Abstract
Main conclusion
Medicinal and aromatic plants are known to produce secondary metabolites that find uses as flavoring agents, fragrances, insecticides, dyes and drugs. Biotechnology offers several choices through which secondary metabolism in medicinal plants can be altered in innovative ways, to overproduce phytochemicals of interest, to reduce the content of toxic compounds or even to produce novel chemicals. Detailed investigation of chromatin organization and microRNAs affecting biosynthesis of secondary metabolites as well as exploring cryptic biosynthetic clusters and synthetic biology options, may provide additional ways to harness this resource.
Plant secondary metabolites are a fascinating class of phytochemicals exhibiting immense chemical diversity. Considerable enigma regarding their natural biological functions and the vast array of pharmacological activities, amongst other uses, make secondary metabolites interesting and important candidates for research. Here, we present an update on changing trends in the biotechnological approaches that are used to understand and exploit the secondary metabolism in medicinal and aromatic plants. Bioprocessing in the form of suspension culture, organ culture or transformed hairy roots has been successful in scaling up secondary metabolite production in many cases. Pathway elucidation and metabolic engineering have been useful to get enhanced yield of the metabolite of interest; or, for producing novel metabolites. Heterologous expression of putative plant secondary metabolite biosynthesis genes in a microbe is useful to validate their functions, and in some cases, also, to produce plant metabolites in microbes. Endophytes, the microbes that normally colonize plant tissues, may also produce the phytochemicals produced by the host plant. The review also provides perspectives on future research in the field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A variety of organic compounds are synthesized by plants, which are chiefly classified as primary and secondary metabolites. Primary metabolites are required for basic processes like photosynthesis, respiration, growth and development. Secondary metabolites are other phytochemicals, which are specifically accumulated and are not present merely as intermediates of chemical processes. These compounds are very diverse and distribution of specific types of secondary metabolites is often restricted to taxonomically related species. Though precise functions of secondary metabolites in plant metabolism and physiology are as yet unclear, they are believed to play various roles in interactions of plants with their environment, like (a) providing protection to plants against pathogens (Schwekendiek et al. 2007; Naoumkina et al. 2008) (b) providing protection against abiotic stresses like UV radiation (Xu et al. 2008) (c) attractants for pollinators (Kessler and Baldwin 2007; González-Teuber and Heil 2009) (d) signal molecules (Xu et al. 2009) etc.
The major reason for interest in plant secondary metabolites stems from their overwhelming diversity. They appear to be a never-ending source of novel chemical structures with a variety of pharmacological activities. Nearly 100,000 such metabolites have been isolated from higher plants (Verpoorte et al. 1999; Afendi et al. 2012). Several of these chemicals are used as flavoring agents, fragrances, insecticides, dyes and drugs. Since time immemorial plants and their products have also been used as traditional medicines for treatment of common ailments (Crozier et al. 2006); an estimate suggests that up to 70,000 species of plants are used in folk medicine (Farnsworth and Soejartto 1991). In India about 7,500 plant species are used in ethnomedicines (Shankar and Majumdar 1997). In China, about 1,000 medicinal plants are commonly used as traditional medicine (He and Sheng 1997). Advances in chemistry and pharmacology have validated or vitiated the claims of traditional medicines and have discovered the active principles. About 50 % of all US-FDA approved drugs introduced in the market are natural products or their analogues (Vuorelaa et al. 2004). However, often the raw material could be limiting and its exploitation may be surrounded by ecological concerns. One of the key objectives of plant biotechnology is to develop eco-friendly ways of large-scale production of pharmacologically active compounds. Moreover, the enormous biosynthetic potential of plants is yet to be exploited completely and biotechnology could be used to generate novel chemical compounds, with enhanced or newer bioactivities, through activation of silent or cryptic metabolic clusters.
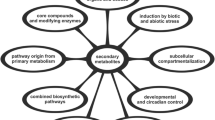
Powerful molecular tools have been used to exploit microbial biochemistry to produce novel compounds (Prather and Martin 2008). This in part, could be attributed to lesser complexity, clustering of genes involved in a pathway, lower redundancy, easy amenability to genetic intervention and availability of genome sequences of a large number of microbes. Biotechnological interventions have also played a major role in improvement of crop yields and quality. Crops have also been engineered to produce valuable enzymes, heterologous proteins and antibodies (Desai et al. 2010). Despite such progress in plant molecular biology, only limited application of biotechnology has been seen in medicinal and aromatic plants (MAPs). In MAPs, generally there is paucity of available molecular information and standardized protocols for transgenesis and marker-assisted selection are also not readily available. In contrast, for most crop species, presence of large EST libraries, genome sequences for at least some of the crops and standardized protocols for transgenesis have played a role in employing biotechnology for improvement of crop yields. However, for MAPs, use of hairy root cultures and bioreactors for production of secondary metabolites have become popular (Srivastava and Srivastava 2007). Moreover, reducing time and costs of de novo genome and EST sequencing have made it possible to unravel the molecular secrets of secondary metabolite production by MAPs. Here we present an update of biotechnological applications in plant secondary metabolism (Fig. 1). Supplementary Table 1 summarizes the available biotechnologies for few pharmacologically important plant secondary metabolites obtained from MAPs.
Bioprocessing
Plant secondary metabolites are usually produced in lesser quantities; often they get accumulated in specific plant organs, at distinct developmental stages, or, on an exposure to a specific stress, or in a particular agro-geo-climatic zone (Chemler and Koffas 2008). Many such metabolites like taxol, artemisinin, forskolin etc. are very difficult to synthesize chemically, and the process is economically unviable (Hashimoto et al. 1988; Corey et al. 1988; Heinstein and Chang 1994; Bouwmeester et al. 2006). Industrial scale plant tissue culture presents itself as a commercially viable alternative for production of phytochemicals, considering the (1) increasing demand for metabolites of interest, (2) long-time scales required for certain slow growing plants, (3) continuously reducing land availability for large-scale cultivation of plants, and (4) destruction of wild populations of medicinal plants through blatant exploitation.
Suspension culture
Suspension cultures are fast growing and amenable to continuous culture in a chemostat. For establishing plant cell suspension cultures, the undifferentiated plant cell culture or callus is generally transferred into liquid medium and agitated on a rotary shaker. However, plant cell cultures growing in such environments show propensity towards production of certain compounds only (Smetanska 2008). Few biosynthetic pathways, such as those involved in production of cinnamic acid derivatives, anthraquinones, berberines, shikonins, anthocynanins etc., express very well in suspension cultures (Chattopadhyay et al. 2002; Chiang and Abdullah 2007). Many times these compounds get spontaneously accumulated in suspension cell cultures, often at concentrations much higher than found in intact plants, even without any efforts for medium engineering. In contrast, other compounds such as morphinan alkaloids, tropane alkaloids (e.g. hyoscyamine and scopolamine), quinoline alkaloids, dimeric monoterpene indole alkaloids (e.g. vinblastine and vincristine) etc. are expressed only in traces in suspension cultures (Berlin 1997). Large-scale efforts to increase their expression through medium engineering and use of elicitors have not yielded results that can lead to commercial exploitation of tissue cultures for production of these compounds.
Organ culture
Organ culture has been explored for production of such phytochemicals that are not expressed by suspension culture. Morphinan alkaloids of Papaver somniferum L. (Papaveraceae), dimeric indole alkaloid (anhydrovinblastine—a direct precursor of vinblastine and vincristine) of Catharanthus roseus (L.) G. Don (Apocynaceae), sesquiterpene lactone (artemisinin) of Artemisia annua L. (Asteraceae), for instance, are produced in better quantities in shoot culture (Endo et al. 1987; Liu et al. 2003; Tisserat and Berhow 2009). Similarly root cultures produce better amounts of tropane alkaloids, such as hyoscyamine and scopolamine, as compared to suspension cultures (Berlin 1997; Saito and Mizukami 2002).
Hairy root culture
Genetic transformation of plant cells using Agrobacterium rhizogenes results in differentiation into hairy roots. These roots can be excised from the infection site, bacteria can be removed using antibiotic treatment, and then the hairy roots can be cultured indefinitely in liquid medium. Hairy roots cultures have several advantages, such as (1) high growth rate, (2) genotypic and phenotypic stability over long culture periods, (3) they do not require exogenous supply of plant growth regulators, and (4) produce high levels of secondary metabolites (Srivastava and Srivastava 2007). Expression levels of genes contained in the inserted T-DNA have been correlated to the amount of secondary metabolites produced. The variability in different insertion lines can be used to select for the lines that are better producers. Apart from phytochemicals, hairy roots have also been explored, though with limited success, for the production of heterologous proteins (Tzfira and Citovsky 2008).
Process optimization
Like any other plant tissue culture process, the use of suspension cells and organ cultures, requires addition of auxin and cytokinins in specific ratio, to either promote dedifferentiation into suspension cells, or in a different ratio for differentiation into specific plant organs. However, if the products (phytochemicals) are to be used as crude extract for human/animal consumption, it is desirable to avoid addition of plant growth regulators. Media optimization also plays a crucial role in production of secondary metabolites. For certain secondary metabolites, a media that supports maximum biomass production, may not actually result in corresponding higher yields of secondary metabolites. For instance, addition of nitrogen-rich medium resulted in 25–30 % increase in biomass but only marginal (2–9 %) increase in shikonin concentration by Lithospermum erythrorhizon Siebold & Zucc. (Boraginaceae) culture (Srinivasan and Ryu 1993). The nature of carbon source used could also affect biomass as well as production of secondary metabolites. Glucose and/or sucrose are considered good carbon sources for biomass increase in plant cell culture, but not for hairy root culture (Giri and Narasu 2000). Carbon–nitrogen ratio also affects secondary metabolite biosynthesis. Nitrogen is usually supplied as ammonia and nitrates. Ratios of these two, as well as total nitrogen content in the medium could affect secondary metabolite production differently in different plant cell cultures. Alternating carbon- and nitrogen-rich media enhanced shikonin production in cultures of L. erythrorhizon, while betacyanin production was markedly increased in cultures of Phytolacca americana L. (Phytolaccaceae) with increase in total nitrogen supply (Sakuta et al. 1987; Srinivasan and Ryu 1993). Similarly phosphate limitation also reduced anthraquinone synthesis by cultures of Galium mollugo L. (Rubiaceae) (Wilson and Balague 1985). Production of secondary metabolites by plant cell cultures is also greatly potentiated by addition of biotic (of biological origin) or abiotic (chemical or physical) elicitors (Karuppusamy 2010). Use of abiotic elicitors is thought to induce production of phytoallexins and release of biotic elicitors from plant cell walls (Davis et al. 1986). Sequential treatment of commonly used elicitors such as methyl jasmonate (abiotic), salicylic acid (abiotic) and yeast extract (biotic) at 24 h intervals was found to enhance the accumulation of dihydrosanguinarine (2.5 times) and sanguinarine (5.5 times) in Eschscholzia californica Cham. (Papaveraceae) suspension culture (Cho et al. 2008). Addition of sodium vanadate and vanadyl sulfate was found to increase the production of coumarins in the suspension culture of Angelica archangelica L. (Apiaceae) (Siatka and Kasparová 2007). Use of UV-B light leads to increase in production of catharanthine in Catharanthus roseus cell suspension culture and flavonoid production in Passiflora quadrangularis L. (Passifloraceae) callus culture (Ramani and Chelliah 2007; Antognoni et al. 2007). Addition of filtered and autoclaved mycelial extract of Verticillium dahliae increased the production of artemisinin from hairy root cultures of A. annua without affecting the growth and morphology of hairy roots (Wang et al. 2000a, b). Electric current also appears to be a good elicitor for secondary metabolite production. Pea hairy roots treated with 30–100 mA of electric current produced 13 times higher amounts of (+)-pisatin compared to the non-elicited controls. Similarly seedlings, intact roots or cell suspension cultures of Trigonella foenum-graecum L. (Fabaceae), Medicago truncatula Gaertn. (Fabaceae), Arabidopsis thaliana (L.) Heynh. (Brassicaceae), Trifolium pratense L. (Fabaceae) and Cicer arietinum L. (Fabaceae) also produced increased levels of secondary metabolites in response to electro-elicitation (Kaimoyo et al. 2008).
Several types of bioreactors have been used for production of secondary metabolites by plant cell cultures. The general aspects that need to be taken care of during bioreactor design are (1) low shear mixing for efficient nutrient transport without sedimentation or clumping of cells, (2) optimal aeration with low shear stress, (3) sterility of the process, and (4) introduction of light for phototrophic cultures. Stirred reactor, rotating drum reactor, fluidized bed reactor, airlift reactor, etc. have been used for both suspension cells and hairy root cultures. Bubble column and aerated reactors were found to be more suitable for organ cultures. Souret et al. (2003) compared terpenoid gene expression patterns and artemisinin production from hairy root cultures of A. annua in shake flask, bubble column reactor and mist reactor. They found that bubble column reactor supported more biomass production while more artemisinin was produced in mist reactor. Moreover, root samples from different regions of the same reactor showed considerable differences in expression of terpenoid pathway genes. Scale-up to bioreactors as well as the choice of bioreactor continues to remain a challenge. Mathematical models have been used to evaluate the process parameters against productivity and provide optimal conditions using different types of bioreactors (Rizvi 2012).
Molecular elucidation of plant secondary metabolite pathways
Molecular elucidation in broad sense consists of finding out the precise chemical route of metabolite biosynthesis, enzymes catalyzing the biosynthetic reactions, genes encoding the biosynthetic enzymes and regulatory factors that control secondary metabolite biosynthesis. Identification of genes involved in plant secondary metabolite biosynthesis is a very important component of biotechnology of MAPs. The availability of molecular information with regards to production and regulation of plant secondary metabolites enables the biotechnologist to rationally tinker with the biosynthetic machinery. Approaches used to study secondary metabolite pathways have changed considerably over time, with the availability of new molecular tools and technological advances. These approaches may be roughly divided into pre-genomic era and post-genomic era approaches.
Precursor labeling
Labeling experiments and retro biosynthetic studies generally precede the identification of enzymes and genes involved in secondary metabolite biosynthesis. These are used to trace the precise chemical route of biosynthesis. For instance, terpenoids that contribute one-third of all known secondary metabolites were shown to be produced by condensation of C5 units—IPP (isopentenyl pyrophosphate) and DMAPP (dimethylallyl pyrophosphate) (Poulter et al. 1981). Earlier it was thought that only the cytosolic mevalonate (MEV) pathway produces IPP, the universal precursor of all terpenoids. However, with the use of 13C labeled intermediates, it was shown that in microorganisms and plants, certain terpenoids are produced not from mevalonate pathways, but from another pathway also producing IPP/DMAPP (Rohmer 1999). Now it is well established that cytosolic mevalonate pathway provides precursors for synthesis of sesquiterpenes (C15) and triterpenes (C30) while plastidial methylerythritol phosphate (MEP) pathway provides precursors for synthesis of monoterpenes (C10), diterpenes (C20) and tetraterpenes (C40) (Dudareva et al. 2005).
Pre-genomic era approaches
Biochemical approach
Biochemical approach for molecular dissection of secondary metabolite pathways has been very useful in the pre-genomic era. Here, once the chemical route of metabolite biosynthesis is known, a hypothetical scheme is laid, based on the plausible reaction mechanisms. Enzyme activity is detected in cell free systems and then one proceeds for activity-guided purification of the enzyme using various chromatographic techniques. The purified protein (enzyme) is sequenced, degenerate primers are designed and partial cDNA is amplified using polymerase chain reaction (PCR). The sequence of partial cDNA is used to design RACE (Rapid amplification of cDNA ends) primers and full-length cDNA is cloned. Heterologously expressed protein is checked for the enzyme activity against purified substrates. Phenylalanine aminomutase that catalyzes the first committed step in taxol side-chain biosynthesis was cloned from Taxus chinensis Roxb. (Taxaceae) using this approach (Steele et al. 2005). Similarly, phenylalanine ammonia lyase that catalyzes the first committed step in phenylpropanoid biosynthesis was cloned from Pinus taeda L. (Pinaceae) using this method (Whetten and Sederoff 1992).
Positional cloning, tagging and expression libraries
Another pre-genomic era approach, involves positional cloning of biosynthetic pathway genes. This approach starts with creation of mutants that are defective in secondary metabolite synthesis, mainly those metabolites whose deficiency results in score able phenotypes, such as color, aroma and flavor. Mutants are classified into complementation groups and map-based cloning of gene ensues. The open reading frames in the cloned DNA are expressed in heterologous system and assayed for enzyme activity. To cite an illustration, these methods were used in discovery of an alternative pathway for formation of β-carotene in plant chloroplasts. Two mutations that affect tomato pigmentation: Beta, a dominant mutation that increases β-carotene and old gold, a recessive mutation that stops β-carotene synthesis and increases lycopene production, were analyzed. Positional cloning and further molecular analysis revealed that Beta encoded a lycopene β-cyclase that converts lycopene to carotene. Old gold was found to be a null allele of Beta (Ronen et al. 2000).
Alternatively, a functionally expressed cDNA library is screened for the requisite enzyme activity against purified substrates. Once the expected enzyme activity is detected, the clone is sequenced. This approach has been used for cloning of several cytochrome P450 enzymes, that catalyze various steps in many secondary metabolite pathways (Schoendorf et al. 2001).
Homology based cloning
Once large number of sequences were accumulated using the above-mentioned methods, it became evident, that related enzymes share considerable sequence homology both at protein and DNA level, at least in conserved domains, which could be used for designing degenerate primers and cloning of related genes in new plant species. This approach considerably reduced the time required for cloning of secondary metabolite pathway genes, and has been successfully employed for several important plant secondary metabolites. To exemplify, β-caryophyllene synthase of A. annua which converts farnesyl diphosphate to β-caryophyllene, was cloned using this approach (Cai et al. 2002). Similarly, Engprasert et al. (2004) aligned the protein sequences of geranylgeranyl diphosphate synthase and identified regions of high homology. Degenerate primers were used to amplify partial GGPP synthase gene and eventually full-length cDNA was cloned from Coleus forskohlii (Willd.) Briq. (Lamiaceae).
Post-genomic era approaches
Differential expression analysis, EST libraries, NGS
In the post-genomics era, the reducing costs of DNA sequencing, availability of large-scale proteomics platforms and development of better bioinformatics tools have changed the outlook and approaches to understand the plant secondary metabolite pathways at a molecular level. Often the secondary metabolites are synthesized in specific plant organs, for instance, leaf trichomes are sites of synthesis of several secondary metabolites (Lommen et al. 2006). This property is exploited for conducting a differential expression-based transcriptomics study. Treatment with biotic or abiotic elicitors that induce production of specific secondary metabolites could also present an opportunity for conduction of a differential expression study. Suppression subtractive hybridization is one such method, following which the EST library is sequenced. Recently the advances in massively parallel sequencing techniques (next generation sequencing platforms like Roche 454®, Illumina Solexa® etc.) have considerably reduced the time required for sequencing of differentially expressed transcriptomes. Wherever genomic resources preexist, a microarray-based differential expression study may be conducted. Differentially expressed RNAs (or proteins, in case of comparison of 2D PAGE profiles) are analyzed by bioinformatic tools. Genes, which could be involved in the biosynthesis of secondary metabolite in question, are identified on the basis of homology. Further their expression pattern helps to predict with some degree of certainty, whether they could be involved in secondary metabolite biosynthesis. Once the genes are predicted, one generally goes for fishing out the full-length cDNA and heterologous expression followed by in vitro enzyme activity determination. To further prove the role of the gene-of-interest in secondary metabolite biosynthesis, knock-out or knock-down lines may be created using transgenesis and then accumulation of preceding intermediate may be tested, as per the proposed biosynthetic pathway. These methods have been employed for characterizing several secondary metabolite pathway genes such as those involved in the production of anti-cancer compounds—vincristine and vinblastine in C. roseus (Rischer et al. 2006; Miettinen et al. 2014). Co-expression of secondary metabolite or essential oil components with specific ESTs have also been used for associating functions of genes with metabolites (Fang et al. 2014; Mahajan et al. 2015). Transcriptome sequencing on Roche 454® platform was recently used to better understand the regulation of artemisinin (anti-malarial) metabolism in A. annua (Soetaert et al. 2013).
Functional genomics
Reverse genetics has become a popular tool for functional genomics and could be utilized for molecular elucidation of secondary metabolite pathways. Once an EST library or a plant genome is sequenced (and ORFs predicted), genome-scale approaches, mostly utilizing the power of RNA interference-based knock-down, can be employed to find the function of genes (Alonso and Ecker 2006). However, many times the secondary metabolite of interest is produced by an exotic plant species or in some cases by trees, where these methods may not be viable due to unavailability of transgenesis protocols and unreasonable time scales.
Metabolic engineering of plant secondary metabolism
A thorough understanding of biosynthetic machinery and regulatory aspects of plant secondary metabolism are critical for rational metabolic engineering. The biosynthetic processes in a cell are highly networked and several possible fates are possible with tens or hundreds of interactions at each step of a biosynthetic pathway often leading to unpredictable results in metabolic engineering. A systems biology approach, integrating information from metabolomics, proteomics and transcriptomics enables the biotechnologist to engineer the metabolic pathways with higher chances of predictable results (Yang et al. 2014). The main objectives of metabolic engineering of secondary metabolite pathways are to produce novel metabolites, to over produce selective metabolites, to reduce the percentage of toxic and unwanted chemicals and to engineer the biosynthetic apparatus into a microorganism for cheaper, large-scale production of plant secondary metabolites (refer Table 1 for examples).
Overexpression of the enzyme catalyzing the rate-limiting step in a pathway is often used as a strategy to increase the metabolic flux through a pathway. For instance, overexpression of strictosidine synthase, an early enzyme in alkaloid biosynthetic pathway, in C. roseus cells leads to increased accumulation of alkaloids (Whitmer et al. 1998). Scopolamine, a medicinally important compound produced by several solanaceous species, is produced by the oxidation of hyoscyamine to scopolamine. Hyocyamine 6β-hydroxylase catalyzes the oxidative reactions that lead to conversion of hyocyamine to scopolamine (Hashimoto et al. 1987). Simultaneous introduction and overexpression of hyocyamine 6β-hydroxylase and putrescine N-methyltransferase in transgenic Hyoscyamus niger L. (Solanaceae) hairy root line resulted in almost nine times higher yields of scopolamine as compared to wild type (Zhang et al. 2004). Similarly, higher levels of anthocyanins and flavonoids are desirable in food products, since these have antioxidant activity. Chalcone isomerase (CHI) is an early enzyme of flavonoid biosynthesis. Overexpression of CHI (cloned from Petunia) in tomato plants led to a 78-fold increase of flavonoid levels compared to control (Muir et al. 2001). Overexpression of farnesyl diphosphate synthase in A. annua led to 2–3 fold increase in artemisinin production (Chen et al. 2000). Another approach for increasing metabolic flux is to inhibit competitive pathways. Blocking a competitive branch of monoterpenoid metabolic network, that converts pulegone, (a common precursor of menthol and menthofuran) to menthofuran, resulted in increased accumulation of menthol. This was achieved by making transgenics expressing antisense gene for menthofuran synthase (Mahmoud and Croteau 2001). Inhibition of specific steps in a metabolic pathway has also been attempted to allow accumulation of preceding intermediate. Forsythia plants produce lignans, such as matairesinol using pinoresinol as precursor. Pinoresinol is converted to matairesinol by pinoresinol/lariciresinol reductase (PLR) and secoisolariciresinol dehydrogenase. Down-regulation of PLR expression, using an RNAi construct led to a complete loss of matairesinol and a 20-fold accumulation of pinoresinol in its glucoside form, compared to the controls (Kim et al. 2009). In P. somniferum, reduction of codeinone reductase, an enzyme encoded by a multigene family was achieved by silencing the entire gene family using a chimeric small hairpin RNA construct. This led to accumulation of precursor alkaloid (S)-reticuline in the transgenic plants, at the expense of morphine, codeine, oripavine and thebaine (Allen et al. 2004). As an alternative, increased production of a secondary metabolite is also possible by engineering the regulatory mechanism of secondary metabolite biosynthesis. For instance, silencing of DET1 regulatory gene using RNAi, in tomato fruits, resulted in increased apocarotenoid and flavonoid content (Davuluri et al. 2005). Overexpression of maize regulatory gene leaf color (Lc), in transgenic apples resulted in increased flavonoid content (Li et al. 2007). In maize suspension cells, overexpression of transcription factors C1 and R led to increased accumulation of anthocyanins (Grotewold et al. 1998). Heterologous overexpression of maize C1 and R genes in Arabidopsis resulted in increased pigmentation in normally pigmented tissues and induced pigmentation even in non-pigmented tissues (Lloyd et al. 1992). Overexpression of ORCA3, a transcription factor, in C. roseus results in upregulation of the genes of terpenoid indole alkaloid (TIA) pathway. ORCA3 directly interacts with the jasmonate and elicitor response element (JERE) in the upstream promoter region of strictosidine synthase (TIA pathway enzyme) and induces its increased expression. Transgenic suspension cells of C. roseus, simultaneously overexpressing ORCA3 and G10H (encoding a cytochrome P450 enzyme; not responsive to Orca3 overexpression) led to threefold increase in accumulation of alkaloids (van der Fits and Memelink 2000).
Removal or reduction in quantity of toxic chemicals is also an important goal of metabolic engineering. Nornicotine, which is a precursor for a carcinogen, is produced by N-demethylation of nicotine. In transgenic tobacco plants, RNAi-induced silencing of CYP82E4 gene (encoding the enzyme that catalyzes this N-demethylation) was employed to suppress the production of nornicotine (Gavilano et al. 2006). ZntA gene of Escherichia coli encodes a lead/cadmium/zinc transporting ATPase. Ectopic expression of E. coli ZntA gene in Arabidopsis plants led to reduction in cellular levels of these heavy metals and improved resistance against lead and cadmium (Lee et al. 2003). In Coffee plants, CaXMT1, CaMXMT1 (theobromine synthase) and CaDXMT1 (caffeine synthase), enzymes successively add methyl groups to xanthosine converting it into caffeine. In sensitive individuals higher caffeine content could cause palpitations, increased blood pressure and insomnia. Transgenic coffee plants expressing RNAi constructs against these genes resulted in coffee with up to 70 % reduction in caffeine content (Ogita et al. 2003).
Sometimes metabolic engineering efforts may unpredictably yield novel compounds. Action of two multifunctional cytochrome P450 enzymes (CYPs) and a specific UDPG-glucosyltransferase catalyze the production of dhurrin from tyrosine. Overexpression of CYP79A1, first enzyme of the pathway, in Arabidopsis resulted in the formation of p-hydroxybenzylglucosinolates, which are normally not found in this plant species (Bak et al. 1999). In E. californica, RNAi-mediated suppression of berberine bridge forming enzyme leads to accumulation of reticuline which is a precursor of isoquinoline alkaloids. As an obvious outcome the products of this pathway, such as sanguinarine, were considerably reduced. However, laudanine, a methylated derivative of berberine accumulated in the transgenic plants (Fujii et al. 2007).
Exploring endophytes for production of plant secondary metabolites
Microbes that colonize host plant tissues without any apparent adverse effect (in contrast to pathogens), and survive in a mutualistic/commensal association are called endophytes. It is estimated that each plant may harbor one or more endophytic species (Tan and Zou 2001). The host plants, in their respective agro-geo-climatic zones provide unique niches to these microbes. In culture medium, outside their host plant species, these endophytes often produce bioactive compounds, which sometimes are the same as those produced by the host plant species. The power of this approach was first demonstrated by the discovery of taxol producing fungal endophyte Taxomyces andreanae, isolated from the host tree Taxus brevifolia Nutt. (Taxaceae) (Strobel et al. 1993). After this discovery, many other fungal endophytes isolated from various species of Taxus as well as from other trees were shown to produce taxanes (Pulici et al. 1996; Strobel et al. 1996; Bashyal et al. 1999; Wang et al. 2000a, b). Another plant metabolite, torreyanic acid, a potential anti-cancer agent, was found to be produced from an endophytic fungus Pestalotiopsis microspora isolated from the endangered tree Torreya taxifolia Arn. (Taxaceae) (Lee et al. 1996). Endophyte isolated from Podophyllum peltatum L. (Berberidaceae) was reported to produce podophyllotoxin (Eyberger et al. 2006). Dysoxylum binectariferum (Roxb.) Hook.f. ex Bedd., (Meliaceae) is an endangered tree known for production of rohitukine, which is a precursor of flavopiridol (drug approved for treatment of chronic lymphocytic leukemia in EU) (Mahajan et al. 2014). An endophyte, Fusarium proliferatum, isolated from D. binectariferum, has been reported to produce rohitukine (Mohana Kumara et al. 2012). Microbes, due to their small generation time and high growth rates are desirable for industrial production of metabolites. Despite the discovery of high value plant secondary metabolites produced from endophytic fungi, to date there appears to be no report of commercial exploitation of these fungi for industrial scale production. It has been observed that after a few generations, the amounts of plant secondary metabolite produced by the endophytic microbe growing in culture medium, reduces to a great extent. For instance, a sharp attenuation in the production of camptothecin was noted from the first to seventh generation subculture of camptothecin-producing endophyte isolated from Camptotheca acuminata Decne. (Cornaceae) (Kusari et al. 2009). Horizontal transfer of genetic material (DNA or RNA) between the host plant and the endophytic microbe has been proposed to explain the production of phytochemicals by endophytes. Gene encoding 10-deacetylbaccatin-III-10-O-acetyl transferase was found to be present in the endophytic fungus Cladosporium cladosporioides MD2 isolated from Taxus media Rehder (Taxaceae). It shared 99 % identity with the homologous gene in host tree (T. media) and about 97 % identity with homologous genes in other species of Taxus (Zhang et al. 2009). In the endophytic fungus P. microspora, it has been observed that repeats of telomeric sequence 5′-TTAGGG-3′ are added to the termini of foreign transforming DNA and they replicate independently of the chromosomal DNA (Long et al. 1998). It is possible that these segments of DNA are lost or become silent during sub culturing, in the absence of any selective pressure. The endophytic microbe and host plant cells share an intimate and complex relationship. More research into this relationship and the biology of endophytic microbes may help to understand the phenomenon of attenuation in a better way.
Rewiring microbial biochemistry to produce plant secondary metabolites
An important component of elucidation of plant secondary metabolite pathways(s) is to clone and express the putative gene in a microbe (E. coli or Saccharomyces cerevisiae) and determine its biochemical activity on pure substrates so as to assign its role in the plant pathway. Co-expression of more than one gene of a pathway in a microbe results into a primitive metabolic cluster. For instance, an artificial curcuminoid biosynthetic pathway was constructed in E. coli by co-expressing phenylalanine ammonia-lyase (PAL) from the yeast Rhodotorula rubra, 4-coumaroyl:CoA ligase (4CL) from L. erythrorhizon and curcuminoid synthase (CUS) from rice (Oryza sativa L.; Poaceae), which resulted in the production of curcuminoids by the recombinant E. coli (Hwang et al. 2003). Recombinant E. coli cultures expressing 4-coumaroyl CoA ligase (4CL) from A. thaliana and stilbene synthase (STS) cloned from Arachis hypogaea L. (Fabaceae), converted the externally added precursor 4-coumaric acid to resveratrol (>100 mg/L) and externally added caffeic acid to piceatannol (>10 mg/L) (Watts et al. 2006).
Heterologous expression of a complete biosynthetic pathway of a complex plant secondary metabolite in a microbial host is considerably tough and tricky, compared to the above examples. Sometimes there may be missing links in a biosynthetic pathway; enzyme activities encoded by genes that have not yet been identified or cloned. Involvement of multiple enzymatic steps, which sometimes may not necessarily function in a linear fashion, makes cloning of multiple genes and their functional co-expression difficult in a microbial host. Even after overcoming most of these difficulties, and functionally expressing all the genes of a secondary metabolite biosynthetic pathway, the biggest challenge is optimization of enzyme activities to make the process economically feasible. For instance, biosynthesis of FDA approved, anti-cancer chemotherapeutic agent, paclitaxel (taxol) involves about 19 steps (Croteau et al. 2006). Reconstitution of first five committed steps of taxol biosynthesis in budding yeast for production of taxadien-5α-acetoxyl-10β-ol resulted in trace amounts of taxadien-5α-ol while taxadiene was produced only at a concentration of 1 mg/L (Dejong et al. 2006). Using regulatory proteins to inhibit competitive pathways, combinatorial biosynthesis and codon optimization of the cloned pathway genes, helped to increase the yield of taxadiene by 40 folds (Engels et al. 2008).
Lycopene, a bright red-colored carotenoid found in fruits and vegetables, is an antioxidant well known for its preventive activity against several types of cancers (Palozza et al. 2011; Takeshima et al. 2014). Heterologous expression of geranylgeranyl diphosphate synthase, phytoene synthase and phytoene desaturase in E. coli resulted in the production of lycopene (Bartley et al. 1999). An effective mutation screening method was used to identify targets for further increasing the production of lycopene in Blakeslea trispora fungus (Wang et al. 2013).
Advanced precursor of another FDA approved molecule—artemisinin, was produced in S. cerevisiae by heterologous reconstitution of a part of the artemisinin pathway from A. annua, involving five enzymes CYP71AV1, CPR1, CYB5, ADH1 and ALDH1. Notably, the engineered yeast produced artemisinic acid to a concentration of 25 g/L. The artemisinic acid produced by fermentation can be chemically converted to artemisinin at a much lower price compared to extraction from plants (Ro et al. 2006; Paddon et al. 2013).
Microbes have thus immensely contributed in biotechnology of plant secondary metabolism by providing a model for its elucidation and as biosynthetic factories for production of phytochemicals.
Conclusion and Future perspectives
Bioprocessing of plant cultures holds great promise for production of phytochemicals. It provides an alternative method for production of phytochemicals on a large scale, in an economically viable and ecology friendly manner. Traditionally, brute-force methods have been used for selecting a better cell line for production of phytochemical of interest (Thomas et al. 2006). Combining bioprocessing with genetic engineering could help in making the tissue culture processes more productive. This however, requires a better understanding of the biosynthetic pathway and its regulation. It is known that certain pathways express better in suspension culture while others may not express at all (Berlin 1997; Chattopadhyay et al. 2002; Chiang and Abdullah 2007). This may be regulated by certain transcription factors that do not express well in undifferentiated cells (Xu et al. 2012; Patra et al. 2013). Alternatively, epigenetic changes like DNA methylation (Cazzonelli et al. 2010) or expression of certain microRNAs (Mahajan et al. 2011) may regulate the transcription of biosynthetic pathway enzymes. A detailed understanding of these regulatory mechanisms may help to rationally tinker with the secondary metabolite biosynthetic pathways. It is presumed that transport proteins might be playing a critical role in accumulation of secondary metabolites, at high concentration, in specialized cells or in a specific cell organelle (Brodelius and Pedersen 1993; Roytrakul and Verpoorte 2007). These proteins present another avenue for metabolic engineering of plant secondary metabolism. This needs to be done in conjunction with development of easier and faster plant transformation methods for medicinal plants. In cases where competitive pathways have to be inhibited, use of chemical inhibitors may be explored (Demain 1998; Sergeant et al. 2009; Craney et al. 2012). Alternatively, the use virus-induced gene silencing (VIGS) may be explored for silencing the targeted genes, to circumvent the need for development of elaborate transgenesis protocols (Huang et al. 2012). Construction of hybrid pathways, in engineered micro-organisms, using a combination of genes from different plant systems as well as other microbes, may be useful for optimizing/enhancing the yields of phytochemicals produced by the microbes. Use of genes from other systems that encode enzyme with analogous activities but are not sensitive to feedback inhibition may also be an option. Such innovative, synthetic biology approaches may also result in production of novel chemical scaffolds. Further, sharing of engineered microbial strains amongst researchers may aid the prospects of producing advanced/novel phytochemicals in microbes. Exploring the use of endophytes for phytochemicals has been criticized due to the unresolved mystery of attenuation of phytochemical production after a few generations (Kusari et al. 2009). However, there are reports where certain chemical activators were able to restore phytochemical production in otherwise attenuated endophytic cultures (Li et al. 1998). Use of such chemicals may also be explored. Analyzing the changes in genome sequence, epigenetic structure and transcription over successive generations may help to understand the reasons due to which endophytes loose the ability to produce phytochemicals after a few generations in culture. This may, in future, help to harness this resource of naturally engineered microbial strains that produce phytochemicals of interest, in a more meaningful manner. Further, bioinformatics tools for discovery, molecular understanding and methods for activation of silent or cryptic metabolic clusters are available for microbial species (Olano et al. 2014; Seyedsayamdost 2014), however, this remains a relatively unexplored area in plants. Discovery and activation of such silent metabolic clusters in plants, may result in production of novel phytochemicals.
Author contribution
SGG conceived the idea, wrote the manuscript and prepared the figure. VM prepared the tables. YSB provided critical inputs during the preparation of this manuscript and carried out proofreading of the manuscript before submission.
Abbreviations
- 4CL:
-
4-Coumaroyl:CoA ligase
- CHI:
-
Chalcone isomerase
- CUS:
-
Curcuminoid synthase
- CYPs:
-
Cytochrome P450 enzymes
- DMAPP:
-
Dimethylallyl pyrophosphate
- EST:
-
Expressed sequenced tag
- IPP:
-
Isopentenyl pyrophosphate
- mA:
-
Milli ampere (electric current)
- MAP:
-
Medicinal and aromatic plants
- MEP:
-
Methylerythritolphosphate pathway
- MEV:
-
Mevalonate pathway
- ORF:
-
Open reading frame
- PAL:
-
Phenylalanine ammonia-lyase
- PLR:
-
Pinoresinol/lariciresinol reductase
- STS:
-
Stilbene synthase
- T DNA:
-
Transfer deoxyribose nucleic acid
- TIA:
-
Terpenoid indole alkaloid pathway
- US-FDA:
-
United States Food and Drug Administration
- UV:
-
Ultra violet
- VIGS:
-
Virus induced gene silencing
References
Afendi FM, Okada T, Yamazaki M et al (2012) KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol 53:e1. doi:10.1093/pcp/pcr165
Allen RS, Millgate AG, Chitty JA et al (2004) RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22:1559–1566. doi:10.1038/nbt1033
Alonso JM, Ecker JR (2006) Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet 7:524–536. doi:10.1038/nrg1893
Antognoni F, Zheng S, Pagnucco C et al (2007) Induction of flavonoid production by UV-B radiation in Passiflora quadrangularis callus cultures. Fitoterapia 78:345–352. doi:10.1016/j.fitote.2007.02.001
Bak S, Olsen CE, Petersen BL et al (1999) Metabolic engineering of p-hydroxybenzylglucosinolate in Arabidopsis by expression of the cyanogenic CYP79A1 from Sorghum bicolor. Plant J 20:663–671. doi:10.1046/j.1365-313X.1999.00642.x
Bartley GE, Scolnik PA, Beyer P (1999) Two Arabidopsis thaliana carotene desaturases, phytoene desaturase and zeta-carotene desaturase, expressed in Escherichia coli, catalyze a poly-cis pathway to yield pro-lycopene. Eur J Biochem 259:396–403
Bashyal B, Li YJ, Strobel G et al (1999) Seimatoantlerium nepalense, an endophytic taxol producing coelomycete from Himalayan yew (Taxus wallachiana). Mycotaxon 72:33–42
Berlin J (1997) Secondary products from plant cell cultures. In: Rehm H-J, Reed G (eds) Biotechnology: products of secondary metabolism, vol 7. Wiley, New York, pp 593–640
Bouwmeester HJ, Bertea CM, Kraker JW, Franssen MCR (2006) Research to improve artemisinin production for use in the preparation of anti-malarial drugs. In: Bogers RJ, Craker LE, Langer D (eds) Medicinal and aromatic plants. Springer Netherlands, Amsterdam, pp 275–290
Brodelius P, Pedersen H (1993) Increasing secondary metabolite production in plant-cell culture by redirecting transport. Trends Biotechnol 11:30–36. doi:10.1016/0167-7799(93)90072-H
Cai Y, Jia J-W, Crock J et al (2002) A cDNA clone for beta-caryophyllene synthase from Artemisia annua. Phytochemistry 61:523–529
Cankar K, Jongedijk E, Klompmaker M et al (2014) (+)-Valencene production in Nicotiana benthamiana is increased by down-regulation of competing pathways. Biotechnol J. doi:10.1002/biot.201400288
Cazzonelli CI, Roberts AC, Carmody ME, Pogson BJ (2010) Transcriptional control of SET domain group 8 and carotenoid isomerase during Arabidopsis development. Mol Plant 3:174–191. doi:10.1093/mp/ssp092
Chattopadhyay S, Farkya S, Srivastava AK, Bisaria VS (2002) Bioprocess considerations for production of secondary metabolites by plant cell suspension cultures. Biotechnol Bioprocess Eng 7:138–149. doi:10.1007/BF02932911
Chemler JA, Koffas MAG (2008) Metabolic engineering for plant natural product biosynthesis in microbes. Curr Opin Biotechnol 19:597–605. doi:10.1016/j.copbio.2008.10.011
Chen D-H, Ye H-C, Li G-F (2000) Expression of a chimeric farnesyl diphosphate synthase gene in Artemisia annua L. transgenic plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci 155:179–185. doi:10.1016/S0168-9452(00)00217-X
Chiang L, Abdullah MA (2007) Enhanced anthraquinones production from adsorbent-treated Morinda elliptica cell suspension cultures in production medium strategy. Process Biochem 42:757–763. doi:10.1016/j.procbio.2007.01.005
Cho H-Y, Son SY, Rhee HS et al (2008) Synergistic effects of sequential treatment with methyl jasmonate, salicylic acid and yeast extract on benzophenanthridine alkaloid accumulation and protein expression in Eschscholtzia californica suspension cultures. J Biotechnol 135:117–122. doi:10.1016/j.jbiotec.2008.02.020
Corey EJ, Jardine P da S, Rohloff JC (1988) Total synthesis of (+/−)-forskolin. J Am Chem Soc 110:3672–3673. doi:10.1021/ja00219a059
Craney A, Ozimok C, Pimentel-Elardo SM et al (2012) Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem Biol 19:1020–1027. doi:10.1016/j.chembiol.2012.06.013
Croteau R, Ketchum REB, Long RM et al (2006) Taxol biosynthesis and molecular genetics. Phytochem Rev 5:75–97. doi:10.1007/s11101-005-3748-2
Crozier A, Jaganath IB, Clifford MN (2006) Phenols, Polyphenols and Tannins: An overview. In: Crozier A, Clifford MN, Ashihara H (eds) Plant Second Metab. Occur. Struct. Role Hum. Diet. Blackwell Publishing Ltd., Oxford, pp 1–24
Davis KR, Darvill AG, Albersheim P (1986) Several biotic and abiotic elicitors act synergistically in the induction of phytoalexin accumulation in soybean. Plant Mol Biol 6:23–32. doi:10.1007/BF00021303
Davuluri GR, van Tuinen A, Fraser PD et al (2005) Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol 23:890–895. doi:10.1038/nbt1108
Dejong JM, Liu Y, Bollon AP et al (2006) Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol Bioeng 93:212–224. doi:10.1002/bit.20694
Demain AL (1998) Induction of microbial secondary metabolism. Int Microbiol 1:259–264
Desai PN, Shrivastava N, Padh H (2010) Production of heterologous proteins in plants: strategies for optimal expression. Biotechnol Adv 28:427–435. doi:10.1016/j.biotechadv.2010.01.005
Dudareva N, Andersson S, Orlova I et al (2005) The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc Natl Acad Sci USA 102:933–938. doi:10.1073/pnas.0407360102
Endo T, Goodbody A, Misawa M (1987) Alkaloid production in root and shoot cultures of Catharanthus roseus. Planta Med 53:479–482. doi:10.1055/s-2006-962777
Engels B, Dahm P, Jennewein S (2008) Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards taxol (paclitaxel) production. Metab Eng 10:201–206. doi:10.1016/j.ymben.2008.03.001
Engprasert S, Taura F, Kawamukai M, Shoyama Y (2004) Molecular cloning and functional expression of geranylgeranyl pyrophosphate synthase from Coleus forskohlii Briq. BMC Plant Biol 4:18. doi:10.1186/1471-2229-4-18
Eyberger AL, Dondapati R, Porter JR (2006) Endophyte fungal isolates from Podophyllum peltatum produce podophyllotoxin. J Nat Prod 69:1121–1124. doi:10.1021/np060174f
Fang L, Hou Y, Wang L et al (2014) Myb14, a direct activator of STS, is associated with resveratrol content variation in berry skin in two grape cultivars. Plant Cell Rep 33:1629–1640. doi:10.1007/s00299-014-1642-3
Farnsworth N, Soejartto D (1991) Global importance of medicinal plants. In: Akerele O, Heywood V, Synge H (eds) Conservation of medicinal plants. Cambridge University Press, Cambridge, pp 25–51
Floss DS, Hause B, Lange PR et al (2008) Knock-down of the MEP pathway isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 inhibits formation of arbuscular mycorrhiza-induced apocarotenoids, and abolishes normal expression of mycorrhiza-specific plant marker genes. Plant J 56:86–100. doi:10.1111/j.1365-313X.2008.03575.x
Fujii N, Inui T, Iwasa K et al (2007) Knockdown of berberine bridge enzyme by RNAi accumulates (S)-reticuline and activates a silent pathway in cultured California poppy cells. Transgenic Res 16:363–375. doi:10.1007/s11248-006-9040-4
Gavilano LB, Coleman NP, Burnley L-E et al (2006) Genetic engineering of Nicotiana tabacum for reduced nornicotine content. J Agric Food Chem 54:9071–9078. doi:10.1021/jf0610458
Giri A, Narasu ML (2000) Transgenic hairy roots. Recent trends and applications. Biotechnol Adv 18:1–22
González-Teuber M, Heil M (2009) Nectar chemistry is tailored for both attraction of mutualists and protection from exploiters. Plant Signal Behav 4:809–813
Grotewold E, Chamberlin M, Snook M et al (1998) Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10:721–740
Gu X-C, Chen J-F, Xiao Y et al (2012) Overexpression of allene oxide cyclase promoted tanshinone/phenolic acid production in Salvia miltiorrhiza. Plant Cell Rep 31:2247–2259. doi:10.1007/s00299-012-1334-9
Hashimoto T, Kohno J, Yamada Y (1987) Epoxidation in vivo of hyoscyamine to scopolamine does not involve a dehydration step. Plant Physiol 84:144–147
Hashimoto S, Sakata S, Sonegawa M, Ikegami S (1988) A total synthesis of (±)-forskolin. J Am Chem Soc 110:3670–3672. doi:10.1021/ja00219a058
He S, Sheng N (1997) Utilization and conservation of medicinal plants in China with special reference to Atractylodes lancea. In: Bodeker G, Bhat KKS, Burley J, Vantomme P (eds) Medicinal plants for forest conservation and health care. Non-wood forest products 11. FAO, Rome, pp 109–115
Heinstein PF, Chang CJ (1994) Taxol. Annu Rev Plant Physiol Plant Mol Biol 45:663–674. doi:10.1146/annurev.pp.45.060194.003311
Huang C, Qian Y, Li Z, Zhou X (2012) Virus-induced gene silencing and its application in plant functional genomics. Sci China Life Sci 55:99–108. doi:10.1007/s11427-012-4280-4
Hwang EI, Kaneko M, Ohnishi Y, Horinouchi S (2003) Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl Environ Microbiol 69:2699–2706. doi:10.1128/AEM.69.5.2699-2706.2003
Kadomura-Ishikawa Y, Miyawaki K, Noji S, Takahashi A (2013) Phototropin 2 is involved in blue light-induced anthocyanin accumulation in Fragaria × ananassa fruits. J Plant Res 126:847–857. doi:10.1007/s10265-013-0582-2
Kaimoyo E, Farag MA, Sumner LW et al (2008) Sub-lethal levels of electric current elicit the biosynthesis of plant secondary metabolites. Biotechnol Prog 24:377–384. doi:10.1021/bp0703329
Karuppusamy S (2010) A review on trends in production of secondary metabolites from higher plants by in vitro tissue, organ and cell cultures. J Med Plants Res 3:1222–1239
Kessler D, Baldwin IT (2007) Making sense of nectar scents: the effects of nectar secondary metabolites on floral visitors of Nicotiana attenuata. Plant J 49:840–854. doi:10.1111/j.1365-313X.2006.02995.x
Kim HJ, Ono E, Morimoto K et al (2009) Metabolic engineering of lignan biosynthesis in Forsythia cell culture. Plant Cell Physiol 50:2200–2209. doi:10.1093/pcp/pcp156
Kusari S, Zühlke S, Spiteller M (2009) An endophytic fungus from Camptotheca acuminata that produces camptothecin and analogues. J Nat Prod 72:2–7. doi:10.1021/np800455b
Lee JC, Strobel GA, Lobkovsky E, Clardy J (1996) Torreyanic acid: a selectively cytotoxic quinone dimer from the endophytic fungus Pestalotiopsis microspora. J Org Chem 61:3232–3233. doi:10.1021/jo960471x
Lee J, Bae H, Jeong J et al (2003) Functional expression of a bacterial heavy metal transporter in Arabidopsis enhances resistance to and decreases uptake of heavy metals. Plant Physiol 133:589–596. doi:10.1104/pp.103.021972
Lewis RS, Jack AM, Morris JW et al (2008) RNA interference (RNAi)-induced suppression of nicotine demethylase activity reduces levels of a key carcinogen in cured tobacco leaves. Plant Biotechnol J 6:346–354. doi:10.1111/j.1467-7652.2008.00324.x
Li JY, Sidhu RS, Ford EJ et al (1998) The induction of taxol production in the endophytic fungus- Periconia sp from Torreya grandifolia. J Ind Microbiol Biotechnol 20:259–264. doi:10.1038/sj.jim.2900521
Li H, Flachowsky H, Fischer TC et al (2007) Maize Lc transcription factor enhances biosynthesis of anthocyanins, distinct proanthocyanidins and phenylpropanoids in apple (Malus domestica Borkh.). Planta 226:1243–1254. doi:10.1007/s00425-007-0573-4
Li W, Wang B, Wang M et al (2014) Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation. J Integr Plant Biol 56:364–372. doi:10.1111/jipb.12136
Liu C-Z, Guo C, Wang Y, Ouyang F (2003) Factors influencing artemisinin production from shoot cultures of Artemisia annua L. World J Microbiol Biotechnol 19:535–538. doi:10.1023/A:1025158416832
Lloyd AM, Walbot V, Davis RW (1992) Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258:1773–1775
Lommen WJM, Schenk E, Bouwmeester HJ, Verstappen FWA (2006) Trichome dynamics and artemisinin accumulation during development and senescence of Artemisia annua leaves. Planta Med 72:336–345. doi:10.1055/s-2005-916202
Long DM, Smidansky ED, Archer AJ, Strobel GA (1998) In vivo addition of telomeric repeats to foreign DNA generates extrachromosomal DNAs in the taxol-producing fungus Pestalotiopsis microspora. Fungal Genet Biol 24:335–344. doi:10.1006/fgbi.1998.1065
Mahajan V, Mahajan A, Pagoch SS et al (2011) microRNA mediated regulation of plant secondary metabolism: an in silico analysis. J Nat Sci Biol Med 2:44
Mahajan V, Sharma N, Kumar S et al (2014) Production of rohitukine in leaves and seeds of Dysoxylum binectariferum: an alternate renewable resource. Pharm Biol 4:1–5. doi:10.3109/13880209.2014.923006
Mahajan V, Rather IA, Awasthi P et al (2015) Development of chemical and EST-SSR markers for Ocimum genus. Ind Crops Prod 63:65–70
Mahmoud SS, Croteau RB (2001) Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc Natl Acad Sci USA 98:8915–8920. doi:10.1073/pnas.141237298
Miettinen K, Dong L, Navrot N et al (2014) The seco-iridoid pathway from Catharanthus roseus. Nat Commun 5:3606. doi:10.1038/ncomms4606
Mohana Kumara P, Zuehlke S, Priti V et al (2012) Fusarium proliferatum, an endophytic fungus from Dysoxylum binectariferum Hook.f, produces rohitukine, a chromane alkaloid possessing anti-cancer activity. Antonie Van Leeuwenhoek 101:323–329. doi:10.1007/s10482-011-9638-2
Muir SR, Collins GJ, Robinson S et al (2001) Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat Biotechnol 19:470–474. doi:10.1038/88150
Naoumkina MA, He X, Dixon RA (2008) Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol 8:132. doi:10.1186/1471-2229-8-132
Ogita S, Uefuji H, Yamaguchi Y et al (2003) Producing decaffeinated coffee plants. Nature 423:823. doi:10.1038/423823a
Olano C, García I, González A et al (2014) Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol 7:242–256. doi:10.1111/1751-7915.12116
Paddon CJ, Westfall PJ, Pitera DJ et al (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496:528–532. doi:10.1038/nature12051
Palozza P, Simone RE, Catalano A, Mele MC (2011) Tomato lycopene and lung cancer prevention: from experimental to human studies. Cancers (Basel) 3:2333–2357. doi:10.3390/cancers3022333
Patra B, Schluttenhofer C, Wu Y et al (2013) Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim Biophys Acta Gene Regul Mech 1829:1236–1247. doi:10.1016/j.bbagrm.2013.09.006
Poulter CD, Wiggins PL, Le AT (1981) Farnesylpyrophosphate synthetase. A stepwise mechanism for the 1′-4 condensation reaction. J Am Chem Soc 103:3926–3927. doi:10.1021/ja00403a054
Prather KLJ, Martin CH (2008) De novo biosynthetic pathways: rational design of microbial chemical factories. Curr Opin Biotechnol 19:468–474. doi:10.1016/j.copbio.2008.07.009
Pulici M, Sugawara F, Koshino H et al (1996) Pestalotiopsins A and B: new caryophyllenes from an endophytic fungus of Taxus brevifolia. J Org Chem 61:2122–2124. doi:10.1021/jo951736v
Ramani S, Chelliah J (2007) UV-B-induced signaling events leading to enhanced-production of catharanthine in Catharanthus roseus cell suspension cultures. BMC Plant Biol 7:61. doi:10.1186/1471-2229-7-61
Rischer H, Oresic M, Seppänen-Laakso T et al (2006) Gene-to-metabolite networks for terpenoid indole alkaloid biosynthesis in Catharanthus roseus cells. Proc Natl Acad Sci USA 103:5614–5619. doi:10.1073/pnas.0601027103
Rizvi Z (2012) Application of artificial neural networks for predicting maximum in vitro shoot biomass production of safed musli (Chlorophytum borivilianum). J Med Diagn Methods 01:1–6. doi:10.4172/scientificreports.464
Ro D-K, Paradise EM, Ouellet M et al (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940–943. doi:10.1038/nature04640
Rohmer M (1999) The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants†. Nat Prod Rep 16:565–574. doi:10.1039/a709175c
Ronen G, Carmel-Goren L, Zamir D, Hirschberg J (2000) An alternative pathway to beta -carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Natl Acad Sci USA 97:11102–11107. doi:10.1073/pnas.190177497
Roytrakul S, Verpoorte R (2007) Role of vacuolar transporter proteins in plant secondary metabolism: Catharanthus roseus cell culture. Phytochem Rev 6:383–396. doi:10.1007/s11101-006-9022-4
Saito K, Mizukami H (2002) Plant cell cultures as producers of secondary compounds. In: Oksman-Caldentey K-M, Barz WH (eds) Plant biotechnology and transgenic plants. CRC Press, New York, pp 66–91
Sakuta M, Takagi T, Komamine A (1987) Effects of nitrogen source on betacyanin accumulation and growth in suspension cultures of Phytolacca americana. Physiol Plant 71:459–463. doi:10.1111/j.1399-3054.1987.tb02884.x
Schoendorf A, Rithner CD, Williams RM, Croteau RB (2001) Molecular cloning of a cytochrome P450 taxane 10 beta-hydroxylase cDNA from Taxus and functional expression in yeast. Proc Natl Acad Sci USA 98:1501–1506. doi:10.1073/pnas.98.4.1501
Schwekendiek A, Spring O, Heyerick A et al (2007) Constitutive expression of a grapevine stilbene synthase gene in transgenic hop (Humulus lupulus L.) yields resveratrol and its derivatives in substantial quantities. J Agric Food Chem 55:7002–7009. doi:10.1021/jf070509e
Sergeant MJ, Li J-J, Fox C et al (2009) Selective inhibition of carotenoid cleavage dioxygenases: phenotypic effects on shoot branching. J Biol Chem 284:5257–5264. doi:10.1074/jbc.M805453200
Seyedsayamdost MR (2014) High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci USA 111:7266–7271. doi:10.1073/pnas.1400019111
Shankar D, Majumdar B (1997) Beyond the biodiversity convention: the challenges facing the biocultural heritage of India’s medicinal plants. In: Bodeker G, Bhat KKS, Burley J, Vantomme P (eds) Medicinal plants for forest conservation and health care. Non-wood forest products 11. FAO, Rome, pp 87–99
Sharafi A, Sohi HH, Mousavi A et al (2013) Metabolic engineering of morphinan alkaloids by over-expression of codeinone reductase in transgenic hairy roots of Papaver bracteatum, the Iranian poppy. Biotechnol Lett 35:445–453. doi:10.1007/s10529-012-1080-7
Shkryl YN, Veremeichik GN, Bulgakov VP, Zhuravlev YN (2011) Induction of anthraquinone biosynthesis in Rubia cordifolia cells by heterologous expression of a calcium-dependent protein kinase gene. Biotechnol Bioeng 108:1734–1738. doi:10.1002/bit.23077
Siatka T, Kasparová M (2007) Effect of vanadium compounds on the growth and production of coumarins in the suspension culture of Angelica archangelica L. Ceska Slov Farm 56:230–234
Smetanska I (2008) Production of secondary metabolites using plant cell cultures. Adv Biochem Eng Biotechnol 111:187–228. doi:10.1007/10_2008_103
Soetaert SSA, Van Neste CMF, Vandewoestyne ML et al (2013) Differential transcriptome analysis of glandular and filamentous trichomes in Artemisia annua. BMC Plant Biol 13:220. doi:10.1186/1471-2229-13-220
Song J, Wang Z (2011) RNAi-mediated suppression of the phenylalanine ammonia-lyase gene in Salvia miltiorrhiza causes abnormal phenotypes and a reduction in rosmarinic acid biosynthesis. J Plant Res 124:183–192. doi:10.1007/s10265-010-0350-5
Souret FF, Kim Y, Wyslouzil BE et al (2003) Scale-up of Artemisia annua L. hairy root cultures produces complex patterns of terpenoid gene expression. Biotechnol Bioeng 83:653–667. doi:10.1002/bit.10711
Srinivasan V, Ryu DD (1993) Improvement of shikonin productivity in Lithospermum erythrorhizon cell culture by alternating carbon and nitrogen feeding strategy. Biotechnol Bioeng 42:793–799. doi:10.1002/bit.260420702
Srivastava S, Srivastava AK (2007) Hairy root culture for mass-production of high-value secondary metabolites. Crit Rev Biotechnol 27:29–43. doi:10.1080/07388550601173918
Steele CL, Chen Y, Dougherty BA et al (2005) Purification, cloning, and functional expression of phenylalanine aminomutase: the first committed step in taxol side-chain biosynthesis. Arch Biochem Biophys 438:1–10. doi:10.1016/j.abb.2005.04.012
Strobel G, Stierle A, Stierle D, Hess WM (1993) Taxomyces andreanae, a proposed new taxon for a bulbilliferous hyphomycete associated with Pacific yew (Taxus brevifolia). Mycotaxon 47:71–80
Strobel G, Yang X, Sears J et al (1996) Taxol from Pestalotiopsis microspora, an endophytic fungus of Taxus wallachiana. Microbiology 142:435–440
Takeshima M, Ono M, Higuchi T et al (2014) Anti-proliferative and apoptosis-inducing activity of lycopene against three subtypes of human breast cancer cell lines. Cancer Sci 105:252–257. doi:10.1111/cas.12349
Tan RX, Zou WX (2001) Endophytes: a rich source of functional metabolites (1987 to 2000). Nat Prod Rep 18:448–459. doi:10.1039/b100918o
Thomas J, Raj Kumar R, Mandal AKA (2006) Metabolite profiling and characterization of somaclonal variants in tea (Camellia spp.) for identifying productive and quality accession. Phytochemistry 67:1136–1142. doi:10.1016/j.phytochem.2006.03.020
Tian Q, Wang X, Li C et al (2013) Functional characterization of the poplar R2R3-MYB transcription factor PtoMYB216 involved in the regulation of lignin biosynthesis during wood formation. PLoS ONE 8:e76369. doi:10.1371/journal.pone.0076369
Tisserat B, Berhow M (2009) Production of pharmaceuticals from papaver cultivars in vitro. Eng Life Sci 9:190–196. doi:10.1002/elsc.200800100
Tzfira T, Citovsky V (2008) Agrobacterium: from biology to biotechnology. Springer, New York, p 750
Van der Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289:295–297
Verpoorte R, van der Heijden R, ten Hoopen HJG, Memelink J (1999) Metabolic engineering of plant secondary metabolite pathways for the production of fine chemicals. Biotechnol Lett 21:467–479. doi:10.1023/A:1005502632053
Vuorelaa P, Leinonenb M, Saikkuc P et al (2004) Natural products in the process of finding new drug candidates. Curr Med Chem 11:1375–1389
Wang H, Ye HC, Li GF, Liu BYZK (2000a) Effect of fungal elicitors on cell growth and artemisinin accumulation in hairy root cultures of Artemisia annua. Acta Bot Sin 42:905–909
Wang J, Li G, Lu H et al (2000b) Taxol from Tubercularia sp. strain TF5, an endophytic fungus of Taxus mairei. FEMS Microbiol Lett 193:249–253
Wang Q, Luo W, Gu Q-Y et al (2013) Enhanced lycopene content in Blakeslea trispora by effective mutation-screening method. Appl Biochem Biotechnol 171:1692–1700. doi:10.1007/s12010-013-0468-8
Watts KT, Lee PC, Schmidt-Dannert C (2006) Biosynthesis of plant-specific stilbene polyketides in metabolically engineered Escherichia coli. BMC Biotechnol 6:22. doi:10.1186/1472-6750-6-22
Whetten RW, Sederoff RR (1992) Phenylalanine ammonia-lyase from loblolly pine: purification of the enzyme and isolation of complementary DNA clones. Plant Physiol 98:380–386
Whitmer S, Canel C, Hallard D et al (1998) Influence of precursor availability on alkaloid accumulation by transgenic cell Line of Catharanthus roseus. Plant Physiol 116:853–857
Wilson G, Balague C (1985) Biosynthesis of anthraquinone by cells of Galium mollugo L. grown in a chemostat with limiting sucrose or phosphate. J Exp Bot 36:485–493. doi:10.1093/jxb/36.3.485
Xu C, Sullivan JH, Garrett WM et al (2008) Impact of solar ultraviolet-B on the proteome in soybean lines differing in flavonoid contents. Phytochemistry 69:38–48. doi:10.1016/j.phytochem.2007.06.010
Xu M, Dong J, Wang H, Huang L (2009) Complementary action of jasmonic acid on salicylic acid in mediating fungal elicitor-induced flavonol glycoside accumulation of Ginkgo biloba cells. Plant Cell Environ 32:960–967. doi:10.1111/j.1365-3040.2009.01976.x
Xu K, Liu J, Fan M et al (2012) A genome-wide transcriptome profiling reveals the early molecular events during callus initiation in Arabidopsis multiple organs. Genomics 100:116–124. doi:10.1016/j.ygeno.2012.05.013
Yan P, Keji J, Juan L et al (2012) Effects of over-expression of allene oxide cyclase on camptothecin production by cell cultures of Camptotheca acuminata. Afr J Biotechnol 11:6535–6541
Yang D, Du X, Yang Z et al (2014) Transcriptomics, proteomics, and metabolomics to reveal mechanisms underlying plant secondary metabolism. Eng Life Sci. doi:10.1002/elsc.201300075
Yuan Y, Wu C, Liu Y et al (2013) The Scutellaria baicalensis R2R3-MYB transcription factors modulate flavonoid biosynthesis by regulating GA metabolism in transgenic tobacco plants. PLoS ONE 8:e77275. doi:10.1371/journal.pone.0077275
Zhang L, Ding R, Chai Y et al (2004) Engineering tropane biosynthetic pathway in Hyoscyamus niger hairy root cultures. Proc Natl Acad Sci USA 101:6786–6791. doi:10.1073/pnas.0401391101
Zhang P, Zhou P-P, Yu L-J (2009) An endophytic taxol-producing fungus from Taxus media, Cladosporium cladosporioides MD2. Curr Microbiol 59:227–232. doi:10.1007/s00284-008-9270-1
Zhang S, Ma P, Yang D et al (2013) Cloning and characterization of a putative R2R3 MYB transcriptional repressor of the rosmarinic acid biosynthetic pathway from Salvia miltiorrhiza. PLoS ONE 8:e73259. doi:10.1371/journal.pone.0073259
Acknowledgments
SGG acknowledges the financial support for this work from CSIR 12th FYP project ‘PMSI’ (BSC0117) of Council of Scientific and Industrial Research (CSIR).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gandhi, S.G., Mahajan, V. & Bedi, Y.S. Changing trends in biotechnology of secondary metabolism in medicinal and aromatic plants. Planta 241, 303–317 (2015). https://doi.org/10.1007/s00425-014-2232-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2232-x