Abstract
Roots of kudzu (Pueraria lobata) are a rich source of isoflavone O- and C-glycosides. Although O-glycosylation of (iso)flavonoids has been well characterized at the molecular level, no plant isoflavonoid C-glycosyltransferase genes have yet been isolated. To address the biosynthesis of kudzu isoflavonoids, we generated 6,365 high-quality expressed sequence tags (ESTs) from a subtraction cDNA library constructed using RNA from roots that differentially accumulate puerarin. The ESTs were clustered into 722 TCs and 3,913 singletons, from which 15 family I glycosyltransferases (UGTs) were identified. Hierarchical clustering analysis of the expression patterns of these UGTs with isoflavone synthase (IFS) in a range of tissues identified UGTs with potential functions in isoflavone glycosylation. The open reading frames of these UGTs were expressed in E. coli for functional analysis, and one was shown to preferentially glycosylate isoflavones at the 7-O-position. In addition, ESTs corresponding to chalcone synthase, chalcone reductase, chalcone isomerase (CHI) and 2-hydroxyisoflavanone dehydratase were identified. Recombinant CHI proteins had high activities with both 6′-deoxy- and 6′-hydroxy chalcones, typical of Type II CHIs. Establishment of this EST database and identification of genes associated with kudzu isoflavone biosynthesis and glycosylation provide a new resource for metabolic engineering of bioactive kudzu isoflavones.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pueraria lobata (Willd.) Ohwi is a member of the Leguminosae family commonly known as “kudzu”. Puerariae Radix, the dried root of the kudzu plant, has been used for centuries in Chinese herbal medicine for prevention of cardiovascular disease and rehabilitation of patients with stroke. The major secondary metabolites accumulating in kudzu roots are isoflavones such as daidzein, genistein, formononetin and their glucosides. Puerarin, the 8-C-glucoside of daidzein (Rong et al. 1998), is considered the main active principal of Puerariae Radix (Oshima et al. 1988). Interest in the use of puerarin for treatment of blood circulation associated disorders has increased in recent years (Liang et al. 2005; Yeung et al. 2006; Wu et al. 2007), and acute administration of kudzu root extract or puerarin leads to improved glucose tolerance in rodent models of metabolic syndrome and to improved insulin sensitivity (Meezan et al. 2005). The hydrolysable 7-O-glucoside of daidzein, daidzin, may have an opposite effect on glucose tolerance (Meezan et al. 2005). C-glycosyl-isoflavones can also play roles in plant defense against insect predation and pathogen infection (Byrne et al. 1996; Cortes-Cruz et al. 2002; Ni et al. 2008).
Several reports have described the cloning of genes encoding flavonoid O-glycosyltransferases (Jones et al. 2003; Achnine et al. 2005; Sawada et al. 2005; Kim et al. 2006; Li et al. 2007; Modolo et al. 2007; Pang et al. 2008). Recently, plant C-glycosyltransferases involved in the detoxification of 2,4,6-trinitrotoluene (TNT) (Gandia-Herrero et al. 2008) or formation of C- glycosyl flavones in cereals (Brazier-Hicks et al. 2009) have been cloned. However, little is known of the enzymes that C-glycosylate isoflavones. By analogy with the formation of daidzin from daidzein catalyzed by a 7-O-glucosyltransferase utilizing UDP-glucose as sugar donor (Modolo et al. 2007), it would be logical to presume that daidzein is also the precursor for puerarin biosynthesis catalyzed by an isoflavone 8-C-glucosyltransferase. However, early in vivo labeling studies showed that the glycosylation reaction in C-glycosyl flavone biosynthesis likely occurs prior to flavone formation (Wallace and Grisebach 1973; Inoue and Fujita 1977). Thus, 14C labeled 4′,5,7-trihydroxyflavanone (naringenin) was incorporated in a parallel manner into apigenin (flavone) 7-O-glucoside and apigenin 8-C-glucoside (vitexin) and into luteolin 7-O-glucoside and luteolin 8-C-glucoside (orientin) (Wallace and Grisebach 1973). However, radioactivity from 14C labeled apigenin and luteolin was only detected in O-glycosylated and O-methylated flavones, but not in C-glycosylflavones (Wallace et al. 1969). These findings were rationalized when it was shown that 2-hydroxyflavanones were the substrates for glycosylation leading to C-glycosyl flavones with enzyme preparations in vitro (Kerscher and Franz 1987), and this was confirmed for the recently cloned cereal flavonoid C-glycosyltransferases (Brazier-Hicks et al. 2009).
Precursor labeling studies also suggest a non-classical route to C-glycosyl-isoflavones. Thus, 14C-labeled chalcone (isoliquiritigenin), but not isoflavone (daidzein), was efficiently incorporated into puerarin in kudzu roots, supporting the idea that the C-glycosylation reaction of puerarin biosynthesis occurs prior to isoflavone formation (Fig. 1). Further competitive feeding studies with isoliquiritigenin and liquiritigenin suggested that C-glycosylation in kudzu might take place at the chalcone rather than the flavanone level (Inoue and Fujita 1977) (Fig. 1). The key step in isoflavone biosynthesis is the migration of the aryl group (B-ring) of a flavanone precursor from C-2 to the adjacent C-3 to generate the isoflavone skeleton. This enzymatic process is a two-step reaction (Kochs and Grisebach 1986; Hashim et al. 1990; Crombie and Whiting 1992). The first step is the 2-hydroxylation of the C-ring associated with the aryl migration to generate 2-hydroxyisoflavanone, catalyzed by a cytochrome P-450-dependent monooxygenase, generally but misleadingly called isoflavone synthase (IFS). cDNAs encoding this enzyme have been cloned from several plant species (Akashi et al. 1999; Steele et al. 1999; Jung et al. 2000). The second step is the dehydration of 2-hydroxyisoflavanone to yield isoflavone, catalyzed by a 2-hydroxyisoflavanone dehydratase (HID). cDNAs encoding HID have been cloned and characterized from licorice (Glycyrrhiza echinata) and soybean (Glycine max) (Akashi et al. 2005). 2-HID has also been purified to apparent homogeneity from kudzu cell cultures, and shown to convert 2,7,4′-trihydroxyisoflavanone to daidzein with kinetic parameters similar to those of the soybean HID (Hakamatsuka et al. 1998; Akashi et al. 2005). However, the kudzu HID has not been characterized at the DNA sequence level.
Potential biosynthetic pathways to puerarin, the daidzein 8-C-glycoside in kudzu. Isoliquiritigenin could be glycosylated at the 3′-C position by a chalcone glycosyltransferase (GT) to form a chalcone C-glycoside, which is then isomerized to liquiritigenin C-glycoside by chalcone isomerase (CHI). The latter could then be converted to puerarin by the sequential actions of isoflavone synthase (IFS) and trihydroxyflavanone C-glycoside dehydratase (HID). Alternatively, the trihydroxyisoflavanone could be glycosylated by a C-GT to form a trihydroxyisoflavanone C-glycoside, which is then converted to puerarin by HID. In the simplest model, daidzein is the direct substrate for puerarin formation catalyzed by a C-GT
It is noteworthy that the 4′-O-methylation of isoflavones occurs at the level of the 2-hydroxyisoflavanone prior to the action of HID (Akashi et al. 2000; Akashi et al. 2003). It is therefore possible that the 8-C-glycosylation in the formation of puerarin may also occur at the level of 2-hydroxyisoflavanone rather than chalcone as previously suggested, since neither CHI nor IFS have been reported to use glycosylated conjugates as substrates, and C-glycosylated chalcones have not been described as kudzu metabolites.
Analysis of expressed sequence tags (EST) has greatly accelerated gene discovery and facilitated global gene expression profiling, especially in non-model plant species. Examples include the discovery of many genes involved in secondary metabolism, including the biosynthesis of monoterpenes and sequiterpenes (Lange et al. 2000; Shimada et al. 2004), monoterpene indole alkaloids (Murata et al. 2006), diterpenes (Brandle et al. 2002), carotenoids (Jako et al. 2002) and prenylflavonoids (Nagel et al. 2008). This approach has been particularly successful for the discovery of natural product glycosyltransfersases (Achnine et al. 2005; Tian et al. 2006; Richman et al. 2005; Modolo et al. 2007). However, there are currently no EST or genomic resources available from Pueraria lobata.
We here apply an EST-based functional genomics approach to identify genes involved in the biosynthesis of isoflavones in kudzu, with special emphasis on glycosyl transfer reactions.
Materials and methods
Plant materials
Field-collected kudzu plants were obtained from approximately 3 miles southwest of the Samuel Roberts Noble Foundation, Ardmore, Oklahoma. Seeds of commercial kudzu plants were purchased from Kudzu Kingdom Division of Suntop Inc., PO Box 98, Kodak, TN 37764, USA.
Plants were maintained in the greenhouse and propagated by cuttings. Morphological features of the two kudzu lines are shown in Supplemental Fig. 1. Seed and plant materials are available to researchers on request from the authors.
Chemicals
Flavonoid and isoflavonoid acceptor substrates were purchased from Indofine (Hillsborough, NJ). Naringenin chalcone was purchased from Apin Chemicals (Oxon, UK). 2, 4′,7-Trihydroxyisoflavanone was produced by incubation of liquirtigenin (7, 4′-dihydroxyflavanone) with yeast microsomes containing recombinant Medicago truncatula IFS, as described previously (Liu et al. 2006). All other chemicals were from Sigma-Aldrich (St. Louis, MO).
HPLC analysis of secondary metabolites from young kudzu roots
One gram of young root tissues was ground to powder in liquid nitrogen and extracted overnight with 5 ml of acetone at 4°C. The extract was centrifuged at 3,500 rpm for 30 min. The supernatant was dried under a stream of nitrogen, the residues were resuspended in methanol, and an aliquot was analyzed by reverse-phase HPLC (Hewlett Packard 1100 system) on a 5-μm C18 column (250 × 4.6 mm, Waters spheroisorb 5-μm ODS2) using the solvent gradient described previously (He et al. 2008).
cDNA library construction
Total RNA was isolated from roots of the field-collected and commercial kudzu plants separately using an RNeasy plant mini kit according to the manufacturer’s instructions (QIAGEN, Valencia, CA). cDNA was synthesized from the two RNA preparations using the Super Smart PCR cDNA synthesis kit (Clontech, Mountain View, CA). cDNA subtraction was performed with the commercial kudzu root cDNA as the driver using the PCR-select cDNA subtraction kit according to the manufacturer’s protocol (Clontech). The cDNA pools were cloned into pGEM T-easy vectors (Promega, Madison, WI).
DNA template preparation and sequencing
The cDNA library was plated onto LB-ampicillin plates containing IPTG and X-gal, and white colonies were picked into 384-well blocks containing 120 μl TB-ampicillin medium. The cultures were allowed to grow overnight in a HiGro incubator shaker. Plasmid DNA was prepared using a Biomek FX workstation using standard protocols. Sequencing was performed on a 3730 sequencer using BigDye3.1 (Applied Biosystems, Foster City, CA).
Cloning and expression of kudzu glycosyltransferases
Glycosyltransferase ESTs, which were not full length, were extended by 5′-or 3′-rapid amplification of the cDNA ends (RACE) using the Smart RACE cDNA amplification kit (Clontech). Primers for the cDNA end amplification are listed in Supplemental Table 1. Full-length clones were obtained by RT-PCR based on the RACE sequence information. Full-length cDNAs of GT03H14, GT03H24, GT04F14 and GT07O02 were cloned into the protein expression vector pET28a with restriction enzyme adaptors, and all other UGTs were introduced into pENTR/D-TOPO vector to create entry clones. The insert in entry clones was then transferred into the destination vector pDEST17 by LR recombination reaction (Invitrogen, Carlsbad, CA) for protein expression in E. coli. Primers for the restriction enzyme- and Gateway-based cloning of the glycosyltransferases are listed in Supplemental Table 2. Protein was expressed in E. coli BL21(DE3) pLysS (Novagen, Madison, WI) with 0.5 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG) induction overnight at 16°C and purified using a MagneHis protein purification kit (Promega).
Assay of O-glycosyltransferase activity
Enzyme reactions were performed with 1–3 μg of enzyme in 50-μl reaction volumes containing 50 mM Tris–HCl pH 7.5, 5 mM UDP-glucose and 250 μM acceptor substrate at 30°C for 3 h. The reactions were stopped with 10-μl (240 mg/ml) trichloroacetic acid (TCA) and the products were analyzed by HPLC as described previously (He et al. 2008). For kinetic analysis of UGT04F14, a 50-μl reaction mix containing 50 mM Tris–HCl, pH 7.5, 5 mM UDP-glucose, 0–500 μM genistein and 1.5 μg of enzyme was incubated at 30°C for 1 h. The reaction was stopped with TCA prior to HPLC analysis. Kinetic parameters were determined by hyperbolic regression analysis (http://homepage.ntlworld.com/john.easterby/hyper32.html).
Cloning and expression of chalcone isomerase
Kudzu chalcone isomerase 2 (CHI2) was identified from the EST collection of the kudzu cDNA library described above. Kudzu chalcone isomerase 1 (CHI1) was cloned based on the cDNA sequence previously reported (D63577) (Terai et al. 1996). Full-length CHI 1 and CHI 2 were PCR amplified with the primer pairs: (CHI 1) 5′- CACCATGGCGGCAGCAGCAGC-3′ and 5′-TCAGACTATAATGCCGTGGCT-3′; and (CHI 2) 5′-CACCATGGCCACTCCAGCATCC-3′ and 5′- CTAAGGATTGTTGGCCTCTTTGAG-3′. The full length clones were then introduced into pENTR/D-TOPO vector to create entry clones, which were introduced into destination vector pDEST17 by LR recombination reaction for protein expression in E. coli (Invitrogen, Carlsbad, CA). The recombinant protein was expressed in E. coli BL21 (DE3) pLysS (Novagen) after induction with 0.5 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG) overnight at 16°C. Protein was purified using a MagneHis protein purification kit (Promega).
Assay of CHI activities
CHI assays were performed at 25°C in a 50-μl reaction containing 50 mM Tris–HCl, pH 7.5, 1% ethanol, 250–500 μM substrate and 1–5 μg protein. The reactions were stopped by the addition of 50 μl methanol, vortexed and centrifuged for 15 min at 13,000 rpm. The supernatant was used for HPLC assay as described above.
Real-time quantitative RT-PCR
Total RNA was isolated from different developmental stages of roots, nodes and internodes and treated with Turbo DNA-free DNase I (Ambion, Austin, TX). RNA integrity was evaluated with an Agilent 2100 Bioanalyzer. cDNA synthesis was performed using the DNaseI-treated RNAs and Superscript III reverse transcriptase (Invitrogen).
The PCR primers for real-time qRT-PCR were designed using Primer Express Software 3.0 for Real-Time PCR from Applied Biosystems (Supplemental Table 3) using the default parameters. PCR reactions were performed in an optical 384-well plate with an ABI PRISM 7900 HT sequence detection system (Applied Biosystems). Reactions contained 5 μl 2× Power SYBR Green Master Mix reagent (Applied Biosystems), 2 μl of a 1:20 dilution of cDNA, and 2 μl of gene-specific primer pair (1 μM) in a final volume of 10 μl. PCR reactions were performed as described (Czechowski et al. 2005). Data were collected and analyzed using SDS 2.2.1 Software (Applied Biosystems). PCR efficiency was estimated using LinRegPCR software (Ramakers et al. 2003) and transcript levels were determined by relative quantification (Pfaffl 2001) using the actin gene from the kudzu EST collection as a reference. Transcript cluster analysis was performed using Spotfire Software (TIBCO Spotfire).
Amino acid sequence alignment
Deduced amino acid sequences of glycosyltransferases from kudzu and Arabidopsis thaliana were aligned using the ClustalW algorithm (Thompson et al. 1994).
Accession numbers
The cDNA sequence data for this article can be found in the GenBank/EMBL data libraries under the following accession numbers: GT01K01 (HQ219038), GT02J01 (HQ219039), GT03H14 (HQ219040), GT03H24 (HQ219041), GT04F14 (HQ219042), GT07O02 (HQ219043), GT10J15 (HQ219044), GT12D15 (HQ219045), GT12P06 (HQ219046), GT14A05 (HQ219047), GT14K13 (HQ219048), GT14M03 (HQ219049), GT18P15 (HQ219050), GT19J14 (HQ219051), GT21C20 (HQ219052), IFS (HQ219053), HID (HQ219054) and CHI2 (HQ219055). The kudzu EST sequences have been deposited in dbEST (http://www.ncbi.nlm.nih.gov/dbEST/) with dbEST ID #s: 71178728-71185092 (GenBank accession ID #s: HO703606-HO709970).
Results and discussion
Isoflavone accumulation in kudzu root and stem tissues
Kudzu roots produce many secondary metabolites, with isoflavone C- and O- glucosides predominating. We obtained seeds of kudzu from a commercial source, and also collected root and stem cuttings from a plant growing wild in a field hedge SW of Ardmore, Oklahoma. HPLC analysis indicated that young roots derived from the field-collected kudzu isolate produced significant amounts of puerarin in addition to other isoflavone O-glucosides (e.g., the 7-O-glucosides of daidzein, genistein and biochanin A), whereas no puerarin was detected in roots of the commercially obtained kudzu material (Fig. 2a, b). The commercial kudzu line was morphologically very similar to the field-collected material, but could possibly be a different Pueraria species. It did, however, accumulate the major isoflavone 7-O-glucosides characteristic of kudzu roots (Fig. 2a, b).
Kudzu grows as a vine, and cuttings of field-collected kudzu established in the greenhouse produced long runners with distinct nodes and internodes (Fig. 3a, b). Analysis of these tissues indicated that puerarin levels increased with developmental age, with lowest levels in the first (youngest) internode to highest in the most mature (6th) node (Fig. 3). The higher levels in nodes than in internodes are likely due to the presence of adventitious root initials in the nodal regions (Fig. 3c), as puerarin levels are high in roots. We also detected a number of isoflavone aglycones (daidzein, genistein, formononetin) and their O-glycosides in the runners; their concentrations followed a similar developmental pattern to that of puerarin (Fig. 3c, d).
Production of puerarin in the stems of kudzu vines (field-collected isolate). a Vines growing in the greenhouse. b Runner showing the different internodes. c Close-up of a nodal region (node 6) showing adventitious roots. d HPLC profiles showing isoflavonoids in successive nodes e As above, for successive internodes
Generation of a kudzu root cDNA library and EST collection
The above-described field-collected and commercial kudzu roots provided material for a subtractive library approach toward the identification of genes involved in puerarin biosynthesis. A subtraction library with the commercial kudzu root cDNA as the driver was therefore constructed, to enrich for genes involved in C-glycosyl isoflavone biosynthesis (see “Materials and methods” section). To generate the ESTs, the inserts of 7,466 randomly picked cDNA clones were partially sequenced with M13 forward primer. The 6,365 high-quality sequences (average 538 bp in length) were clustered into 722 tentative consensus sequences (TCs) and 3,913 singletons to yield 4,635 apparent unigenes. These unisequences were annotated by BLAST search against the NCBI NR database, and 2,023 of the ESTs (43.6%) had significant sequence similarity to genes with assigned functions, 1,485 ESTs (32%) showed sequence similarity to genes with unknown function and 1,127 ESTs (24.3%) had no database match (Supplemental Fig. 2a). Gene ontology analysis showed that the largest group of ESTs was involved in catalytic activity (30%), and 2% were associated with transcription regulation activity (Supplemental Fig. 2b), which will provide a resource for future regulatory gene discovery. Only 32 ESTs were predicted to be involved in (iso)flavonoid biosynthesis based on our current understanding of the pathway (Tian et al. 2008); each gene was represented by only a single EST except for chalcone synthase (5 ESTs), chalcone reductase (10 ESTs), isoflavone synthase (3 ESTs) and isoflavone reductase (9 ESTs).
Genes of isoflavone biosynthesis in kudzu roots
Analysis of the kudzu root EST collection allowed us to identify gene sequences corresponding to most of the enzymes of isoflavone biosynthesis, including CHI, IFS and HID (Table 1). The kudzu IFS is most closely related to soybean IFS (AF195798) with 92.7% amino acid sequence identity. qRT-PCR confirmed that IFS transcripts were highly expressed in kudzu root and runner tissues that actively produce puerarin (Fig. 4). IFS is the entry point enzyme in the biosynthesis of isoflavonoids, and its over-expression has led to de novo or increased accumulation of isoflavone glycosides in various plant species (Deavours and Dixon 2005; Liu et al. 2002; Yu et al. 2000, 2003). IFS from kudzu provides a useful tool for metabolic engineering of puerarin.
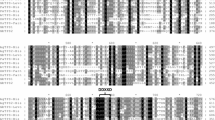
Hierarchical cluster analysis of transcript levels of kudzu glycosyltransferases in relation to isoflavone synthase in different kudzu tissues at different developmental stages. Tissues were harvested from internodes 1–6 and nodes 1–6 with 1 = youngest, roots at 6 days, 17 days, 3 weeks and 5 weeks, and young leaves. Transcript levels were determined by qRT-PCR, and clustering was performed using Spotfire software (TIBCO Spotfire)
Blast search of the kudzu full-length HID sequence (obtained by RACE from the kudzu EST collection) showed that it had 87% amino acid identity to soybean HID, 60% identity to licorice HID and possessed the residues of the putative catalytic triad (Thr, Asp and His) and the oxyanion hole (His–Gly–Gly) as reported (Akashi et al. 2005) (Supplemental Fig. 3a). The kudzu HID gene is expressed in roots, but also in stems (Supplemental Fig. 3b). Recently, it was reported that overexpression of soybean HID in hairy roots of Lotus japonicus led to accumulation of daidzein and genistein at comparable levels to those found in transgenic Arabidopsis or alfalfa over-expressing IFS (Liu et al. 2002; Deavours and Dixon 2005), but that over-expression of licorice IFS did not lead to accumulation of isoflavones (Shimamura et al. 2007). The authors concluded that HID was a critical determinant for isoflavone biosynthesis in L. japonicus hairy root cultures. Cloning of the HID from kudzu may therefore provide an additional tool for metabolic engineering of isoflavonoids, including the C-glucosides. It is interesting to note that there was only one copy of HID in the genome of the field-collected kudzu material (data not shown). This implies that, if C-glycosylation takes place at the level of 2-hydroxyisoflavanone (see below), the kudzu HID would be able to use both 8-C-glucosyl 2,4′,7-trihydroxyisoflavanone and 2,4′,7-trihydroxyisoflavanone as substrates in the formation of puerarin and daidzein, respectively. Likewise, soybean HID can use both 4′-hydroxylated and 4′-methoxylated 2-hydroxyisoflavanones as substrates, whereas licorice HID is specific for 2,7-dihydroxy-4′-methoxyisoflavanone. The copy numbers of HID in these two latter species have not been reported (Akashi et al. 2005).
CHI catalyzes the second committed step in (iso)flavonoid biosynthesis, the isomerization of 6′-deoxychalcone (4,2′,4′-trihydroxychalcone) or 6′-hydroxychalcone (4,2′,4′,6′-tetrahydroxychalcone) into corresponding 5-deoxyflavanone (7,4′-dihydroxyflavanone) or 5-hydroxyflavanone (5,7,4′-trihydroxyflavanone, naringenin) (Fig. 1). Type I CHIs, generally found in non-leguminous plants, catalyze conversion of only 6′-hydroxychalcones to the corresponding 5-hydroxyflavanones, whereas Type II CHIs, mostly found in leguminous plants, catalyze isomerization of both 6′-deoxy- and 6′-hydroxy chalcones. Amino acid sequence identities of around 70–80% are observed within the Type I and Type II CHI groups, whereas the between-group identities are around 50% (Shimada et al. 2003). The deduced amino acid sequences of kudzu CHIs 1 and 2 were most closely related to soybean CHI 1A and 1B1 (Ralston et al. 2005), and clustered in the Type II CHI group. Kudzu CHI 1 had 93.6% amino acid identity to soybean CHI 1A, and kudzu CHI 2 had 93.2% identity to soybean CHI 1B1 (Supplemental Fig. 4). Sequence alignment showed that both kudzu CHIs had identical active site residues to those in the Medicago sativa CHI crystal structure (Supplemental Fig. 5) (Jez et al. 2000).
The substrate specificities of both kudzu CHIs were determined by activity assay in vitro. Both recombinant CHIs were highly expressed as soluble proteins in E. coli and were purified using the MagneHis tag procedure. Ten chalcone substrates were used for enzyme activity determination. Both CHI1 and 2 had similar substrate preferences to soybean CHI 1A, and isomerized six out of the ten chalcones tested, including both 6′-deoxy and 6′-hydroxy chalcones (Table 2, Supplemental Fig. 6a, b). Thus, both kudzu CHI1 and 2 are Type II CHIs.
qRT-PCR analysis showed that both CHIs had very similar tissue-specific expression pattern and were highly expressed in 17-day, 24-day and 5-week-old kudzu roots, with CHI1 being expressed approximately sixfold higher than CHI 2 (Supplemental Fig. 7a, b).
Identification of glycosyltransferases from the kudzu EST collection
Using keyword searching from the kudzu EST database, 15 family 1 glycosyltransferases (UGTs) were identified (Table 3). Full-length cDNAs of these UGTs were obtained by 5′ and 3′ RACE. Deduced amino acid sequence alignments of the 15 UGTs confirmed the presence of the conserved PSPG box (UDP-binding domain) at the C-termini (Supplemental Fig. 8a, b). The sequence identity between the kudzu UGTs was from 22 to 80%. Phylogenetic analysis of the kudzu enzymes with representative Arabidopsis UGTs showed that 11 out of the 15 kudzu UGTs clustered into just two groups of Arabidopsis UGTs (five in Group D and six in Group E) (Supplemental Fig. 9).
Group D UGTs are considered to be related to stress or defense responses. For example, expression of UGT73B3 and UGT73B5 is induced by salicylic acid in Arabidopsis, and T-DNA tagged mutants (ugt73b3 and ugt73b5) exhibit decreased resistance to the bacterial pathogen Pseudomonas syringae pv tomato-AvrRpm1 (Langlois-Meurinne et al. 2005). UGT73B3 and UGT73B4 are active in vitro toward hydroxybenzoic acids and analogs of salicylic acid (Lim et al. 2002), while UGT73C1 and UGT73C5 have been characterized as glycosylating the plant hormones trans-zeatin or brassinosteroid (Hou et al. 2004; Poppenberger et al. 2005). However, UGT73C6 was identified as a UDP-glucose: flavonol-3-O-rhamnoside-7-O-glucosyltransferase (Jones et al. 2003). Interestingly, UGT73B4 and UGT73C1 are capable of forming both O- or C-glucosidic bonds on 2,4,6-trinitrotoluene (TNT) (Gandia-Herrero et al. 2008). Similarly, the urdamycin glycosyltransferase (UrdGT2) of antibiotic biosynthesis from the soil bacterium Streptomyces fradiae displays both C- and O-glycosylation activity (Durr et al. 2004). The kudzu UGTs clustering in Group D (GT02J01, GT14A05, GT03H14, GT14K13, GT18P15) were more related to each other than to the Arabidopsis UGTs in this Group (Supplemental Fig. 9).
Arabidopsis Group E includes the UGT 71 and 72 families (Ross et al. 2001). Some members of the 71 family have been shown to glycosylate abscisic acid and caffeic acid (Lim et al. 2003; Priest et al. 2005). Members of the Arabidopsis UGT72B subfamily can detoxifiy pollutants such as 3,4-dichloroaniline and 2,4,5-trichlorophenol with bifunctional O- and N-glycosylation activities (Brazier-Hicks and Edwards 2005; Brazier-Hicks et al. 2007), whereas members of the UGT72E subfamily may be involved in lignin monomer glycosylation (Lim et al. 2005; Lanot et al. 2006) and UGT72L1 from the model legume Medicago trucnatula, preferentially O-glycosylates (-)-epicatechin as a key step in the biosynthesis of proanthocyanidins (Pang et al. 2008). The kudzu UGTs clustering in Group E include GT04F14, GT12D15, GT07O02, GT01K01, GT19J14 and GT03H24 (Supplemental Fig. 9). GT04F14, GT12D15 and GT07O02 are in the UGT72B subfamily and GT01K01 is closely related to the UGT72E subfamily, while GT03H24 falls in the UGT71 family.
Kudzu GT10J15 clusters in the Arabidopsis UGT Group A that is relatively small and includes UGT91C1, which is a member of a separate clade of sugar branch-forming glycosyltransferases (Frydman et al. 2004; Morita et al. 2005; Bowles et al. 2005). Further phylogenetic analysis of GT10J15 showed that it also clusters with sugar branch-forming glycosyltransferases (Supplemental Fig. 10). Its amino acid sequence is most closely related (74% similarity) to that of a UDP-glucose: anthocyanidin 3-O-glucoside-2″-O-glucosyltransferase from Ipomoea nil (Supplemental Table 4).
Supplemental Table 4 shows the mostly closely related functionally characterized plant UGTs to each of the 15 kudzu UGTs. The amino acid sequence similarities between the kudzu UGTs and the rice flavone C-GT (Brazier-Hicks et al. 2009) vary from 18% (GT10J15) to 33% (GT19J14).
Cluster analysis of expression patterns for identification of candidate isoflavone glycosyltransferases
To search for UGTs with potential functions in isoflavone glycosylation, we performed hierarchical cluster analysis of expression profiles (determined by qRT-PCR) to determine which UGTs were most tightly co-expressed with IFS. Tissues examined included different developmental stages of internodes, nodes, root and young leaves. Cluster analysis showed that IFS was expressed in all tissues examined, but with about five- to tenfold higher expression in root tissues compared to other tissues (Fig. 4). This expression pattern reflects that of isoflavone conjugate accumulation, which is highest in roots. GT12P06, GT01K01, GT04F14, GT02J01, GT07O02 and GT10J15 transcripts were highly expressed in roots compared to the other tissues, and the overall expression patterns of GT12P06 and GT01K01 were more than 97% identical to that of IFS, although their expression levels were much lower. GT04F14, GT02J01, GT07O02 and GT10J15 exhibited more than 85% similarity in transcript expression pattern to IFS, with expression levels of between 8 and 15% that of IFS. GT14A05, GT14K13 and GT14M03 were primarily expressed in young leaves, GT03H24 transcripts were highly expressed in all tissues except young leaves, and GT18P15 and GT19J14 transcripts had similar expression patterns, primarily in root tissues but also high in young leaves (Fig. 4). Together, these results identify 6 out of the 15 UGTs as being candidates for isoflavone O- and C-glycosyltransferases, and these were therefore prioritized for functional analysis.
Functional identification of kudzu UGTs
The open reading frames of the kudzu UGTs were cloned into the protein expression vector pDEST17 with an N-terminal His tag, and the proteins were expressed in E. coli BL21(DE3) pLysS cells and purified using MagneHis beads. Three of the UGTs, GT03H24, GT12D15 and GT21C20, did not produce protein when expressed in E. coli and were not analyzed further. Enzyme activities of the remaining UGTs were determined with a range of (iso)flavonoid substrates (Supplemental Fig. 10) and UDP-glucose as sugar donor. Neither GT12P06 nor GT01K01, the two UGTs with closest expression pattern to that of IFS, showed activity with any of the substrates listed in Supplemental Fig. 11.
GT04F14 showed broad substrate specificity and was active with isoflavone, flavone, flavonol and coumarin substrates (Table 3). All isoflavones tested were glycosylated on the 7-hydroxyl group, suggesting high regiospecificity with this class of flavonoid. GT04F14 also showed very high activities with the coumarins esculetin and scopoletin (>60% conversion in the standard assay) as reported for many plant UGTs (Lim et al. 2003). A regiospecificity switching event for glycosylation of esculetin on the 6- or 7-hydroxyl group occurred between glycosyltransferase families 71 and 72 in Group E, with family 71 conjugating on the 7-OH and family 72 preferring the 6-OH (Lim et al. 2003). Since GT04F14 clusters in the family 72 clade, it is likely that it also glycosylates the 6-OH of esculetin. Kinetic parameters for GT04F14 were determined with the isoflavone genistein. The Km value was 39.11 μM and the efficiency (Kcat/Km) was 70.23 M−1 S−1, values which are quite comparable to those for other plant UGTs (He et al. 2006; Tian et al. 2006). However, GT04F14 showed no activity with potential intermediates of puerarin biosynthesis, namely the chalcone isoliquiritigenin, the flavanone liquiritigenin (or naringenin), or 2,7,4′-trihydroxyisoflavanone. Since GT04F14 has quite diverse substrate specificity, it may have potential as a biocatalyst in biological fermentation systems for regioselective synthesis of diverse natural product glycosides (Lim et al. 2004; He et al. 2008).
Most of the other recombinant UGTs showed no activity with the substrates in Supplemental Fig. 11, GT03H14 exhibited weak activity toward liquiritigenin (1.4% conversion in the standard assay), GT07O02 was active toward 6,7,4′-trihydroxyisoflavone (4.1% conversion) and GT14A05 catalyzed glycosylation of luteolin on the 7-hydroxyl and of quercetin on the 3- and 4′-hydroxyls (about 1% conversion). Since GT10J15 is closely related to sugar-branching glycosyltransferases, isoflavone and flavonol glucosides such as daidzein 7-O-glucoside, genistein 7-O-glucoside and kaempferol 3-O-glucoside were examined as potential substrates, but no activity was detected.
Conclusions and perspectives
This study reports an EST analysis of transcripts expressed in roots of Pueraria lobata. The 4,635 unit ESTs provide a significant resource for comparative genomic studies between plant species for gene discovery. Genes encoding several key enzymes of isoflavone biosynthesis, including CHS, CHR, CHI, IFS and HID, were identified in the EST library and will provide tools for subsequent metabolic engineering of kudzu isoflavone glycosides.
The CHIs identified here showed broad substrate specificity similar to Type II CHIs from leguminous plants (Shimada et al. 2003). It is likely that kudzu CHI 1 and 2 will function as typical Type II CHIs in converting chalcone rather than chalcone C-glycoside to flavanone in vivo. Furthermore, none of the UGTs we have characterized are active with chalcone as substrate. We therefore feel that it is unlikely that C-glycosylation occurs at the chalcone stage during puerarin biosynthesis, although this was suggested by early labeling studies (Inoue and Fujita 1977). Subsequent to this previous hypothesis, it was shown that 4′-O-methylation during isoflavone biosynthesis occurs at the level of the trihydroxyisoflavanone product of IFS, not the isoflavone itself (Akashi et al. 2003), and this explains why labeled trihydroxychalcone but not daidzein is incorporated into the pterocarpan medicarpin (Dewick and Martin 1979). Similarly, 6- or 8-C-glycosylation of flavones occurs at the corresponding trihydroxyflavanone stage (Brazier-Hicks et al. 2009). We therefore propose that 8-C-glycosylation during puerarin biosynthesis likely takes place at the trihydroxyisoflavanone stage. This is supported by the observation that crude extracts from kudzu roots can convert 2,7,4′-trihydroxyisoflavanone to an unidentified intermediate of lower retention time (greater hydrophilicity), and that this compound disappears on exposure of extracts to weak acid, with a subsequent increase in the level of puerarin (Supplemental Fig. 12). The unstable intermediate is probably the 8-C-glucoside of the trihydroxyisoflavanone, which then dehydrates to puerarin at lower pH. However, this must remain a hypothesis until we can demonstrate the exact chemical nature of the unstable intermediate and show that it can also be formed by a recombinant kudzu UGT. To date, we have been unable to show glycosylation of trihydroxyisoflavanone by any of the UGTs described in the present paper.
Although the in vitro enzyme activities of many of the kudzu UGTs have not been determined, their in vivo biological functions are likely associated with secondary metabolism. GT04F14 showed 7-O-glycosylation regiospecificity toward several isoflavones in vitro. Since isoflavone 7-O- glycosides are major metabolites accumulated in kudzu roots, it is possible that GT04F14 is involved in isoflavone glycosylation in vivo. It is interesting that GT04F14 shows high activity toward fisetin (>60% conversion rate), since fisetin has been reported to enhance long-term memory and protect the nervous system (Maher et al. 2006; Maher 2008). This UGT could potentially be used as a biocatalyst for production of fisetin glycosides by fermentation.
Plant natural products often have more than one sugar attached at different positions, or even have branched-sugar modifications. The latter have significant biological functions in enhancing bitterness (Frydman et al. 2004), sweetness of Stevia rebaudiana (Brandle and Telmer 2007) or flower color (Kondo et al. 1987). The glycosyltransferases catalyzing the secondary metabolite branch-sugar modifications have evolved into a group distinct from other glycosyltransferases, and only a few of their cDNA have been cloned and functionally characterized (Frydman et al. 2004; Richman et al. 2005, Morita et al. 2005; Sawada et al. 2005; Noguchi et al. 2008). Kudzu GT10J15 is likely an additional member to this underexploited group of glycosyltransferases, since it has high amino acid similarity to a functionally characterized sugar-branching glycosyltranferase (Morita et al. 2005) and is phylogenetically clustered in this group.
References
Achnine L, Huhman DV, Farag MA, Sumner LW, Blount JW, Dixon RA (2005) Genomic-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J 41:875–887
Akashi T, Aoki T, Ayabe S (1999) Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol 121:821–828
Akashi T, Sawada Y, Aoki T, Ayabe S (2000) New scheme of the biosynthesis of formononetin involving 2, 7, 4′-trihydroxyisoflavanone but not daidzein as the methyl acceptor. Biosci Biotechnol Biochem 64:2276–2279
Akashi T, Sawada Y, Shimada N, Sakurai N, Aoki T, Ayabe S (2003) cDNA cloning and biochemical characterization of S-adenosyl-l-methionine: 2, 7, 4′-trihydroxyisoflavanone 4′-O-methyltransferase, a critical enzyme of the legume isoflavonoid phytoalexin pathway. Plant Cell Physiol 44:103–112
Akashi T, Aoki T, Ayabe SI (2005) Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol 13:882–891
Bowles DJ, Isayenkova J, Lim EK, Poppenberger B (2005) Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol 8:254–263
Brandle JE, Telmer PG (2007) Steviol glycoside biosynthesis. Phytochemistry 68:1855–1863
Brandle JE, Richman A, Swanson AK, Chapman BP (2002) Leaf ESTs from Stevia rebaudiana: a resource for gene discovery in diterpene synthesis. Plant Mol Biol 50:613–622
Brazier-Hicks M, Edwards R (2005) Functional importance of the family 1 glucosyltransferases UGT72B1 in the metabolism of xenobiotics in Arabidopsis thaliana. Plant J 42:556–566
Brazier-Hicks M, Offen WA, Gershater MC, Revett TJ, Lim EK, Bowles D, Davies GJ, Edwards R (2007) Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci USA 104:20238–20243
Brazier-Hicks M, Evans KM, Gershaater MC, Puschmann H, Steel PG, Edwards R (2009) The C-glycosylation of flavanoids in cereals. J Biol Chem 284:17926–17934
Byrne PF, McMullen MD, Snook ME, Musket TA, Theuri JM, Widstrom NW, Wiseman BR, Coe EH (1996) Quantitative trait loci and metabolic pathways: genetic control of the concentration of maysin, a corn earworm resistance factor, in maize silks. Proc Natl Acad Sci USA 93:8820–8825
Cortes-Cruz M, Snook M, McMullen MD (2002) The genetic basis of C-glycosyl flavone B-ring modification in maize (Zea mays L.) silks. Genome 46:182–194
Crombie L, Whiting DA (1992) The mechanism of the enzymic induced flavanone–isoflavone change. Tetrahedron Lett 33:3663–3666
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Deavours BE, Dixon RA (2005) Metabolic engineering of isoflavonoid biosynthesis in alfalfa. Plant Physiol 138:2245–2259
Dewick PM, Martin M (1979) Biosynthesis of pterocarpan, isoflavan and coumestan metabolites of Medicago sativa: chalcone, isoflavone and isoflavanone precursors. Phytochemistry 18:597–602
Durr C, Hoffmeister D, Wohlert S, Ichinose K, Weber M, Mulert U, Thorson JS, Bechthold A (2004) The glycosyltransferase UrdGT2 catalyzes both C- and O-glycosidic sugar transfers. Angew Chem Int Ed 43:2962–2965
Frydman A, Weisshaus O, Bar-Peled M, Huhman DV, Sumner LW, Marin FR, Lewin-Sohn E, Fluhr R, Dreesel J, Eyal Y (2004) Citrus fruit bitter flavors: isolation and functional characterization of the gene CM1, 2RhaT encoding a 1, 2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. Plant J 40:88–100
Gandia-Herrero F, Lorenz A, Larson T, Graham IA, Bowles D, Rylott EL, Bruce N (2008) Detoxification of the explosive 2, 4, 6-trinitrotoluene in Arabidopsis: discovery of bifunctional O- and C-glucosyltransferases. Plant J 56:963–974
Hakamatsuka T, Mori K, Ishida S, Ebizuka Y, Sankawa U (1998) Purification of 2-hydroxyisoflavanone dehydratase from the cell cultures of Pueraria lobata. Phytochemistry 49:497–505
Hashim MF, Hakamatsuka T, Ebizuka Y, Sankawa U (1990) Reaction mechanism of oxidative rearrangement of flavanone in isoflavone biosynthesis. FEBS Lett 271:219–222
He XZ, Wang XQ, Dixon RA (2006) Mutational analysis of the Medicago glycosyltransferase UGT71G1 reveals residues that control regioselectivity for (iso)flavonoid glycosylation. J Biol Chem 281:34441–34447
He XZ, Li WS, Blount JW, Dixon RA (2008) Regioselective synthesis of plant (iso)flavone glycosides in Escherichia coli. Appl Microbiol Biotechnol 80:253–260
Hou B, Lim EK, Higgins GS, Bowles DJ (2004) N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem 279:47822–47832
Inoue T, Fujita M (1977) Biosynthesis of puerarin in pueraria root. Chem Pharm Bull 25:3226–3231
Jako C, Coutu C, Roewer I, Reed DW, Pelcher LE, Covello PS (2002) Probing carotenoid biosynthesis in developing seed coats of Bixa orellana (Bixaceae) through expressed sequence tag analysis. Plant Sci 163:141–145
Jez JM, Bowman ME, Dixon RA, Noel JP (2000) Structure and mechanism of the evolutionary unique plant enzyme chalcone isomerase. Nat Struct Biol 7:786–791
Jones P, Messner B, Nakajima J, Schaffner A, Saito K (2003) UGT73C6 and UGT78D1 glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem 278:43910–43918
Jung W, Yu O, Lau SMC, O’Keefe DP, Odell J, Fader G, McGonigle B (2000) Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol 18:208–212
Kerscher F, Franz G (1987) Biosynthesis of vitexin and isovitexin: Enzymatic synthesis of the C-glucosylflavones vitexin and isovitexin with an enzyme preparation from Fagopyrum esculentum M. seedlings. Z Naturforsch 42c:519–524
Kerscher F, Gerhard F (1988) Isolation and some properties of an UDP-glucose: 2-hydroxyflavanone-6(or 8)-C-glucosyltransferase from Fagopyrum esculentum M. cotyledons. J Plant Physiol 132:110–115
Kim JH, Kim BG, Ko JH, Lee Y, Hur HG, Lim Y, Ahn JH (2006) Molecular cloning, expression, and characterization of a flavonoid glycosyltransferase from Arabidopsis thaliana. Plant Sci 170:897–903
Kochs G, Grisebach H (1986) Enzymic synthesis of isoflavones. Eur J Biochem 155:311–318
Kondo T, Kawai T, Kamura H, Goto T (1987) Structure determination of heavenly blue anthocyanin, a complex monomeric anthocyanin from the morning glory Ipomoea tricolor, by means of the negative Noe method. Tetrahedron Lett 28:2273–2276
Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R (2000) Probing essential oil biosynthesis and secretion by functional evaluation of expression sequence tags from mint glandular trichomes. Proc Natl Acad Sci USA 97:2934–2939
Langlois-Meurinne M, Gachon CMM, Saindrenan P (2005) Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol 139:1890–1901
Lanot A, Hodge D, Jackson RG, George GL, Elisa L, Lim EK, Vaistij FE, Bowles DJ (2006) The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J 48:286–295
Li L, Modolo LV, Escamilla-Trevino LL, Achnine L, Dixon RA, Wang X (2007) Crystal structure of Medicago truncatula UGT85H2: insights into the structural basis of a multifunctional (iso)flavonoid glycosyltransferases. J Mol Biol 370:951–963
Liang WW, Yu HR, Wang HX (2005) Research progress of pharmacology and clinics about puerarin on treating coronary disease. J Jin Zhou Med Coll 26(5):56–59
Lim EK, Doucet CJ, Li Y, Elias L, Worrall D, Spencer SP, Ross J, Bowles D (2002) The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J Biol Chem 277:586–592
Lim EK, Baldauf S, Li Y, Elias L, Worrall D, Spencer SP, Jackson RG, Taguchi G, Ross J, Bowles DJ (2003) Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 13:139–145
Lim EK, Ashford DA, Hou B, Jackson RG, Bowles DJ (2004) Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotech Bioeng 87:623–631
Lim EK, Jackson RG, Bowles DJ (2005) Identification and characterization of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Lett 579:2802–2806
Liu CJ, Blount JW, Steele CL, Dixon RA (2002) Bottlenecks for metabolic engineering of isoflavone glycoconjugates in Arabidopsis. Proc Natl Acad Sci USA 99:14578–14583
Liu CJ, Deavours BE, Richard SB, Ferrer JL, Blount JW, Huhman DV, Dixon RA, Noel JP (2006) Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses. Plant Cell 18:3656–3669
Maher P (2008) The flavonoid fisetin promotes nerve cell survival from trophic factor withdrawal by enhancement of proteasome activity. Arch Biochem Biophys 476:139–144
Maher P, Akaishi T, Abe K (2006) Flavonoid fisetin promotes ERK-dependent long-term potentiation and enhances memory. Proc Natl Acad Sci USA 103:16568–16573
Maxwell CA, Harrison MJ, Dixon RA (1993) Molecular characterization and expression of alfalfa isoliquiritigenin 2′-O-methyltransferase, an enzyme specifically involved in the biosynthesis of an inducer of Rhizobium meliloti nodulation genes. Plant J 4:971–981
Meezan E, Meezan EM, Jones K, Moore R, Barnes S, Prasain JK (2005) Contrasting effects of puerarin and daidzin on glucose homeostasis in mice. J Agric Food Chem 53:8760–8767
Modolo LV, Blount JW, Achnine L, Naoumkina MA, Wang XQ, Dixon RA (2007) A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol Biol 64:499–518
Morita Y, Hoshino A, Kikuchi Y, Okuhara H, Ono E, Tanaka Y, Fukui Y, Saito N, Nitasaka E, Noguchi H, lida S (2005) Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2′-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J 42:353–363
Murata J, Bienzle D, Brandle JE, Sensen CW, De Luca V (2006) Expressed sequence tags from Madagascar periwinkle (Catharanthus roseus). FEBS Lett 580:4501–4507
Nagel J, Culley LK, Lu YP, Liu EW, Matthews PD, Stevens JF, Page JE (2008) EST analysis of hop glandular trichomes identifys an O-methyltransferase that catalyzes the biosynthesis of xanthohumol. Plant Cell 20:186–200
Ni X, Krakowsky MD, Buntin GD, Rector BG, Guo B, Snook ME (2008) Identification of multiple Ear-colonizing insect and disease resistance in CIMMYT maize inbred lines with varying levels of silk maysin. J Econ Entomol 101:1455–1465
Noguchi A, Fukui Y, Luchi-Okada A, Kakutani S, Satake H, Iwashita T, Nakao M, Umezawa T, Ono E (2008) Sequential glucosylation of a furofuran lignan, (+)-sesaminol, by Sesamum indicum UGT71A9 and UGT94D1 glucosyltransferases. Plant J 54:415–427
Oshima Y, Okuyama T, Takahashi K, Takizawa T, Shibata S (1988) Isolation and high performance liquid chromatography (HPLC) of isoflavonoids from the Pueraria root. Planta Med 54:250–254
Pang Y, Peel GJ, Sharma SB, Tang Y, Dixon RA (2008) A transcript profiling approach reveals an epicatechin glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc Natl Acad Sci USA 105:14210–14215
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Poppenberger B, Fujioka S, Soeno K, George GL, Vaistij FE, Hiranuma S, Seto H, Takatsuto S, Adam G, Yoshida S, Bowles D (2005) The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc Natl Acad Sci USA 102:15253–15258
Priest DM, Jackson RG, Ashford DA, Abrams SR, Bowles DJ (2005) The use of abscisic acid analogues to analyze the substrate selectivity of UGT71B6, a UDP-glycosyltransferase of Arabidopsis thaliana. FEBS Lett 579:4454–4458
Ralston L, Subramanian S, Matsuno M, Yu O (2005) Partial reconstruction of flavonoid and isoflavonoid biosynthesis in yeast using soybean Type I and Type II chalcone isomerases. Plant Physiol 137:1375–1388
Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 13:62–66
Richman A, Swanson A, Humphrey T, Chapman R, McGarvey B, Pocs R, Brandle J (2005) Functional genomics uncovers three glucosyltransferases involved in the synthesis of the major sweet glucosides of Stevia rebaudiana. Plant J 41:56–67
Rong H, Stevens JF, Deinzer ML, Cooman LD, Keukeleire DD (1998) Identification of isoflavones in the roots of Pueraria lobata. Planta Med 64:620–627
Ross J, Lim EK, Bowles D (2001) Higher plant glycosyltransferases. Genome Biol 2:3004.1–3004.6
Sawada S, Suzuki H, Ichimaida F, Yamaguchi MA, Iwashita T, Fukui Y, Hemmi H, Nishino T, Nakayama T (2005) UDP-glucuronic acid: anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers. J Biol Chem 280:899–906
Shimada N, Aoki T, Sato S, Nakamura Y, Tabata S, Ayabe S (2003) A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol 131:941–951
Shimada T, Endo T, Fujii H, Hara M, Ueda T, Kita M, Omura M (2004) Molecular cloning and functional characterization of four monoterpene synthase genes from Citrus unshiu Marc. Plant Sci 166:49–58
Shimamura M, Akashi T, Sakura N, Suzuki H, Saito K, Shibata D, Ayabe S, Aoki T (2007) 2-Hydroxyisoflavanone dehydratase is a critical determinant of isoflavone productivity in hair root cultures of Lotus japonicus. Plant Cell Physiol 48:1652–1657
Steele CL, Gijzen M, Qutob D, Dixon RA (1999) Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch Biochem Biophys 367:147–150
Terai Y, Fujii I, Byun SH, Nakajima O, Hakamatsuka T, Ebizuka Y, Sankawa U (1996) Cloning of chalcone-flavanone isomerase cDNA from Pueraria lobata and its overexpression in Escherichia coli. Protein Expr Purif 8:183–190
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weigh matrix choices. Nucleic Acids Res 22:4673–4680
Tian L, Blount JW, Dixon RA (2006) Phenylpropanoid glycosyltransferases from osage orange (Maclura pomifera) fruit. FEBS Lett 580:6915–6920
Tian L, Pang Y, Dixon RA (2008) Biosynthesis and genetic engineering of proanthocyanidins and (iso)flavonoids. Phytochem Rev 7:445–465
Wallace JW, Grisebach H (1973) The in vivo incorporation of a flavanone into C-glycosylflavones. Biochim Biophys Acta 304:837–841
Wallace JW, Mabry TJ, Alston RE (1969) On the biogenesis of flavone O-glycosides and C-glycosides in the Lemnaceae. Phytochemistry 8:93–99
Wu LM, Qiao HX, Li YK, Li LD (2007) Cardioprotective effects of the combined use of puerarin and danshensu on acute ischemic myocardial injury in rats. Phytotherapy Res 21:751–756
Yeung DK, Leung SW, Xu YC, Vanhoutte PM, Man RY (2006) Puerarin, an isoflavonoid derived from Radix puerariae, potentiates endothelium-independent relaxation via the cyclec AMP pathway in porcine coronary artery. Eur J Pharmacol 552:105–111
Yu O, Shi J, Croes RA, Fader GM, McGonigle B, Odell JT (2000) Production of the isoflavones genistein and daidzein in non-legume dicot and monocot tissues. Plant Physiol 124:781–794
Yu O, Shi J, Hession AO, Maxwell CA, McGonigle B, Odell JT (2003) Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 63:73–76
Acknowledgments
We thank Drs. Ji He and Patrick Zhao for processing of ESTs, Dr. Marina Naoumkina for assistance in transcript cluster analysis, Tui Ray for assistance with qRT-PCR, Darla Boydston for assistance with artwork, and Drs. Yongzhen Pang and Rui Zhou for critical reading of the manuscript. This work was supported by the Oklahoma Center for the Advancement of Science and Technology (OCAST) Plant Science Program, and by the Samuel Roberts Noble Foundation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
He, X., Blount, J.W., Ge, S. et al. A genomic approach to isoflavone biosynthesis in kudzu (Pueraria lobata). Planta 233, 843–855 (2011). https://doi.org/10.1007/s00425-010-1344-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1344-1