Abstract
Plants produce a wide variety of compounds called secondary metabolites (SMs), which are extremely important for their survival. SMs have also medicinal applications, but as chemical synthesis is not economically viable, plant extraction is the mainly option. Different biotechnology strategies are applied to improve the yield of bioproduction of these compounds, but commonly without the desired results due the limited knowledge of biosynthetic and regulatory pathways. Maytenus ilicifolia, a traditional Brazilian medicinal plant from Celastraceae family, produces in both root and leaves three main classes of SMs: sesquiterpenics, flavonoids and quinonemethides. In this study, four cDNA libraries were prepared from root and leaf tissues. The de novo transcriptome included 109,982 sequences that capture 92% of BUSCO orthologs, presented an average length of 737bp and a GC content about 42% of. Function annotation analysis identified homology for 44.8% of the transcripts. Moreover, 67,625 sequences were commonly expressed in both tissues, while 1,044 and 1,171 were differentially expressed in root and leaf, respectively. In terms of SM, enzymes involved in “monoterpenoid biosynthesis” and “isoflavonoid biosynthesis" were identified in root while “flavonoid biosynthesis” and “Biosynthesis of alkaloids” in leaf.
Supported by São Paulo Research Foundation (FAPESP) [2013/07600-3; 2016/16970-7]; National Council for Scientific and Technological Development (CNPq) [303757/2017-5]; National Institute for Science and Technology (INCT).
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Compounds produced by plants are categorized into primary and secondary metabolites (SMs). Primary metabolites, such as carbohydrates, lipids, and proteins, are involved in plant development [9] and essential for cell growth [17]. In contrast, SMs (low molecular weight compounds) are multifunctional metabolites produced as an evolutionary adaptation [4, 23]. They are involved in plant defense and environmental communication [4, 8, 23], plant color, taste, and scent [9] and responses to biotic and abiotic stress [15, 16, 19].
The high variety of biological functions of the SMs is explained by diversified chemical structure [4, 24] originated from a restricted and distinct number of metabolic pathways such as the acetate, shikimic acid, mevalonic acid or methylerythritol phosphate pathways [9, 21]. SM are grouped in three classes: terpenes, alkaloids and phenylpropanoids, each one with its respective and unique properties [4, 24]. These compounds are identified in all plant tissues and their formation and gene regulation is usually organ, tissue, cell and also development specific, indicating that a range of transcription factors must cooperate to transcribe secondary metabolism genes, controlling the general machinery of biosynthetic pathways in production, transport and storage [18, 22].
Many SMs are sources of drugs however, as chemical synthesis is uneconomical, isolation from plants still represents the only option [4, 13]. Different biotechnological strategies have been applied to improve the production of these compounds, but often without the desired results due to the lack of knowledge about the biosynthetic routes [13, 21]. Biotechnology techniques such as transcriptome, proteome or metabolomics are used to identify genes and their functions in plant metabolic pathways in order to clarify the mechanisms involved in SMs synthesis [4].
Maytenus ilicifolia Mart ex Reissek (Celastraceae) is a Brazilian native plant known for its variety of therapeutic properties. It has been used as a treatment of several diseases such as gastric ulcer, dyspepsia, stomach acidity, diabetes and cancer [12, 13, 20]. This species produces three main classes of bioactive compounds: alkaloids sesquiterpene pyridines, flavonoids and quinonemethide triterpenes [13] and the mainly products are maitenin, friedelin, fridelanol, pristimerine and terpenes [14]. Additionally, like other members of Celastraceae family, some compounds are synthetized in a specific tissue: quinone methide triterpenoids are accumulated in root bark [1, 13] and flavonoids in leaves [2].
The analysis of differentially expressed transcripts between two tissues can provide a better understanding of genes involved in secondary metabolic pathways [3, 10, 11]. In this context, the aim of the present study was to analyze whole transcriptome of M. ilicifolia and identify genes involved in biosynthesis of SMs by a comparative profiling of root and leaf. This study is the first report of high-throughput analysis (de novo RNA-Seq) of M. ilicifolia transcriptome that provides new insights at molecular knowledge.
2 Methods
2.1 Plant Material and Total RNA Isolation
Leaves of adult specimen of M. ilicifolia from the medicinal plant garden of the Faculty of Pharmaceutical Sciences and leaves and roots of identified seedlings, with approximately 6 months of planting, were harvested and stored in −80 \(^\circ \)C (Fig. 1A). The total RNA from two specimens of roots (from two seedlings) and two specimens of leaves (one leaf from seedling, coinciding with one of the specimens used for root extraction and one leaf from adult specimen) was isolated from 500 mg of material using RNeasy Plant mini kit (Qiagen, USA) according to the manufacturer’s protocol. RNA quantity and quality were evaluated using Nanodrop 1000 spectrophotometer and Agilent 2100 Bioanalyzer. RNA samples with quality ratios greater than 1.8 (260/280 nm and 28S/18S) and RNA integrity number (RIN) greater than 7 were selected for subsequent processes.
Experimental approaches for Maytenus ilicifolia transcriptome study. A. Two samples of leaves (one leaf from adult specimen and one leaf from seedling) - L1 and L2 - and two samples of roots from two independent seedlings (one root sample coinciding with the same specimen of the leaf sample) - R1 and R2 - were collected and stored in −80 \(^\circ \)C for posterior RNA isolation. B. Library preparation and transcriptome sequencing C. Pipeline used for de novo assembly.
2.2 Library Preparation and Sequencing
After isolated from total RNA with magnetic Oligo (dT) particles, mRNA was chemically fragmented. Subsequently, cDNA libraries were prepared using Illumina TruSeq RNA sample preparation v3 kit (Illumina, USA) (Fig. 1B). Quantification and quality assessment of resulting libraries were performed on Agilent 2100 Bioanalyzer. A total of 20 pmol of the libraries was submitted to “single-read” sequencing in HiSeq 2000 platform (leaf of the adult specimen) - FCAV/Unesp - to generate 100bp reads or sequencing in MiSeq equipment (leaf and roots of seedlings) - LAB Multi-FCFAR/Unesp - to generate 75bp paired-end reads (Table 1).
2.3 Quality Control and de novo Assembly
The public server Galaxy (usegalaxy.org) was used to process the high-throughput data. The raw data generated by the sequencing, FASTQ files, were evaluated by the FastQC tool (v0.11.8) for quality before and after filtering and for GC content. Reads were filtered by TrimGalore! (v0.6.3), removing adapter contamination and low-quality sequences (average quality below 25). Initial and final bases were also removed from sequences with “q” value lower than 25 and, finally, in the final FASTQ file of filtered reads, those with a size greater than 50 base pairs remained.
The high-quality data of roots and leaves samples was assembled using Trinity (v2.9.1) on default parameters. The de novo assembly was evaluated by different quality metrics including N50 length and BUSCO v4.1.2 analysis using OrthoDB v10 ‘embryophyta’ database as a reference to access the assembly and annotation completeness. Filtered reads were remapped to the assembled transcriptome in order to obtain, using Salmon tool, an expression matrix reported in transcripts per kilobase million (TPM). This matrix allowed the filtering of transcripts by low expression, considering only those with at minimum 1% of dominant isoform expression, generating the filtered transcriptome.
2.4 Functional Annotation
TransDecoder tool was used to find the probable coding regions of transcripts and the open reading frames (ORFs) with a minimum length of 100 amino acids. Then, functional annotation of the transcripts was performed using BLASTX against Uni-ProtKB/SwissProt databases and uniprot _trEMBL _plants database (E-value<1e−5). Moreover, a homology search based on the BLASTP was performed using the predicted proteins as query against UniProtKB/SwissProt databases (E-value<1e−5). The assignments of Gene Ontology (GO) terms to transcripts were performed based on UniProtKB/SwissProt database to assign unigenes to functional categories. Additionally, the proteins with Enzyme Commission (EC) numbers were mapped onto the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Database using online KEGG Automatic Annotation Server (www.genome.jp/kegg/kaas) to assign pathway information to the transcripts.
2.5 Differential Expression Analysis
Salmon tool was applied to estimate the expression level of transcripts. Each filtered FASTQ file was separately aligned to the filtered transcriptome. Then, the expression level of each transcript was normalized and reported in TPM. To summarize the results and provide statistical tests for tissue comparison, the differential expression analysis was performed using DESeq2 R package and transcript expression difference was considered significant when the adjusted p-value< 0.05.
2.6 Gene Ontology Enrichment and KEGG Analysis
Gene ontology (GO) enrichment analysis for biological process (BP) and molecular function (MF) for the differentially expressed transcripts in each tissue was conducted using topGO R package. Significant GO terms (Fisher’s exact test p-value< 0.01) were visualized using REViGO (revigo.irb.hr) for semantic space reduction. Transcripts associated with Enzyme Commission (EC) numbers were mapped onto the KEGG pathway database.
3 Results and Discussion
3.1 De novo Assembly and Functional Annotation of M. ilicifolia
The single-read leaf cDNA library and the paired-end leaf and root cDNA libraries subjected to full transcriptome sequencing generated about 115 million of raw reads. The detailed information of the read numbers in different samples is provided in Table 1.
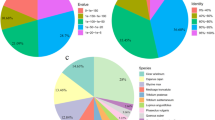
High quality sequencing data, 112,609,211 reads, was used for assembly. The de novo transcriptome generated included 163,780 transcripts (isoforms) with a GC content of 41.8% and the N50 resulting in 1,222bp. The average transcript size was 737 and 22% of them presented more than 1,000bp (Fig. 2A). By considering transcript expression, 15,704 transcripts represented 90% of the total expression data (Ex90) and had an N50 of 1,487bp (Ex90N50). In addition, the assembled transcriptome of M. ilicifolia captured 92.6% of the 1,614 orthologs described for the Virdiplantae database (updated 2020-09-10): 53.0%, 39.6%, 4.2%, and 3.2% of the BUSCO genes were respectively classified as complete single copy, complete duplicate, fragmented and absent. After filtering by low expression, the final transcriptome included 109,982 sequences. These results indicate that the integrity of assembly was high, and the sequencing quality had met the requirements of further analysis.
Aspects of Maytenus ilicifolia transcriptome assembly. A. Size distribution of assembled data. B. Principal component analysis (PCA) on the read counts of root and leaf samples. (L1, L2, R1 and R2 - sample identification described in Fig. 1) C. Venn diagram showing the number of transcripts for each source of sample D. transcriptome traits for each tissue.
Results of PCA analysis revealed the distinct differences in transcript expression patterns among the samples. The first two principal components contain 69.12% of the information grouping different tissues in separate clusters (Fig. 2B). Considering transcripts identified in leaf and root individually, 67,625 isoforms were found in both tissues (Fig. 2C) and showed similar aspects in respect to transcriptome traits (Fig. 2D).
In summary, M. ilicifolia transcriptome had GC content close to 40%, similar values to those reported for Celastraceae family species like staff vine (41.5%) [20] and thunder god vine (37.2%) [22]. Moreover, results of BUSCO analysis captured more than 90% of the orthologs described for the chosen database and the PCA results allowed the confirmation of expression differences in both tissues, root and leaf.
The BLASTX against the uniprot_trEMBL _plants database found 36,625 alignments and revealed that M. ilicifolia predicted transcripts have highest similarity with an organism classified in the same family, Tripterygium sp (47.3%) (Fig. 3A), but homology was find for other family organisms (Fig. 3B). Candidate coding regions in M. ilicifolia transcriptome were identified by TransDecoder and 65,533 ORFs and 46,282 probable coding sequences were predicted. Sequence homology search results against the UniprotKB/SwissProt database by BLASTX (E-value<1e−5, for filtered transcripts) and BLASTP (E-value<1e−5, for predicted protein sequences) were 49,319 (44.8%) and 36,344 (55.5%) aligned transcripts, respectively.
Functional annotation for filtered transcriptome was followed by GO analysis and 43,322 annotated transcripts were categorized into 9,989 GO IDs. The number of transcripts in three main categories of molecular function (MF), biological process (BP) and cellular component (CC) was 41,148, 39,404 and 39,582, respectively. The most dominant GO terms in the MF category were “protein binding,” “ATP binding” and “metal iron binding” (Fig. 3C). In the BP category, “regulation of transcription”, “protein phosphorylation” and “protein ubiquitination” were the most prominent (Fig. 3D). In the CC category, “nucleus”, “plasma membrane” and “integral component of membrane” were the most abundant terms (Fig. 3E).
KEGG annotation analysis was performed to identify active metabolic processes in M. ilicifolia transcriptome. In conclusion, 2,326 transcripts were assigned to 428 KEGG pathways. Considering “Metabolism of terpenoids and polyketides”, the most representative pathway was “Terpenoid backbone biosynthesis (ko00900)”, followed by “Sesquiterpenoid and triterpenoid biosynthesis (ko00909)”, with 216 and 127 mapped sequences that represent 50% and 10% of the orthologous for each pathway, respectively.
In conclusion, the identification of about 40,000 protein accessions indicates that in this study the de novo RNA-Seq and assembly could generate substantial information about M. ilicifolia genes. The functional annotation of transcripts covered a broad range of GO categories and KEGG allowed the identification of transcripts involved in biosynthesis of triterpenoid backbone, as expected for this species.
3.2 Identification of Differentially Expressed Transcripts in Both Tissues
Comparative transcript abundance level revealed significant differential expression of 2,215 transcripts (FDR<0.05) between the transcriptome of both tissues. Levels of expression were represented as log2 ratio of transcripts abundance between leaf and root samples (Fig. 4A), showing the 1,044 differentially expressed transcripts in root and 1,171 in leaf. Working on both tissues, it was observed that a number of transcripts was expressed uniquely in either of the tissues: among differentially expressed transcripts, 424 were exclusively expressed in leaf and 298 in root.
To better characterize the tissue-biased transcriptome profile, topGO package were used to evaluated GO enrichment (p-value<0.01) for the differentially expressed transcripts and further the representative terms were summarized upon removal of redundant using REVIGO. Among the 770 roots differentially expressed annotated transcripts, 568 genes were assigned to 260 GO terms, while in the leaf, from the 902 differentially expressed annotated transcripts, 610 were classified in 265 GO terms.
The GO analysis revealed enrichment for biological processes (BP) in root for “response to ethylene”, “regulation of cellular process” and others (Fig. 3B), while in leaf for “photosynthesis”, “protein-chromophore linkage” and others (Fig. 3C). According to functional analysis terms, leaves and roots of M. illicifoia also differ at levels of molecular function (MF), with transcripts overexpressed in roots being mainly associated with “calcium ion binding”, “iron ion binding” and others (Fig. 4B), while the overexpressed leaf transcripts are associated with “oxidoreductase activity”, “chlorophyll binding” and others (Fig. 4C).
Significant GO terms linked to secondary methabolism were found in 295 differentially expressed transcripts, 164 in root and 131 in leaf. Some terms were found enriched in specific tissue, for example, “2-oxoglutarate-dependent dioxygenase activity” and “response to herbivore” in root and “beta-amyrin synthase activity” and “triterpenoid biosynthetic process” in leaf. Coincident terms like “oxidoreductase activity” were observed in overexpressed transcripts from both tissues (Table 2).
Gene expression differences between root and leaf tissues of Maytenus ilicifolia. A. The values of -log10 adjusted p-value were plotted according to the differential expression between root and leaf (log2 fold change). Differentially expressed root transcripts are high-lighted in brown (left) and differentially expressed leaf transcripts, in green (right). Top ten most represented terms of gene ontology enrichment analysis in biological process (BP) and molecular function (MF) for differentially expressed transcripts for B. root and C. and leaf (Color figure online)
The comparative transcriptome analysis led to the identification of 350 and 487 transcripts associated with Enzyme Commission (EC) numbers in root and leaf, respectively. These tissue-biased transcripts were mapped onto the KEGG pathway database for the “Biosynthesis of plant secondary metabolites map” (ko01060) and related pathways. Enzymes involved in “monoterpenoid biosynthesis” and isoflavonoid biosynthesis” were identified in root overexpressed transcripts while “flavonoid biosynthesis” and “Biosynthesis of alkaloids derived from histidine and purine” in leaf (Table 3).
Taking together, the results of GO enrichment analysis and KEGG mapping of transcripts overexpressed in root or leaf of M. ilicifolia confirmed the well-reported SMs accumulation reveled by other methodological procedures, including flavonoids, triterpenes, and sesquiterpenes in leaves [2], while roots contain terpenes, triterpenes, alkaloids and especially the quinonemethide triterpenes [5, 13, 14].
Finally, from the present study, an extensive transcriptome dataset has been generated from de novo sequencing analyses of M. ilicifolia. The coverage of the transcriptome data is consistent to discover genes involved in the secondary metabolic pathways. Therefore, choosing the root and the leaf for comparative transcriptome analysis facilitated the identification of the genes involved in the organ-specific biosynthesis, an approach widely used for mining and identifying novel genes in biosynthesis of SMs in plants[3, 6, 7, 18, 25, 26].
References
Coppede, J.S., et al.: Cell cultures of Maytenus ilicifolia Mart. Are richer sources of quinone-methide triterpenoids than plant roots in natura. Plant Cell Tissue Organ Cult. (PCTOC) 118(1), 33–43 (2014). 10/f56kq9
De Souza, L.M., Cipriani, T.R., Iacomini, M., Gorin, P.A.J., Sassaki, G.L.: HPLC/ESI-MS and NMR analysis of flavonoids and tannins in bioactive extract from leaves of Maytenus ilicifolia. J. Pharm. Biomed. Anal. 47(1), 59–67 (2008). 10/c4vp7v
Devi, K., Mishra, S.K., Sahu, J., Panda, D., Modi, M.K., Sen, P.: Genome wide transcriptome profiling reveals differential gene expression in secondary metabolite pathway of Cymbopogon winterianus OPEN, 6(21026), 1–11 (2016). 10/f79vzf. Nature Publishing Group
Dziggel, C., Schãfer, H., Wink, M.: Tools of pathway reconstruction and production of economically relevant plant secondary metabolites in recombinant microorganisms. Biotechnol. J. 12(1), 1–14 (2017). 10/f3tn87
Filho, W.B., Corsino, J., Bolzani, V.d.S., Furlan, M., Pereira, A.M.S., França, S.C.: Quantitative determination of cytotoxicFriedo-nor-oleanane derivatives from five morphological types of Maytenus ilicifolia (celastraceae) by reverse-phase high-performance liquid chromatography. Phytochem. Anal. Int. J. Plant Chem. Biochem. Tech. 13(2), 75–78 (2002). https://doi.org/10.1002/PCA.626
Guo, D., Kang, K., Wang, P., Li, M., Huang, X.: Transcriptome profiling of spike provides expression features of genes related to terpene biosynthesis in lavender. Sci. Rep. 10(1), 1–13 (2020). https://doi.org/10.1038/s41598-020-63950-4
Hansen, N.L., et al.: The terpene synthase gene family in Tripterygium wilfordii harbors a labdane-type diterpene synthase among the monoterpene synthase TPS-b subfamily. Plant J. 89(3), 429–441 (2017). 10/f9qghw
Hartmann, T.: 10/bvxmg2
Jan, R., Asaf, S., Numan, M., Lubna, Kim, K.M.: Plant secondary metabolite biosynthesis and transcriptional regulation in response to biotic and abiotic stress conditions. Agronomy 11(5), 1–31 (2021). 10/gmk7dd
Li, W., et al.: De novo leaf and root transcriptome analysis to explore biosynthetic pathway of Celangulin v in Celastrus angulatus maxim. BMC Genomics 20(1), 1–15 (2019). 10/gz2c
Liu, M.H., et al.: Transcriptome analysis of leaves, roots and flowers of Panax notoginseng identifies genes involved in ginsenoside and alkaloid biosynthesis. BMC Genomics 16(1), 1–12 (2015). 10/f69r7v
Mariot, M.P., Barbieri, R.L.: Metabólitos secundários e propriedades medicinais da espinheira-santa (Maytenus ilicifolia Mart. ex Reiss. e M. aquifolium Mart.). Revista Brasileira de Plantas Medicinais 9(3), 89–99 (2007)
Paz, T.A., et al.: Proteome profiling reveals insights into secondary metabolism in Maytenus ilicifolia (Celastraceae) cell cultures producing quinonemethide triterpenes. Plant Cell Tissue Organ Cult. 130(2), 405–416 (2017). 10/gbpzrf
Périco, L.L., Rodrigues, V.P., de Almeida, L.F.R., Fortuna-Perez, A.P., Vilegas, W., Hiruma-Lima, C.A.: Maytenus ilicifolia Mart. ex Reissek pp. 323–335 (2018). https://doi.org/10.1007/978-94-024-1552-0_29
Pradhan, J., Sahoo, S., Lalotra, S., Sarma, R.: Positive impact of abiotic stress on medicinal and aromatic plants. Int. J. Plant Sci. 12(2), 309–313 (2017). 10/gz2g
Ramakrishna, A., Ravishankar, G.A.: Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 6(11), 1720–1731 (2011). 10/fx4rjw
Saddique, M., Kamran, M., Shahbaz, M.: Differential Responses of Plants to Biotic Stress and the Role of Metabolites. Elsevier Inc. (2018). 10/gz2h
Upadhyay, S., Phukan, U.J., Mishra, S., Shukla, R.K.: De novo leaf and root transcriptome analysis identified novel genes involved in Steroidal sapogenin biosynthesis in Asparagus racemosus. BMC Genomics 15(1), 1–13 (2014). 10/gb3gr4
Van Loon, L.C., Rep, M., Pieterse, C.M.: Significance of inducible defense-related proteins in infected plants. Ann. Rev. Phytopathol. 44, 135–162 (2006). 10/csvjsr
Vellosa, J.C., et al.: Antioxidant activity of Maytenus ilicifolia root bark. Fitoterapia 77(3), 243–244 (2006). 10/dzm4t9
Wink, M.: Introduction: biochemistry, physiology and ecological functions of secondary metabolites. Biochem. Plant Second. Metab. Second Ed. 40, 1–19 (2010). 10/b8sdms
Wink, M.: Secondary metabolites: deterring herbivores. In: Encyclopedia of Life Sciences, pp. 1–9, March 2010. 10/c65zd8
Wink, M., Schimmer, O.: Molecular modes of action of defensive secondary metabolites, vol. 39 (2010). 10/cpz4j7
Yang, L., Wen, K.S., Ruan, X., Zhao, Y.X., Wei, F., Wang, Q.: Response of plant secondary metabolites to environmental factors. Molecules 23(4), 1–26 (2018). 10/gdrnqc
Younesi-Melerdi, E., Nematzadeh, G.A., Pakdin-Parizi, A., Bakhtiarizadeh, M.R., Motahari, S.A.: De novo RNA sequencing analysis of Aeluropus littoralis halophyte plant under salinity stress. Sci. Rep. 10(1), 1–14 (2020). 10/gz2m
Zhang, C., Yao, X., Ren, H., Chang, J., Wang, K.: RNA-Seq reveals flavonoid biosynthesis-related genes in pecan (Carya illinoinensis) kernels. J. Agric. Food Chem. 67, 148–158 (2018). 10.gz2n
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Santoni, M.M. et al. (2021). Comparative Transcriptome Profiling of Maytenus ilicifolia Root and Leaf. In: Stadler, P.F., Walter, M.E.M.T., Hernandez-Rosales, M., Brigido, M.M. (eds) Advances in Bioinformatics and Computational Biology. BSB 2021. Lecture Notes in Computer Science(), vol 13063. Springer, Cham. https://doi.org/10.1007/978-3-030-91814-9_1
Download citation
DOI: https://doi.org/10.1007/978-3-030-91814-9_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-91813-2
Online ISBN: 978-3-030-91814-9
eBook Packages: Computer ScienceComputer Science (R0)