Abstract
Main Conclusion
Specific sequences within the leader intron of a soybean polyubiquitin gene stimulated gene expression when placed either within a synthetic intron or upstream of a core promoter.
The intron in the 5′ untranslated region of the soybean polyubiquitin promoter, Gmubi, seems to contribute to the high activity of this promoter. To identify the stimulatory sequences within the intron, ten different sequential intronic sequences of 40 nt were isolated, cloned as tetrameric repeats and placed upstream of a minimal cauliflower mosaic virus 35S (35S) core promoter, which was used to control expression of the green fluorescent protein. Intron fragment tetramers were also cloned within a modified, native intron, creating a Synthetic INtron Cassette (SINC), which was then placed downstream of Gmubi and 35S core promoters. Intron fragment tetramers and SINC constructs were evaluated using transient expression in lima bean cotyledons and stable expression in soybean hairy roots. Intron fragments, used as tetramers upstream of the 35S core promoter, yielded up to 80 times higher expression than the core promoter in transient expression analyses and ten times higher expression in stably transformed hairy roots. Tetrameric intronic fragments, cloned downstream of the Gmubi and 35S core promoters and within the synthetic intron, also yielded increased transient and stable GFP expression that was up to 4 times higher than Gmubi alone and up to 40 times higher than the 35S core promoter alone. These intron fragments contain sequences that seem to act as promoter regulatory elements and may contribute to the increased expression observed with this native strong promoter. Intron regulatory elements and synthetic introns may provide additional tools for increasing transgene expression in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As promoters are the primary regulators of gene expression at the transcriptional level, all organisms are dependent on their activation or deactivation for development and survival (Dean and Schmidt 1995). The mechanisms that are related to the control of gene expression, however, extend well beyond just the proximal promoter sequences and include an array of binding sites for transcription factors and enhancer proteins that increase or decrease gene expression (Potenza et al. 2004; Hernandez-Garcia and Finer 2014). One such repository for transcriptional regulation outside of promoter sequences is the intron.
Introns play important roles in gene regulation at various levels (Le Hir et al. 2003). They are recognized as the facilitators through which different proteins can be obtained from one transcribed genomic sequence through alternative splicing (Maniatis and Tasic 2002). Their role has expanded to include regulation of gene expression [termed intron-mediated enhancement (IME)] through a variety of mechanisms ranging from increased mRNA transcription, stability and accumulation (Rose and Last 1997; Lu et al. 2008) to increased RNA processing and export (Samadder et al. 2008).
To understand the effects of introns on gene expression, efforts have mainly focused on introns found in gene coding sequences, and evaluation of intron activity through deletion analyses of 5′ and 3′ splice sites and branch point sequences, general deletion analyses of entire or partial sequences, and intron translocation into a previously non-intron-containing sequence (Kim et al. 2006; Jeong et al. 2007; Lu et al. 2008). These studies have identified the mechanisms behind IME that are related to intron splicing, position, and orientation; however, the identification of several regulatory elements and the findings that gene expression was increased regardless of intron orientation, position, and splicing indicates that the mechanisms for intron enhancement are not completely defined (Rose 2008; Ibraheem et al. 2010; Rose et al. 2011). In addition, use of introns present in the 5′ untranslated region (UTR) often leads to increased expression of transgenes (Lu et al. 2008; Rose 2008; Ibraheem et al. 2010; Rose et al. 2011). Transcriptional enhancers have been identified in intron sequences by computational methods (Lu et al. 2008; Rose et al. 2011; Ibraheem et al. 2010), but enhancers and other gene regulatory features identified via computational approaches should be validated using gene expression analysis tools (Hernandez-Garcia and Finer 2014).
When used to regulate transgenes, the intron-containing Glycine max polyubiquitin promoter (Gmubi) (Hernandez-Garcia et al. 2009), like many other ubiquitin promoters, shows strong, constitutive expression following introduction, with up to seven times higher expression compared to the widely used Cauliflower Mosaic Virus 35S (35S) promoter. Removal of the 5′ UTR or “leader” intron led to reduced promoter activity (Hernandez-Garcia et al. 2009), while intron translocation or duplication either maintained or increased promoter activity (De La Torre and Finer 2015). Other polyubiquitin genes have also been evaluated and the leader introns appear to contribute to high gene expression levels (Plesse et al. 2001; Wang and Oard 2003; Kamo et al. 2012), which are reduced when the intron is removed, further demonstrating the importance of these leader introns in promoter activity.
Identification of regulatory sequences that affect gene expression has generally been focused on promoter sequences, as most cis-elements that facilitate high levels of expression are located in the proximal promoter region (Hernandez-Garcia and Finer 2014). Although bioinformatics approaches have been used to identify putative elements in the promoter, validation of the elements requires transgene expression approaches following modifications to putative promoter elements (Yang et al. 2013) or introduction of multimeric repeats of rather short sequences of either individual elements or individual components of a regulatory module, placed upstream of a core promoter (Salinas et al. 1992; Rushton et al. 2002; Sawant et al. 2005; Liu et al. 2011; Hernandez-Garcia and Finer 2016). These synthetic promoters, containing element multimers fused with core promoter sequences, are very sensitive to the presence of regulatory elements, and enhancements in gene expression are readily observed. The enhancing effect of particular sequences isolated from introns that are associated with promoters of highly expressed genes and their contribution to promoter activity has not been studied in this manner. The aim of this study was to identify and evaluate intronic sequences from the Gmubi promoter that had a major effect on transgene expression. Tetrameric repeats of specific intron fragments of the Gmubi promoter were generated and placed upstream of a 35S core promoter. These same tetrameric intron fragments (TIFs) were also cloned within a synthetic intron and placed downstream of a 35S core promoter and a Gmubi promoter. Following introduction of synthetic promoter and intron constructs, image analysis of gfp gene expression was used to quantify transgene expression.
Materials and methods
Synthetic promoter and synthetic intron cassette construction
For generation of tetrameric intron fragments (TIFs), ten sequential 40 nt fragments were generated from the intron sequence of the Gmubi promoter (Glyma20g27950.1) (Fig. 1). Complementary oligonucleotides of 45–46 nt (Supplemental Table 1) were designed to anneal to generate intron fragments with terminal restriction sites, which could be used to generate multimeric repeats according to Rushton et al. (2002). In brief, phosphorylated upper and lower strand oligonucleotides of each selected intron sequence was annealed to create intron fragments with an SpeI restriction site at the 5′ end and a XbaI restriction site at the 3′ end. These fragments were then ligated into pFLEV (Hernandez-Garcia et al. 2010a, b, Genbank Accession no. KX156843) upstream of the Cauliflower mosaic virus 35S core promoter (35SCore) regulating a gfp gene sGFP (S65T) (Chiu et al. 1996). Tetrameric intron fragments (TIFs) of each intron element were generated by digesting the vector with either SpeI or XbaI along with BglII and then ligating the fragments, which eliminated the SpeI/XbaI sites from the ligated ends, while leaving the undigested SpeI or XbaI sites for an additional round of multimerization (Fig. 2). This was repeated until tetramers of the intron fragments were recovered upstream of the 35S core promoter (Rushton et al. 2002) to generate the 35S core set of promoters containing each TIF (Fig. 2).
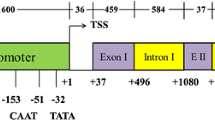
Distribution of intron sequences for generation of 40 nt intron fragments. A conserved sequence consisting of ATT(A/-)(G/C)TTCAGATCCGTT(A/G)A, found in intron fragments 7, 8, 9, 10, and just upstream of the ATG is bolded. The 5′ UTR is highlighted in black, while the predicted intron branch point is highlighted in gray. The predicted splice site is marked with a caret (^) and the ATG translational start is marked with an asterisk (*)
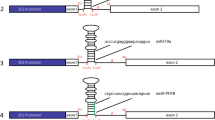
Constructs used for evaluation of TIFs. Schematic for 35SCore construct, 35SCore with tetrameric intron fragments (TIF) upstream of the core promoter, GmubiSINC (synthetic intron cassette) with TIF embedded within a synthetic intron and downstream of the Gmubi promoter, and 35SCoreSINC with TIF embedded within a synthetic intron and downstream of the 35SCore promoter
GmubiSINC (SINC = Synthetic Intron Cassette) was designed to receive the TIFs as an SpeI/XbaI fragment, inserted into a modified Gmubi intron, positioned downstream from its native Gmubi proximal promoter (Fig. 2). GmubiSINC was synthesized from a previously generated Gmubi stuffed intron construction (GmubiIN2X) (De La Torre and Finer 2015), which contained a multiple cloning site from pFLEV (Hernandez-Garcia et al. 2010a, b). Restriction sites BclI/NheI/Acc65I/SpeI and BglII/SalI/XbaI were added by PCR to a pFLEV-derived tetramer using oligos complementary to the 5′ and 3′ end of the tetramer. The resulting PCR product was purified, digested with BclI/BglII and ligated into a similarly digested GmubiIN2X leaving SpeI/XbaI sites for cloning TIFs. GmubiSINC therefore contained the Gmubi proximal promoter, the Gmubi intron with new TIF cloning sites, a gfp coding sequence, and the NOS terminator. The whole synthetic Gmubi intron-containing individual TIFs were subsequently amplified using SINC-XhoI-F and SINC-NcoI-R primers (Supplemental Table 2), and then cloned as a XhoI/NcoI fragment into a XhoI/NcoI digested GFP expression plasmid containing the 35S core promoter to generate 35SCoreSINC (Fig. 2). Successful cloning generated a series of 35S core promoter constructs containing synthetic introns with each TIF. A 35Score core construct containing the synthetic intron without a TIF (35SCoreIN) was also generated by similarly amplifying and cloning the intron sequence without a TIF.
For generation of hairy roots, the binary versions of each construct were created by cloning the promoter, synthetic intron, gfp coding sequence, and NOS terminator into pCAMBIA1300 (CAMBIA, Canberra, Australia). Fragments from GmubiSINC were PCR amplified using the primers GmubiSINC-PstI-F and GmubiSINC-Eco1-R (Supplemental Table 2), digested with PstI and EcoRI and then cloned into PstI/EcoRI-digested pCAMBIA1300. Fragments from the 35SCoreSINC constructs were directly transferred by digesting with PstI and EcoRI and ligating into pCAMBIA1300. All sequences and plasmids generated were confirmed by DNA sequencing. For stable expression in soybean hairy roots, the pCAMBIA1300 constructs were introduced into Agrobacterium rhizogenes K599 using the freeze–thaw method (Chen et al. 1994).
Transient expression analysis in lima bean cotyledons
Transient expression analysis of constructions was conducted using lima bean cotyledonary tissues as previously described (Hernandez-Garcia et al. 2010a, b; Chiera et al. 2007). Lima bean (Phaseolus lunatus L. “Henderson Bush”) seeds were surface sterilized in 4% (v/v) bleach solution with agitation for 20 min and then rinsed with sterile distilled water until the bleach odor was gone. Seeds were then placed between moistened paper towels in Magenta GA7 containers (16 h light; 40 μE m−2 s−1, 25 °C) for 4 days. Two hours prior to bombardment, cotyledons were excised from the germinating seeds and placed on a growth regulator-free culture medium (OMS) containing Murashige and Skoog salts (Murashige and Skoog 1962), B5 vitamins (Gamborg et al. 1968), 3% sucrose, and 0.2% Gelrite™ (Aceto Corporation, Lake Success, NY), pH 5.7. DNA constructs were precipitated unto tungsten particles as previously described (Chiera et al. 2007; Hernandez-Garcia et al. 2010a, b) and introduced into the adaxial surface of the lima bean cotyledonary tissue using a particle inflow gun (Finer et al. 1992). The bombarded cotyledons were then returned to OMS medium for monitoring of GFP expression using an automated image collection system consisting of an MZFLIII dissecting microscope (Leica, Heerbrugg, Switzerland) equipped with a GFP2 filter set, a Spot-RT CCD digital camera (Diagnostic Instruments Inc., Sterling Heights, MI), and a robotic platform (Arrick Robotics Inc., Hurst, TX). Images of each expressing cotyledon were collected hourly for 100 h. Quantitative analysis of GFP expression levels was done using Image J (Rasband 1997) as previously described (Chiera et al. 2007; Hernandez-Garcia et al. 2010a, b). At least six replications per construct were conducted with three replications per experiment. Expression values from 35SCoreSINC constructs with TIFs were calculated as the percent expression of the 35S core promoter at peak GFP expression. Expression values from the GmubiSINC constructs were calculated as the percent expression of the Gmubi promoter at peak GFP expression.
Stable expression evaluation using soybean hairy roots
Soybean hairy roots were generated and analyzed as previously described (Hernandez-Garcia et al. 2010a, b). Soybean (Glycine max cv. Williams 82) seeds were surface sterilized and placed in Magenta GA7 containers for germination as described above for the lima bean seeds. A. rhizogenes K599 cultures containing the promoter derivatives in pCAMBIA1300 were grown overnight in yeast extract peptone (YEP) medium containing 100 mg l−1 kanamycin. Agrobacterium cultures without the binary vector were grown in YEP without antibiotics. Cotyledons (5 days old) were excised and wounded on the abaxial side using a scalpel previously immersed in Agrobacterium. Inoculated cotyledons were placed abaxial side up on sterile moistened filter paper (P5 Fisherbrand, Fisher Scientific, Pittsburgh, PA). After 4 days of co-culture, cotyledons were transferred to OMS medium containing 400 mg l−1 Timentin for hairy root induction. After 2 weeks, roots were excised (~2 cm from the tip) and transferred to OMS medium containing 400 mg l−1 Timentin for an additional 4 days and then transferred to OMS medium containing 400 mg l−1 Timentin and 35 mg l−1 hygromycin for selection. Root tips were examined for GFP expression using the microscope and camera previously described for transient GFP detection, but without active robotic components (Hernandez-Garcia et al. 2010a, b). GFP quantification of 1600 × 1200 pixel images of individual root tips was conducted with the Image J software as previously described (Hernandez-Garcia et al. 2010a, b). For each construct, 15–20 independent hairy root events were analyzed over at least two experiments and statistical analyses were conducted using SAS (SAS Institute Inc, Cary, NC).
IMEter score
The enhancing ability of various forms of synthetic introns/intron fragments was evaluated using IMEter (Parra et al. 2011). IMEter scores were calculated by inputting sequences of the Gmubi intron, the synthetic tetrameric intron fragments alone (TIFs), or the synthetic intron cassettes containing a tetrameric intron fragment within (SINCTIFs). “Glycine max” (soybean) was selected as the species in the database to evaluate the enhancing ability of the intron/intron fragments.
Results
Transient expression with 35SCore constructs
Transient GFP expression analysis of the 35SCore constructions with tetrameric intron fragments (TIFs) upstream of the core promoter showed GFP fluorescence within 2–3 h of DNA introduction for most constructs (Fig. 3). All introductions displayed a similar pattern of expression and a common peak expression time point at 18 h, regardless of promoter strength (Fig. 3). All but one of the 35SCore constructs containing a TIF regulated GFP expression at least two times higher than the minimally expressing 35SCore promoter construct without a TIF. 35SCoreTIF7 and 35SCoreTIF10 were distinctly higher than the other TIFs, showing the highest expression, which was up to 69× and 84× higher than the 35SCore promoter alone, respectively. 35SCoreTIF8, 35SCoreTIF9 and 35SCoreTIF6 also gave high expression, with peak expression levels of approximately 12×, 10× and 7× higher than the 35SCore (Fig. 3). The remaining elements had similar expression values ranging between two and six times higher than the 35SCore, with the exception of 35SCoreTIF4, which showed the lowest expression reaching only 50% of peak 35SCore expression values.
Transient expression profiles for 35SCore constructs. GFP transient expression analysis of 35SCore constructs with tetrameric intron fragments (TIFs) upstream of the 35SCore promoter. GFP expression is shown as the percentage of peak GFP expression of the 35SCore promoter. Internal legends are shown from the highest to lowest expression. a The highest expressing constructs—note the y-axis maximum of 9000% expression of the 35SCore promoter. b The lowest expressing constructs—note the y-axis maximum of 1000% expression of the 35SCore promoter
Stable expression with 35SCore constructs
In stably transformed hairy roots containing the 35SCore constructs, most of the TIFs gave rise to similar GFP expression intensity when compared to the 35SCore promoter construct (Fig. 4). The highest expression in transgenic hairy roots was seen for 35SCoreTIF7 and 35SCoreTIF9 showing a 10×–15× increase in GFP expression levels compared to the 35SCore promoter construct. 35SCoreTIF10 and 35SCoreTIF8 displayed slightly higher levels of GFP expression compared to the remaining TIFs, but 35SCoreTIF8 was not significantly different from the 35SCore without a TIF (Tukey’s test, p ≤ 0.05, Fig. 4).
Transient expression with SINC constructs
The time course of transient GFP expression for all of the GmubiSINC constructs was very similar to the Gmubi promoter alone, with peak expression 20–30 h post-bombardment (Fig. 5). The Gmubi promoter alone showed much higher base expression levels compared to the 35SCore promoter, and expression with the GmubiSINC constructs was easy to visualize without the aid of automated image collection (Fig. 5). All of the GmubiSINC constructs showed the same or higher transient expression levels compared to the Gmubi promoter alone with four GmubiSINC constructs showing much higher to slightly higher expression. GmubiSINCTIF7 and GmubiSINCTIF9 both gave rise to exceptionally high levels of transient GFP expression, approximately six and five times higher than the Gmubi promoter alone (Fig. 5a). These two GmubiSINC constructs showed sustained high GFP expression, 100 h after bombardment. GmubiSINCTIF10 and GmubiSINCTIF8 had up to two times higher expression of GFP compared to Gmubi, while the remaining constructs showed similar levels of GFP expression to the Gmubi promoter (Fig. 5).
Transient expression profiles for GmubiSINCTIF constructs. GFP transient expression analysis (a, b graphs on left) for GmubiSINCTIF constructs introduced into lima bean cotyledons. GFP expression is shown as the percentage of peak GFP expression of the Gmubi promoter. Internal legends are shown from highest to lowest expression. a The highest expressing constructs—note the y-axis maximum of 600% expression of the Gmubi promoter. b The lowest expressing constructs—note the y-axis maximum of 100% expression of the Gmubi promoter. Right panel images show GFP expression in foci in lima bean cotyledons at peak time points for all GmubiSINCTIF constructs and for the 35SCore promoter construct
The time course of transient GFP expression was very similar for all of the 35SCoreSINC constructs with a peak expression at 16–20 h post-bombardment (Fig. 6). The intensities of GFP expression were dependent on which TIF was inserted into the synthetic intron. The highest transient GFP expression was seen for 35SCoreSINCTIF7, which was 40 times higher than the 35SCore promoter. Many of the other 35SCoreSINC constructs also gave increased expression ranging from 2× to 22× higher expression than the 35SCore promoter. A 35SCore construct with the non-synthetic Gmubi intron (35SCoreIN) placed downstream of the promoter gave relatively low expression, but it was higher than the 35SCore promoter alone, 35SCoreSINCTIF1 and 35SCoreSINCTIF4 (Fig. 6).
Transient expression profiles for 35SCoreSINC constructs. Transient GFP expression analysis for 35SCoreSINC constructs bombarded into lima bean cotyledons. GFP is expressed as a percentage of peak GFP expression of the 35S Core promoter. a The highest expressing constructs—note the y-axis maximum of 4000% expression of the 35SCore promoter. b The lowest expressing constructs—note the y-axis maximum of 2000% expression of the Gmubi promoter
Stable expression with SINC constructs
GFP expression in stably transformed soybean hairy roots generated with the GmubiSINC constructs was similar for most of the constructs. Only GmubiSINCTIF7 and GmubiSINCTIF9 showed significantly higher expression than the other Gmubi constructs, reaching 1.5×–2× higher expression compared to Gmubi alone (Tukey’s test, p ≤ 0.05; Fig. 7). Other high expressing constructs, which were not significantly different from Gmubi included GmubiSINCTIF8 and GmubiSINCTIF10.
Activity of GmubiSINC constructs in hairy roots. GFP expression analysis in stably transformed soybean hairy roots containing GmubiSINC constructs with TIFs. GFP intensity was quantified by ImageJ and is presented as grayscale mean ± SE. Columns with the same letter are not significantly different at p ≤ 0.05 (Tukey’s test)
For stable GFP expression in soybean hairy roots transformed with the 35SCoreSINC constructs, inclusion of TIFs yielded significantly higher expression compared to 35SCore promoter alone (Fig. 8). Inclusion of the Gmubi intron with the 35SCore construction (35SCoreIN) gave higher expression in hairy roots, but it was not significantly different from the 35SCore promoter. Variation in GFP expression among hairy roots events could be high, resulting in somewhat large standard error values in GFP expression intensity (Fig. 8). The highest GFP expression from the 35SCoreSINC constructs was obtained with 35SCoreSINCTIF7, which gave a 26× increase in GFP expression, followed by 35SCoreSINCTIF9, which showed a 21× increase, when compared with GFP expression obtained in hairy roots transformed with the 35SCore promoter. The remaining 35SCoreSINC constructs with TIFs showed from 12× to 20× higher GFP expression compared to the 35SCore promoter.
Activity of 35SCoreSINC constructs in hairy roots. GFP expression analysis in stably transformed soybean hairy roots containing 35SCoreSINC constructs with TIFs. GFP intensity was quantified by ImageJ software and is presented as grayscale mean ± SE. Columns with the same letter are not significantly different at p ≤ 0.05 (Tukey’s test)
IMEter score
The sequence of TIF1 yielded the highest IMEter score, followed by TIF10, TIF7 and TIF4 (Supplemental Table 3), while TIF5 and TIF9 showed no predicted intron enhancing ability, with IMEter scores of zero. For TIFs embedded in the synthetic intron cassette (SINCTIFs), the order of IMEter scores from high to low was similar to those of the TIFs alone, with the highest scores for SINCTIF10 and SCINTIF1 and the lowest for SINCTIF5 and SINCTIF9 (Supplemental Table 4). The predicted gene expression enhancing ability of the original Gmubi intron fell behind SINCTIF10, SINCTIF1, SINCTIF7 and SINCTIF4 (Supplemental Table 4).
Discussion
Most of the tetrameric intron fragments (TIFs) stimulated gene expression multiple fold compared to the constructs without the TIF (Figs. 3, 4, 5, 6, 7, 8). This increase was observed when the TIF was placed upstream of a core promoter or within a synthetic intron cassette (SINC). The large increase in GFP expression from placement of the TIFs upstream of the 35S minimal promoter (Figs. 3, 4) suggests that these fragments contain sequences that act like cis-regulatory elements (Rushton et al. 2002; Sawant et al. 2005; Liu et al. 2011; Hernandez-Garcia and Finer 2014). The highest increase in expression from upstream TIFs was always observed with fragments from the 3′ end of the intron (TIFs 7–10). The presence of promoter cis-regulatory elements within all or most of these intron fragments was not predicted by screening with plant cis-regulatory element databases (data not shown). Visual scanning of the four intronic sequences that gave the greatest increase in expression revealed a relatively large repeating sequence among these four fragments that has not been previously reported as a promoter regulatory element (Fig. 1, bold) [ATT(A/-)(G/C)TTCAGATCCGTT(A/G)A].
When the ten TIFs were evaluated upstream of the 35S core promoter, most showed increases in transient expression (Fig. 3). All TIFs led to increased expression in soybean hairy roots (Fig. 4), but only TIF9, TIF7 and TIF10 were significantly higher than the 35Score. Difficulty in detection of significant differences in expression seemed to result from the variability in expression levels among hairy roots events from the introduction of the same TIF. A high variability in gene expression from promoter introductions in hairy roots has been reported previously and resulted from variation in copy number among hairy root events (Hernandez-Garcia et al. 2010a, b). Although the trend of TIFs 7–10 providing the highest expression was clear, the specific TIF that gave the highest expression was different when the expression was quantified using the two different expression validation tools.
When TIFs were placed downstream of either a core 35S promoter or larger Gmubi promoter and within the synthetic intron cassette (SINC), GFP expression increased (Figs. 5, 6, 7, 8), consistent with the results obtained with upstream placement. Tissues containing the GmubiSINC constructs showed up to four times higher GFP expression compared to the highly expressing Gmubi promoter alone (Hernandez-Garcia et al. 2010a, b). The CaMV35SCoreSINC constructs showed 2–40 times higher GFP expression compared to the 35S core promoter. Higher fold increases with minimal or core promoter constructs are common as the baseline expression of core promoter constructs is so low (Figs. 3, 4, 5, 6, 7, 8) and the increases in expression are therefore more pronounced (Rushton et al. 2002; Sawant et al. 2005; Liu et al. 2011; Hernandez-Garcia and Finer 2014). Core promoters are often used as an element validation tool, because they are so sensitive to the presence of cis-regulatory elements. Interestingly, a large increase in gene expression was still observed when the full-length Gmubi promoter was used with TIFs in a synthetic intron. This suggests that further increases in the expression of highly active promoters are still possible by including additional regulatory sequences within synthetic introns. Similar to what was observed with the upstream placement of TIFs, sequences from the 3′ region of the intron gave significantly higher expression than sequences from the 5′ end when used as a TIF in a synthetic intron. Although polyubiquitin promoters from a variety of different plants have been extensively characterized (Plesse et al. 2001; Wang and Oard 2003; Sivamani and Qu 2006; Hernandez-Garcia et al. 2009; Kamo et al. 2012), most of these studies have simply shown that the leader intron in the 5′ UTR contributes in some way to high expression. In addition, studies on intron-mediated enhancement (IME) of gene expression have largely focused on how splicing affects gene expression, or the effects resulting from removal of all of parts of the intron (Rose 2002; Kim et al. 2006; Jeong et al. 2007; Lu et al. 2008). Using different derivatives of the same Gmubi promoter, De La Torre and Finer (2015) recently showed increased gene expression either from translocation of the intron upstream of the promoter or from generation of a stuffed intron, where a large fragment of the intron was inserted into the whole intron. In that study, it seemed that the intron contained enhancers or other cis-regulatory elements, but specific intron sequences were not further evaluated. In this present study, we were able to not only confirm which parts of the intron contributed to increased transgene expression, but showed that a synthetic intron cassette could serve as an additional tool to study intron sequences in the context of the intron. Although different types of synthetic promoters have been generated to study unique promoter sequences or proximal promoter-based cis-acting elements (Salinas et al. 1992; Sawant et al. 2005; Cazzonelli and Velten 2007; Mehrotra and Mehrotra 2010), we believe that we are the first to use this approach for the evaluation of sequences within a synthetic intron.
The mechanism behind the enhancement of gene expression from leader introns within the 5′ UTR may differ from the role of the introns found in gene coding sequences or the 3′ UTR (Rose et al. 2011; Chung et al. 2006). Chung et al. (2006) found that 5′ UTR introns from Arabidopsis were more frequently located proximal to the end of the UTR or closer to the transcriptional start site. These introns were also longer, possibly serving as spacers between the promoter and coding sequences. Evaluation of the IMEter scores for all TIFs or SINC constructs containing different TIFs showed high IMEter scores for TIF10 (SINCTIF10) and TIF7 (SINCTIF7), which also gave high gene expression in our expression assays (Supplemental Tables 3, 4; Figs. 3, 4, 5, 6, 7, 8). However, the IMEter scores for TIF1 (SINCTIF1), TIF4 (SINCTIF4), and TIF9 (SINCTIF9) were similarly high (Supplemental Tables 3, 4), but these TIFs did not yield comparably high levels of gene expression enhancement (Figs. 3, 4, 5, 6, 7, 8). The IMEter scoring system was established using introns from Arabidopsis thaliana, where the IME signals are preferentially located at the 5′ end of the intron (Rose 2004; Rose et al. 2008; Parra et al. 2011), which may be not always be the case for enhancing introns of other species. Leader introns in the promoters from the very highly expressing soybean GmScream promoter family also showed no correlation of IMEter score with high gene expression (Zhang et al. 2015).
IME could be facilitated at the transcriptional and post-transcriptional levels. Generation of hybrid introns containing ends from a COR15a intron with embedded sequences from the UBQ10 or ATPK1 intron in normal and reverse orientations led to increased expression, and the increase seemed to act at the DNA rather than the RNA level (Rose et al. 2011). In this study, we propose that the increase in gene expression mediated by the Gmubi intron is mostly due to transcriptional regulation because the intronic sequences increased expression when placed upstream of a minimal core promoter, outside of their native intron environment. In addition, use of multiple copies of intronic sequences within synthetic introns also led to increased gene expression, but the expression profile and level of gene expression between the placement of TIFs upstream of the intron (Figs. 3, 4) and within the intron (Figs. 6, 8) were not very different. Evaluation of intron sequences within synthetic introns and upstream of promoters should prove useful in both basic and applied research by identifying important sequences within the intron and providing additional means for modulating gene expression.
Author contribution statement
TG and CD jointly generated the TIFs, cloned them into the SINC vectors, and performed the transient and stable expression analyses. The concept of developing and evaluating SINCs was initiated by CD and JF. NZ performed analysis of IMEter for all SINC constructs. TG assembled the first completed manuscript draft. JF was the principal investigator on this work, contributed to project design, isolation of the SINC constructs, data analysis, and manuscript drafting. All authors read and approved the final manuscript.
Abbreviations
- 35S:
-
Cauliflower mosaic virus 35S
- 5′ UTR:
-
5′ Untranslated region
- Gmubi:
-
Glycine max polyubiquitin
- MCS:
-
Multiple cloning site
- MS:
-
Murashige and Skoog
- OMS:
-
MS medium containing no plant growth regulators
- pFLEV:
-
Finer laboratory expression vector
- SINC:
-
Synthetic intron cassette
- TIF:
-
Tetrameric intron fragment
References
Cazzonelli C, Velten J (2007) In vivo characterization of plant promoter element interaction using synthetic promoters. Trans Res 17:437–457
Chen H, Nelson RS, Sherwood JL (1994) Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16:664–670
Chiera JM, Bouchard RA, Dorsey SL, Park E, Buenrostro-Nava MT, Ling PP, Finer JJ (2007) Isolation of two highly active soybean (Glycine max (L.) Merr.) promoters and their characterization using a new automated image collection and analysis system. Plant Cell Rep 26:1501–1509
Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as vital reporter in plants. Curr Biol 6:325–330
Chung BYW, Simons C, Firth AE, Brown CM, Hellens RP (2006) Effect of 5´ UTR introns on gene expression in Arabidopsis thaliana. BMC Genom 7:120
De La Torre CM, Finer JJ (2015) The intron and 5′ distal region of the soybean Gmubi promoter contribute to very high levels of gene expression in transiently and stably transformed tissues. Plant Cell Rep 34:111–120
Dean C, Schmidt R (1995) Plant genomes: a current description. Ann Rev Plant Physiol Plant Mol Biol 46:395–418
Finer JJ, Vain P, Jones MW, McMullen MD (1992) Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep 11:323–328
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Hernandez-Garcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci 217:109–119
Hernandez-Garcia CM, Finer JJ (2016) A novel cis-acting element in the GmERF3 promoter contributes to inducible gene expression in soybean and tobacco after wounding. Plant Cell Rep 35:303–316
Hernandez-Garcia CM, Martinelli AP, Bouchard RA, Finer JJ (2009) A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep 28:837–849
Hernandez-Garcia CM, Bouchard RA, Rushton PJ, Jones ML, Chen X, Timko MP, Finer JJ (2010a) High level of transgenic expression of soybean (Glycine max) GmERF and Gmubi gene promoters isolated by a novel promoter analysis pipeline. BMC Plant Biol 10:237
Hernandez-Garcia CM, Chiera JM, Finer JJ (2010b) Robotics and dynamic image analysis for studies of gene expression in plant tissues. J Vis Exp. doi:10.3791/1733
Ibraheem O, Botha CEJ, Bradley G (2010) In silico analysis of cis-acting regulatory elements in 5′ regulatory regions of the sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comp Biol Chem 34:268–283
Jeong YM, Mun JH, Kim H, Lee S, Kim SG (2007) An upstream region in the first intron of petunia actin-depolymerizing factor 1 affects tissue-specific expression in transgenic Arabidopsis (Arabidopsis thaliana). Plant J 50:230–239
Kamo K, Kim A, Park SH, Joung YH (2012) The 5′ UTR-intron of the Gladiolus polyubiquitin promoter GUBQ1 enhances translation efficiency in Gladiolus and Arabidopsis. BMC Plant Biol 12:79
Kim MJ, Kim H, Shin JS, Chung C, Ohlrogge JB, Suh MC (2006) Seed-specific expression of sesame microsomal oleic acid desaturase is controlled by combinatorial properties between negative cis-regulatory elements in the SeFAD2 promoter and enhancers in the 5′-UTR intron. Mol Gen Genom 276:351–368
Le Hir H, Nott A, Moore MJ (2003) How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 28:215–220
Liu W, Mazarei M, Rudis MR, Fethe MH, Stewart CN (2011) Rapid in vivo analysis of synthetic promoters for plant pathogen phytosensing. BMC Biotechnol 11:108
Lu J, Sivamani E, Azhakanandam K, Sammadder P, Xianggan L, Qu R (2008) Gene expression enhancement mediated by the 5′ UTR intron of the rice rubi3 gene varied remarkably among tissues in transgenic rice plants. Mol Genet Genom 279:563–572
Maniatis T, Tasic B (2002) Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418:236–243
Mehrotra R, Mehrotra S (2010) Promoter activation by ACGT in response to salicylic and abscisic acids is differentially regulated by the spacing between two copies of the motif. J Plant Phys 167:1214–1218
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Parra G, Bradnam K, Rose AB, Korf I (2011) Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res 39:5328–5337
Plesse B, Criqui MC, Durr A, Parmentier Y, Fleck J, Genschik P (2001) Effects of the polyubiquitin gene Ubi.U4 leader intron and first ubiquitin monomer on reporter gene expression in Nicotiana tabacum. Plant Mol Biol 45:655–667
Potenza C, Aleman L, Sengupta-Gopalan C (2004) Targeting transgene expression in research, agricultural, and environmental applications: promoters used in plant transformation. In Vitro Cell Dev Biol Plant 40:1–22
Rasband W (1997) ImageJ. U.S. National Institutes of Health, Bethesda. http://imagej.nih.gov/ij/
Rose AB (2002) Requirements for intron-mediated enhancement of gene expression in Arabidopsis. RNA 8:1444–1453
Rose AB (2004) The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J 40:744–751
Rose AB (2008) Intron-mediated regulation of gene expression. In: Reddy ASN, Golovkin M (eds) Nuclear pre-mRNA processing in plants. Current topics in microbiology and immunology, vol 326. Springer, New York, pp 277–290
Rose AB, Last RL (1997) Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J 11:455–464
Rose AB, Elfersi T, Parra G, Korf I (2008) Promoter-proximal introns in Arabidopsis are enriched in dispersed signals that elevate gene expression. Plant Cell 20:543–551
Rose AB, Emani S, Bradnam K, Korf I (2011) Evidence for a DNA-based mechanism of intron-mediated enhancement. Frontiers Plant Sci 2:1–9
Rushton PJ, Reinstadler A, Lipka V, Lippok B, Somssich IE (2002) Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen-and wound-induced signaling. Plant Cell 14:749–762
Salinas J, Oeda K, Chua N-H (1992) Two G-box-related sequences confer different expression patterns in transgenic tobacco. Plant Cell 4:1485–1493
Samadder P, Sivamani E, Lu J, Li X, Qu R (2008) Transcriptional and post-transcriptional enhancement of gene expression by the 5′ UTR intron of rice rubi3 gene in transgenic rice cells. Mol Gen Genom 279:429–439
Sawant SV, Kiran K, Mehrotra R, Chaturvedi CP, Ansari SA, Singh P, Lodhi N, Tuli R (2005) A variety of synergistic and antagonistic interactions mediated by cis-acting DNA motifs regulate gene expression in plant cells and modulate stability of the transcription complex formed on a basal promoter. J Exp Bot 56:2345–2353
Sivamani E, Qu R (2006) Expression enhancement of a rice polyubiquitin gene promoter. Plant Mol Biol 60:225–239
Wang J, Oard JH (2003) Rice ubiquitin promoters: deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep 22:129–143
Yang Z, Patra B, Li R, Pattanaik S, Yuan L (2013) Promoter analysis reveals cis-regulatory motifs associated with the expression of the WRKY transcription factor CrWRKY1 in Catharanthus roseus. Planta 238:1039–1049
Zhang N, McHale LK, Finer JJ (2015) Isolation and characterization of “GmScream” promoters that regulate highly expressing soybean (Glycine max Merr.) genes. Plant Sci 241:189–198
Acknowledgements
Salaries and research support were provided by the United Soybean Board, Bayer CropScience, and by State and Federal funds appropriated to the Ohio State University/Ohio Agricultural Research and Development Center. This work was partly supported by an OARDC Director’s Associateship Award to NZ in 2011–2012. This research was also supported, in part, through The Consortium for Plant Biotechnology Research, Inc. by DOE Prime Agreement No. DEFG36-02G012026. This support does not constitute an endorsement by DOE or by the Consortium for Plant Biotechnology Research, Inc. of the views expressed in this publication. Mention of trademark or proprietary products does not constitute a guarantee or warranty of the product by OSU/OARDC and also does not imply approval to the exclusion of other products that may also be suitable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Grant, T.N.L., De La Torre, C.M., Zhang, N. et al. Synthetic introns help identify sequences in the 5′ UTR intron of the Glycine max polyubiquitin (Gmubi) promoter that give increased promoter activity. Planta 245, 849–860 (2017). https://doi.org/10.1007/s00425-016-2646-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2646-8