.1016/j.tplants.2006.07.003
Faure-Rabasse S, Le Deunff E, Lainé P, Macduff JH, Ourry A (2002) Effects of nitrate pulses on BnNRT1 and BnNRT2 genes: mRNA levels and nitrate influx rates in relation to the duration of N deprivation in Brassica napus L. J Exp Bot 53:1711–1721
Feng H, Yan M, Fan X, Li B, Shen Q, Miller AJ, Xu G (2011) Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot 62:2319–2332. doi:10.1093/jxb/erq403
Glass AD, Kotur Z (2013) A reevaluation of the role of Arabidopsis NRT1.1 in high-affinity nitrate transport. Plant Physiol 163:1103–1106. doi:10.1104/pp.113.229161
Good AG, Shrawat AK, Muench DG (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9:597–605. doi:10.1016/j.tplants.2004.10.008
Harris JM (2015) Abscisic acid: hidden architect of root system structure. Plants 4:548–572. doi:10.3390/plants4030548
Harris JM, Dickstein R (2010) Control of root architecture and nodulation by the LATD/NIP transporter. Plant Signal Behav 5:1365–1369
Heath KD, Stock AJ, Stinchcombe JR (2010) Mutualism variation in the nodulation response to nitrate. J Evol Biol 23:2494–2500. doi:10.1111/j.1420-9101.2010.02092.x
Hichri I, Boscari A, Castella C, Rovere M, Puppo A, Brouquisse R (2015) Nitric oxide: a multifaceted regulator of the nitrogen-fixing symbiosis. J Exp Bot 66:2877–2887. doi:10.1093/jxb/erv051
Ho CH, Tsay YF (2010) Nitrate, ammonium, and potassium sensing and signaling. Curr Opin Plant Biol 13:604–610. doi:10.1016/j.pbi.2010.08.005
Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138:1184–1194. doi:10.1016/j.cell.2009.07.004
Hsu PK, Tsay YF (2013) Two phloem nitrate transporters, NRT1.11 and NRT1.12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiol 163:844–856. doi:10.1104/pp.113.226563
Huang NC, Liu KH, Lo HJ, Tsay YF (1999) Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11:1381–1392
Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109:9653–9658. doi:10.1073/pnas.1203567109
Kiba T, Feria-Bourrellier AB, Lafouge F, Lezhneva L, Boutet-Mercey S, Orsel M, Bréhaut V, Miller A, Daniel-Vedele F, Sakakibara H, Krapp A (2012) The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24:245–258. doi:10.1105/tpc.111.092221
Krapp A, David LC, Chardin C, Girin T, Marmagne A, Leprince AS, Chaillou S, Ferrario-Méry S, Meyer C, Daniel-Vedele F (2014) Nitrate transport and signalling in Arabidopsis. J Exp Bot 65:789–798. doi:10.1093/jxb/eru001
Krouk G, Tillard P, Gojon A (2006) Regulation of the high-affinity NO3 − uptake system by NRT1.1-mediated NO3 − demand signaling in Arabidopsis. Plant Physiol 142:1075–1086. doi:10.1104/pp.106.087510
Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, Gassmann W, Geiger D, Gojon A, Gong JM, Halkier BA, Harris JM, Hedrich R, Limami AM, Rentsch D, Seo M, Tsay YF, Zhang M, Coruzzi G, Lacombe B (2014) A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci 19:5–9. doi:10.1016/j.tplants.2013.08.008
Léran S, Edel KH, Pervent M, Hashimoto K, Corratgé-Faillie C, Offenborn JN, Tillard P, Gojon A, Kudla J, Lacombe B (2015) Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci Signal 8:ra43. doi:10.1126/scisignal.aaa4829
Li JY, Fu YL, Pike SM, Bao J, Tian W, Zhang Y, Chen CZ, Li HM, Huang J, Li LG, Schroeder JI, Gassmann W, Gong JM (2010) The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22:1633–1646. doi:10.1105/tpc.110.075242
Liang Y, Mitchell DM, Harris JM (2007) Abscisic acid rescues the root meristem defects of the Medicago truncatula latd mutant. Dev Biol 304:297–307. doi:10.1016/j.ydbio.2006.12.037
Libault M (2014) The carbon-nitrogen balance of the nodule and its regulation under elevated carbon dioxide concentration. Biomed Res Int 2014:1–7. doi:10.1155/2014/507946
Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK, Tillard P, Lin HL, Wang YY, Tsai CB, Gojon A, Tsay YF (2008) Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20:2514–2528. doi:10.1105/tpc.108.060244
Linkohr BI, Williamson LC, Fitter AH, Leyser HM (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29:751–760
Liu KH, Huang CY, Tsay YF (1999) CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11:865–874
Lodwig EM, Hosie AH, Bourdès A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS (2003) Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422:722–726. doi:10.1038/nature01527
López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6:280–287
Marino D, Frendo P, Ladrera R, Zabalza A, Puppo A, Arrese-Igor C, González EM (2007) Nitrogen fixation control under drought stress. Localized or systemic? Plant Physiol 143:1968–1974. doi:10.1104/pp.106.097139
Medici A, Krouk G (2014) The Primary Nitrate Response: a multifaceted signalling pathway. J Exp Bot 65:5567–5576. doi:10.1093/jxb/eru245
Morère-Le Paven MC, Viau L, Hamon A, Vandecasteele C, Pellizzaro A, Bourdin C, Laffont C, Lapied B, Lepetit M, Frugier F, Legros C, Limami AM (2011) Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legume Medicago truncatula. J Exp Bot 62:5595–5605. doi:10.1093/jxb/err243
Nacry P, Bouguyon E, Gojon A (2013) Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 370:1–29
Näsholm T, Kielland K, Ganeteg U (2009) Uptake of organic nitrogen by plants. New Phytol 182:31–48. doi:10.1111/j.1469-8137.2008.02751.x
Okamoto M, Vidmar JJ, Glass AD (2003) Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol 44:304–317
Orsel M, Filleur S, Fraisier V, Daniel-Vedele F (2002a) Nitrate transport in plants: which gene and which control? J Exp Bot 53:825–833
Orsel M, Krapp A, Daniel-Vedele F (2002b) Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol 129:886–896. doi:10.1104/pp.005280
Ouyang J, Cai Z, Xia K, Wang Y, Duan J, Zhang M (2010) Identification and analysis of eight peptide transporter homologs in rice. Plant Sci 179:374–382
Pellizzaro A, Clochard T, Cukier C, Bourdin C, Juchaux M, Montrichard F, Thany S, Raymond V, Planchet E, Limami AM, Morère-Le Paven MC (2014) The nitrate transporter MtNPF6.8 (MtNRT1.3) transports abscisic acid and mediates nitrate regulation of primary root growth in Medicago truncatula. Plant Physiol 166:2152–2165. doi:10.1104/pp.114.250811
Pellizzaro A, Clochard T, Planchet E, Limami AM, Morère-Le Paven MC (2015) Identification and molecular characterization of Medicago truncatula NRT2 and NAR2 families. Physiol Plant 154:256–269. doi:10.1111/ppl.12314
Planchet E, Rannou O, Ricoult C, Boutet-Mercey S, Maia-Grondard A, Limami AM (2011) Nitrogen metabolism responses to water deficit act through both abscisic acid (ABA)-dependent and independent pathways in Medicago truncatula during post-germination. J Exp Bot 62:605–615. doi:10.1093/jxb/erq294
Planchet E, Verdu I, Delahaie J, Cukier C, Girard C, Morère-Le Paven MC, Limami AM (2014) Abscisic acid-induced nitric oxide and proline accumulation in independent pathways under water-deficit stress during seedling establishment in Medicago truncatula. J Exp Bot 65:2161–2170. doi:10.1093/jxb/eru088
Saito A, Tanabata S, Tanabata T, Tajima S, Ueno M, Ishikawa S, Ohtake N, Sueyoshi K, Ohyama T (2014) Effect of nitrate on nodule and root growth of soybean (Glycine max (L.) Merr.). Int J Mol Sci 15:4464–4480. doi:10.3390/ijms15034464
Salehin M, Huang YS, Bagchi R, Sherrier DJ, Dickstein R (2013) Allelic differences in Medicago truncatula NIP/LATD mutants correlate with their encoded proteins’ transport activities in planta. Plant Signal Behav 8:e22813. doi:10.4161/psb.22813
Siddiqi MY, Glass AD, Ruth TJ, Fernando M (1989) Studies of the regulation of nitrate influx by barley seedlings using 13NO3 −. Plant Physiol 90:806–813
Siddiqi MY, Glass AD, Ruth TJ, Rufty TW (1990) Studies of the uptake of nitrate in barley: I. Kinetics of 13NO3 − Influx. Plant Physiol 93:1426–1432
Signora L, De Smet I, Foyer CH, Zhang H (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 28:655–662
Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, Gentzbittel L, Childs KL, Yandell M, Gundlach H, Mayer KF, Schwartz DC, Town CD (2014) An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15:312. doi:10.1186/1471-2164-15-312
Tian Q, Chen F, Liu J, Zhang F, Mi G (2008) Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J Plant Physiol 165:942–951. doi:10.1016/j.jplph.2007.02.011
Tsay YF, Schroeder JI, Feldmann KA, Crawford NM (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72:705–713
Veereshlingam H, Haynes JG, Penmetsa RV, Cook DR, Sherrier DJ, Dickstein R (2004) nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol 136:3692–3702. doi:10.1104/pp.104.049064
Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107:4477–4482. doi:10.1073/pnas.0909571107
von Wittgenstein NJ, Le CH, Hawkins BJ, Ehlting J (2014) Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evol Biol 14:11. doi:10.1186/1471-2148-14-11
Walch-Liu P, Ivanov II, Filleur S, Gan Y, Remans T, Forde BG (2006) Nitrogen regulation of root branching. Ann Bot 97:875–881. doi:10.1093/aob/mcj601
Wang YY, Tsay YF (2011) Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23:1945–1957. doi:10.1105/tpc.111.083618
Wang R, Liu D, Crawford NM (1998) The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc Natl Acad Sci USA 95:15134–15139
Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132:556–567. doi:10.1104/pp.103.021253
Wang YY, Hsu PK, Tsay YF (2012) Uptake, allocation and signaling of nitrate. Trends Plant Sci 17:458–467. doi:10.1016/j.tplants.2012.04.006
Yendrek CR, Lee YC, Morris V, Liang Y, Pislariu CI, Burkart G, Meckfessel MH, Salehin M, Kessler H, Wessler H, Lloyd M, Lutton H, Teillet A, Sherrier DJ, Journet EP, Harris JM, Dickstein R (2010) A putative transporter is essential for integrating nutrient and hormone signaling with lateral root growth and nodule development in Medicago truncatula. Plant J 62:100–112. doi:10.1111/j.1365-313X.2010.04134.x
Yokoyama T, Kodama N, Aoshima H, Izu H, Matsushita K, Yamada M (2001) Cloning of a cDNA for a constitutive NRT1 transporter from soybean and comparison of gene expression of soybean NRT1 transporters. Biochim Biophys Acta 1518:79–86
Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279:407–409
Zhang H, Forde BG (2000) Regulation of Arabidopsis root development by nitrate availability. J Exp Bot 51:51–59
Zhang H, Jennings A, Barlow PW, Forde BG (1999) Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA 96:6529–6534
Zhang H, Rong H, Pilbeam D (2007) Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J Exp Bot 58:2329–2338. doi:10.1093/jxb/erm114
Zhang C, Bousquet A, Harris JM (2014) Abscisic acid and LATD/NIP modulate root elongation via reactive oxygen species in Medicago truncatula. Plant Physiol 166:644–658. doi:10.1104/pp.114.248542
Acknowledgements
Authors wish to thank Pr. David C. Logan (IRHS, Beaucouzé, France) for critical reading of the manuscript and English language correction. The work was supported by the QUALISEM research program, funded by Région Pays de Loire (France).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pellizzaro, A., Alibert, B., Planchet, E. et al. Nitrate transporters: an overview in legumes. Planta 246, 585–595 (2017). https://doi.org/10.1007/s00425-017-2724-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-017-2724-6