Abstract
Genetic and physiological studies have to-date revealed evidence for five signaling pathways by which the chloroplast exerts retrograde control over nuclear genes. One of these pathways is dependent on product(s) of plastid protein synthesis, for another the signal is singlet oxygen, a third employs chloroplast-generated hydrogen peroxide, a fourth is controlled by the redox state of the photosynthetic electron transport chain, and a fifth involves intermediates and possibly proteins of tetrapyrrole biosynthesis. These five pathways may be part of a complex signaling network that links the functional and physiological state of the chloroplast to the nucleus. Mutants defective in various steps of photosynthesis reveal a surprising diversity in nuclear responses suggesting the existence of a complex signaling network.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cells of plants and eukaryotic algae harbor, in addition to the nucleus, two DNA-containing organelles, the chloroplasts and the mitochondria. It is now widely accepted that these organelles evolved from free-living ancestors of prokaryotic organisms. The vast majority of genetic information of these endosymbionts was transferred to the nuclear genomes of their hosts (Rujan and Martin 2001; Martin et al. 2002). In the case of Chlamydomonas reinhardtii, 72 protein encoding genes are retained by the chloroplast (Maul et al. 2002); whereas in its mitochondria, only eight protein encoding genes are retained (GenBank Accession No. CRU03843).
However, chloroplasts are estimated to harbor about 3,000 proteins (Abdallah et al. 2000). Thus, the vast majority of proteins in this organelle are encoded by the nucleus, translated in the cytosol and subsequently transported into chloroplasts. The transfer of genes from the prokaryotic endosymbionts to the nucleus of the host cell was accompanied by the evolution of host regulatory systems that control the biological functions of the endosymbionts. Thus, nucleus-encoded proteins tightly control essential steps of gene expression within the organelles (Goldschmidt-Clermont 1998; Leon et al. 1998). At the same time, plant cells in the course of evolution had to face the challenge to coordinate the expression of genes distributed between chloroplasts or mitochondria and the nucleus.
In this study, I will focus on retrograde signaling pathways by which chloroplasts exert control over nuclear genes. The first evidence for a role of the chloroplast in the expression of nuclear genes came from mutants of barley and Pelagonium defective in plastidic ribosomes. In these mutants, nucleus encoded but plastid localized enzymes of the Calvin cycle were found to be significantly reduced (Bradbeer and Börner 1978; Hagemann and Börner 1978). Based on these data a “chloroplast control principle” was proposed to regulate a subset of nuclear genes in response to the functional state of the chloroplasts (Bradbeer et al. 1979; Börner 1981; Börner 1986).
Evidence that intact or functional chloroplasts were required for the expression of certain nuclear genes was also deduced from plants missing carotenoids due to mutation or herbicide(norflurazon) application. In such plants, bleaching of green tissue occurs upon exposure to high light intensities. Although the damage inflicted by this treatment apparently is restricted to the chloroplasts (Reiss et al. 1983), the expression of a subset of nuclear genes encoding photosynthesis-related proteins was also found to be decreased. Although it was originally proposed that a single “plastid signal” or “plastid factor” was responsible (Oelmüller and Mohr 1986a, b; Oelmüller 1989; Taylor 1989), it is now clear that the operation of multiple signaling pathways coordinates the expression of nuclear genes with the requirements of plastids (reviewed in Beck 2001; Papenbrock and Grimm 2001; Rodermel 2001; Gray et al. 2003; Surpin et al. 2002; Gray 2003; Pfannschmidt 2003; Pfannschmidt et al. 2003; Beck and Grimm 2005).
However, the route to an understanding of this network will have to be based on the elaboration of individual signaling pathways. Those chloroplast-to-nucleus signaling pathways for which experimental evidence has accumulated, will be discussed herein: one requires plastid protein synthesis, a second is based on chloroplast-generated singlet oxygen, a third uses hydrogen peroxide, a fourth employs the redox poise generated by the photosynthetic electron transfer chains, and a fifth involves tetrapyrrole biosynthesis intermediates. Many excellent contributions have been made that address these signaling processes; I apologize to those, whose work could not be cited due to limitations of space.
Plastid protein synthesis is required for the expression of a set of nuclear genes
In pigment-deficient leaves of the chloroplast-ribosome-deficient barley mutant albostrians, only significantly reduced concentrations were measured for several nuclear encoded proteins which are targeted to plastids (Calvin cycle enzymes; Bradbeer et al. 1979), functionally associated to the plastid (nitrate reductase; Börner et al. 1986), or to the peroxisome (glycolate oxidase, catalase, hydroxy pyruvate reductase; Boldt et al. 1992; Boldt et al. 1997). Using nuclear run-on transcription assays and Northern blot hybridization it was eventually shown that the downregulation occurred at the level of transcription (Hess et al. 1991, 1994). However, the very same experiments with this mutant provided evidence also for the upregulation of certain other genes such as chalcone synthase (Hess et al. 1994), genes for mitochondrial proteins (Hedtke et al. 1999), and of a key player in the context of plastid gene expression, the nucleus-encoded plastid RNA polymerase (Emanuel et al. 2004).
The treatment of plants with chloramphenicol, lincomycin, erythromycin or streptomycin which are all chloroplast-specific inhibitors of translation, resulted in decreased expression levels of nuclear-encoded proteins related to photosynthesis as well (Oelmüller et al. 1986a, b; Adamska 1995; Gray et al. 1995; Sullivan and Gray 1999). Surprisingly, the inhibitors were effective in preventing nuclear gene expression only if applied within the first 2–3 days of seedling development (Oelmüller et al. 1986a, b; Bajracharya et al. 1987; Gray et al. 1995). This suggested that the generation of the plastid signal must involve a product of early plastid gene expression.
Among the gun (genome uncoupled) mutants that retain partial LHCB and RBCS expression following photo-oxidative damage (Susek et al. 1993), gun1 was shown to relieve the repressing effect of inhibition of plastid protein synthesis (Gray et al. 2003; McCormac and Terry 2004). This suggests that GUN1 encodes a component of the signaling pathway affected by inhibition of plastid protein synthesis.
Conceivably, inhibition of plastid protein synthesis may merely prevent the plastids from reaching the developmental stage when plastid-to-nucleus signaling is operational (Hess et al. 1994). The availability of targeted and cell-line-specific knock-outs within chloroplasts of plants beyond the seedling stage offered a new route to address this problem. The application of a recombination-induced knock-out strategy resulted in chimeric plants with cell lines that in their chloroplasts lack the aadA gene conferring resistance to spectinomycin and other cell lines with the aadA gene. In the presence of spectinomycin, chloroplast translation was inhibited in cell lines that lacked the aadA gene. In the organs-affected (leafs and flowers) severe morphological alterations were observed (Ahlert et al. 2003). Evidently, in the absence of chloroplast protein synthesis, the signal(s) needed for proper organ development are not generated.
This collection of data allows the conclusion that plastid protein synthesis generates a signal that is a prerequisite for the expression of a subset of nuclear genes (Fig. 1). These nuclear genes encode plastid constituents, but also proteins found in other cellular compartments. However, whether it is really a product of plastid protein biosynthesis which is directly involved in one form of plastid-to-nucleus signaling has remained unresolved up to the present day. As of date, there is, no indication of how an inhibition of plastidal protein synthesis results in decreased expression of certain nuclear genes.
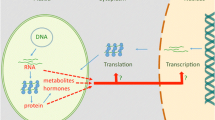
Model depicting five signaling pathways by which the plastid controls nuclear gene expression. D, H and I refer to the three subunits of Mg-chelatase. Signaling from the tetrapyrrole biosynthetic pathway may occur either according to model 1, or model 2, or both. The ROS singlet oxygen (1O2) and hydrogen peroxide (H2O2) are shown to employ different signaling routes
Reactive oxygen species generated in chloroplasts regulate the expression of nuclear genes
In plants, reactive oxygen species (ROS) are produced continuously as byproducts of various metabolic pathways that are localized in different cellular compartments. The principal ROS generated in chloroplasts under high-light stress are singlet oxygen by photosystem (PS)II and the superoxide anion by PSI; the latter rapidly dismutates to hydrogen peroxide (Apel and Hirt 2004). Recent progress in this field is based on the elaboration of conditions that result in the generation of defined ROS within chloroplasts, i.e., singlet oxygen and hydrogen peroxide.
Signaling by singlet oxygen
The specific generation of singlet oxygen in plastids was achieved by irradiation of the Arabidopsis thaliana flu mutant. This mutant accumulates high amounts of protochlorophyllide (Pchld) within its plastids that generates singlet oxygen upon irradiation. The effect of singlet oxygen production on gene expression was analyzed by DNA microarrays that comprised more than 95% of the total Arabidopsis genome. As a result, 70 genes were detected which were specifically upregulated and nine genes which were downregulated (op den Camp et al. 2003). Two arguments for an action of singlet oxygen within the plastids were presented. Firstly, singlet oxygen has an extremely short half life in cells of about 200 ns. Secondly, the distance over which singlet oxygen may act was calculated to be up to 10 nm. Singlet oxygen thus qualifies as a plastid generated signal that could play a specific role in the activation of a genetically determined stress response program.
The flu plants were subjected to a screen for lines that no longer respond to plastid-generated singlet oxygen by growth inhibition. One group of mutants identified was shown to harbor a second site mutation in EXECUTER1. In these mutants, the stress response of Arabidopsis thaliana, caused by the release of singlet oxygen, was abrogated (Wagner et al. 2004). Also singlet oxygen, produced either by irradiation of Pchld or by PSII under high light conditions in the presence of the PSII-specific inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethyl urea (DCMU) that blocks the reduction of plastoquinone, did not activate the stress response program in a mutant defective in EXECUTER1. Thus, this protein either enables plants to perceive singlet oxygen as a signal or is involved in the transduction of this signal. The N-terminal part of EXECUTER1 resembles import signal sequences of nuclear-encoded plastid proteins (Wagner et al. 2004). If the plastid localization of EXECUTER1 would be confirmed, a first important step in the characterization of the signaling pathway that links plastid-generated singlet oxygen to nuclear gene expression is done (Fig. 1).
Singlet oxygen is the principal ROS produced by PSII when carotenoid-deficient plant tissue—either due to mutations or due to the application of herbicides like norflurazon that inhibit carotenoid biosynthesis—is exposed to light (Krieger-Liszkay 2004). These conditions lead to severe damage of the chloroplasts (Reiss et al. 1983), and eventually to the downregulation of some nuclear genes—principally those encoding photosynthesis-related proteins (Oelmüller 1989). However, a comparison of the nuclear genes downregulated by this treatment with those either up- or downregulated by singlet oxygen generated by irradiation of Pchld accumulating flu mutants, revealed no overlap. Therefore, singlet oxygen produced by PSII in the absence of carotenoids possibly activates a signaling pathway which is entirely different from that triggered by singlet oxygen derived from Pchld or PSII in the presence of DCMU (Wagner et al. 2004). Such a separate pathway gains support from the analysis of gun mutants in which genes that normally are downregulated by norflurazon treatment were found to be less affected (Susek et al. 1993). Interestingly, four out of five of these mutants exhibit defects in tetrapyrrole metabolism (see below).
Signaling by hydrogen peroxide
Hydrogen peroxide is another ROS produced by chloroplasts. It accumulates upon a shift of plants, from moderate to high light intensities. Under these conditions, superoxide radicals are formed at PSI due to a hyperreduction of electron carrier chains leading to the reduction of oxygen (Mehler reaction). Superoxide dismutates to hydrogen peroxide and may accumulate in this form (Mullineaux and Karpinski 2002). The induction of the nuclear gene for ascorbate peroxidase (APX2) was linked to plastid-produced hydrogen peroxide accumulation. Evidence for a crucial role of hydrogen peroxide was provided by experiments in which infiltration of leaves with catalase resulted in reduced APX2 expression after high-light treatment (Karpinski et al. 1999). An involvement of the redox state of the plastoquinone and/or QA pools, originally considered as likely candidates for APX2 activation (Karpinski et al. 1997), appeared to be unlikely since APX2 induction was observed in the absence of changes in their redox state (Fryer et al. 2003). However, recent studies with transgenic tobacco plants that either expressed chloroplast-localized catalase or a thylakoid-bound ascorbate peroxidase suggest a participation of both, the redox state of the plastoquinone pool and H2O2 in the expression of a cytosolic ascorbate peroxidase. The initial induction, observed upon exposure to photooxidative stress conditions, was assigned to the reduced state of the plastoquinone pool, while a subsequent increase in mRNA levels correlated with a continuous increase in H2O2 levels (Yabuta et al. 2004).
How and where the plastid generated hydrogen peroxide is sensed has not yet been elucidated. Hydrogen peroxide is thought to diffuse as freely as water across biological membranes and therefore could directly interact with extraplastidic signaling components. This raises the problem as to how cells may differentiate between hydrogen peroxide generated in plastids from hydrogen peroxide produced somewhere else, e.g., as a consequence of pathogen attack at the plasma membrane. The stress reactions induced in plants by pathogens clearly differ from those induced by high light intensities (Bray et al. 2000; Hammond-Kosack and Jones 2000). This suggests differences in the intracellular distribution of H2O2-responsive targets that mediate the specificities of the responses, a prime target being the chloroplasts themselves (Fig. 1).
One may conclude that at least two ROS, singlet oxygen and hydrogen peroxide, generated in chloroplasts, are involved in specific signaling to the nucleus. Both elicit different responses at the level of gene expression and can be assumed to employ different signaling pathways (Fig. 1). The routes by which these ROS communicate with the nucleus remain to be elucidated. Here, the recently identified mutants such as EXECUTER1, blocked in singlet oxygen signaling, offer a promising approach.
Regulation of nuclear genes by the redox poise of photosynthetic electron transfer chains
Environmentally induced changes in the redox state of photosynthetic electron transport components act as signals that regulate expression of genes within the chloroplast (Pfannschmidt et al. 1999) and of a subset of genes within the nucleus. By this way, photosynthesis contributes important information to the regulation of nuclear gene expression that is not sensed by cytosolic photoreceptors. The chloroplast itself serves as a sensor for changes in light quality and quantity and thus can induce physiological acclimation reactions.
A prime example for this type of chloroplast-to-nucleus signaling is the stimulation of LHCB1 transcription in the green alga Dunaliella when high light-acclimated cells were exposed to low light intensities (Escoubas et al. 1995). Clues towards an understanding of the underlying mechanism that mediates this regulatory pattern came from the application of compounds that inhibit the electron transfer from PSII to the cytochrome b6/f complex at different sites. Treatment with the PSII-specific inhibitor DCMU resulted in enhanced LHC gene transcription even in high light. In contrast, the partial inhibition of plastoquinol oxidation with 2,5-di-bromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) caused a repression of transcription at low light intensities (Escoubas et al. 1995; Maxwell et al. 1995; Durnford and Falkowski 1997). These results pointed towards a regulatory role of the redox state of the plastidic plastoquinone pool (Fig. 1). This regulatory system initially was viewed as a general mechanism for the acclimation of the photosynthetic apparatus to changes in light intensity and various forms of environmental stress (Durnford and Falkowski 1997).
However, in higher plants the redox poise of the plastoquinone pool appears to play a minor regulatory role for nuclear genes since, so far, only the plastocyanin gene (PetE), the gene for cytosolic ascorbate peroxidase, both in tobacco, and the ELIP2 gene of Arabidopsis could be shown to respond to changes in its redox state (Pfannschmidt et al. 2001; Kimura et al. 2003; Yabuta et al. 2004). The transcription of other nuclear genes investigated, i.e., those encoding PSI components or nitrate reductase, while also coupled to photosynthetic electron transport, appears not to be controlled by the redox state of the plastidic plastoquinone but rather via different redox systems. Studies with DCMU and DBMIB as well as the analysis of cytb6/f-defective mutants indicated that redox signals involved in the regulation of these genes appear to originate downstream from the cytb6/f complex (Pfannschmidt et al. 2001; Sherameti et al. 2002). LHCII expression, studied in winter rye plants grown under a variety of light and temperature regimens, provides another example. Here, LHCII protein phosphorylation was used to monitor redox changes within chloroplasts. A coregulation between LHCII phosphorylation and LHCII mRNA levels was observed, but no correlation with the reduction state of the plastoquinone pool (Pursiheimo et al. 2001). These results support the concept of a redox sensor downstream from the plastoquinone pool that plays an important role in nuclear gene regulation (Fig. 1). They also point to differences in regulation between green algae and higher plants, since the former, as unicellular organisms, exhibit more dynamic responses to changes in environment and the addition of inhibitors like DCMU and DBMIB.
In addition to transcriptional regulation, chloroplast redox signals may control nuclear genes (Fed-1, PetE) by regulating mRNA stability (Petracek et al. 1997, 1998; Sullivan and Gray 2002).
The route(s) by which the redox state of the thylakoid membrane is signaled to the nucleus, is poorly understood. If the plastoquinone pool is the sensor, it might involve a phosphorylation cascade. Studies with inhibitors of protein phosphatases provided suggestive evidence for a participation of phosphorylated intermediates since the addition of such inhibitors to algal cell cultures prevented the increase in LHC gene expression at low light intensities (Escoubas et al. 1995; Durnford and Falkowski 1997). However, it should be stressed that molecular evidence for signaling components downstream from plastoquinol remains to be elucidated.
A more global analysis of nuclear genes employed DNA arrays with 3,292 Arabidopsis genes. Two thousand six hundred and sixty one of these were predicted to be nuclear-encoded chloroplast proteins while 631 were assumed to encode non-chloroplast proteins (Richly et al. 2003). Changes in mRNA levels were registered for 2,133 of these genes when plants grown with light that preferentially is absorbed by PSI (PSI light) were transferred to PSII light. This shift, resulting in a reduction of thylakoid electron carriers, caused an upregulation of 1,121 genes and a downregulation of 1,012 genes. More detailed studies revealed that 286 of these genes were regulated directly by redox signals from the photosynthetic electron transport chains; 86 were found to be upregulated, 200 were downregulated (Fey et al. 2005). One limitation of this set of data, are the rather small changes in expression levels observed, for most of the genes; they were below a factor of two. The combined data, nonetheless, may be viewed as evidence for a regulatory role of the redox state of photosynthetic electron transport components in the regulation of a significant number of nuclear genes. Since this regulation is dependent on light, the photosynthetic apparatus may be viewed as a photoreceptor that registers the quality and quantity of photons. The plastid molecules that sense this information have, with the exception of the plastoquinone pool, not yet been identified.
Tetrapyrroles regulate nuclear transcription
Physiological and genetic evidence suggests that tetrapyrroles represent one type of plastid signal. In plants, the early steps of tetrapyrrole biosynthesis up to protoporphyrinogen IX (Protogen) take place only in chloroplasts (Fig. 2). A fraction of the Protogen then, by unknown means, is transported into mitochondria to serve as precursor of heme (reviewed in Papenbrock and Grimm 2001). The other fraction remains in the chloroplast to serve as precursor of both, heme and chlorophyll. Protogen first is converted into protoporphyrin IX (Proto). The insertion of Fe2+ into Proto leads to the formation of protoheme, the precursor of heme; Mg2+ insertion into Proto gives rise to Mg-protoporphyrin IX (MgProto), the precursor of chlorophyll (Chl) (Beale 1999).
Scheme of the tetrapyrrole biosynthetic pathway in photosynthetic eukaryotic algae and higher plants. All enzymes of the pathway for chlorophyll synthesis, starting with glutamyl-tRNA synthase, are located within the chloroplast. Shown are the major intermediates and the genes mentioned in the text. Dashed lines indicate multiple steps. The mutational defects and inhibitors addressed are indicated in bold letters
Tetrapyrroles play a regulatory role within chloroplasts. While there is no evidence for their function in plastid gene expression, the role of tetrapyrroles appears to be focused on the control of glutamyl-tRNA reductase activity, the enzyme for the rate-limiting step in porphyrin biosynthesis (Weinstein and Beale 1985; Vothknecht et al. 1996; Meskauskiene et al. 2001; Meskauskiene and Apel 2002).
Porphyrins may also be viewed as attractive candidates for chloroplast molecules in organelle-to-nucleus signaling. They are known to play a regulatory role in yeast and mammals where heme was shown to modulate the DNA-binding activity of certain transcription factors (Hon et al. 1999; Ogawa et al. 2001). However, it has yet to be determined how heme, whose biosynthesis in yeast and mammals takes place in mitochondria, is transported to cytoplasm/nucleus. In plants, mitochondrial to nucleus signaling, possibly involving heme, has not yet been reported.
Signaling role of tetrapyrroles in higher plants
In higher plants, evidence of tetrapyrrole involvement in plastid-to-nucleus signaling came from the analysis of gun mutants of Arabidopsis (Susek et al. 1993; Surpin et al. 2002). With the exception of gun1, four mutants (gun2-5) were shown to have lesions in genes involved in tetrapyrrole synthesis and/or degradation.
Mutant gun5 has a leaky point mutation in the CHLH gene that only weakly impaired Chl synthesis but repression of LHCB expression in the presence of norflurazon in this mutant was partially released. This suggested an involvement of the Mg-chelatase H subunit (CHL H) in regulating plastid-to-nucleus-signaling (Mochizuki et al. 2001). Subsequently, LHCB expression levels in the wild type during norflurazon treatment, were shown to be inversely correlated with steady state levels of MgProto. In gun5 mutant plants subjected to norflurazon treatment, the accumulation of MgProto was reduced relative to the wild type, suggesting a role for MgProto in LHCB regulation (Strand et al. 2003). The finding that incubation of leaf protoplasts with MgProto, but not with porphobilinogen, Proto, or heme, repressed LHCB expression was also consistent with this hypothesis. And so were experiments in which the gun5 mutant was treated with α,α′ dipyridyl, a metal chelator known to inhibit the formation of Pchld from MgProtoMe, which presumably increased MgProto accumulation resulting in a repression of LHCB. Moreover, other mutants defective in MgProto formation, comprising those encoding defective genes for coproporphyrinogen oxidase, porphobilinogen deaminase, as well as the D and H subunits of Mg-chelatase were also non-repressed for LHCB expression when grown with norflurazon. Consequently, mutants with reduced MgProto synthesizing capacity should exhibit the gun phenotype, namely, non-repressed expression of LHCB in the presence of norflurazon (Strand 2004).
The gun4 mutant also appears to fall into this category. A defect in the gun4 gene resulted in reduced Chl accumulation and elevated levels of LHCB mRNA in the presence of norflurazon. While Mg-porphyrin levels for this mutant have not been reported, the plastid-localized GUN4 protein binds Proto/MgProto and activates Mg-chelatase (Larkin et al. 2003).
Also the phenotypes of mutants gun2 (allelic to hy1) and gun3 (allelic to hy2) that are blocked in the biosynthesis of phytochromobilin (Fig. 2) may be explained by a reduced MgProto synthesis. Both mutations inhibit the degradation of heme, and this results in a reduced synthesis of ALA, presumably due to feedback inhibition of glutamyl-tRNA reductase by heme (Terry and Kendrick 1999). As a consequence, those mutations, as shown for gun2, partially block the accumulation of MgProto observed after treatment with norflurazon (Strand et al. 2003).
Other experimental approaches using higher plants also support a correlation between the accumulation of Mg-porphyrins and lower levels of LHCB and RBCS transcripts. Treating etiolated cress seedlings with thujaplicin inhibited PChld synthesis and resulted in the accumulation of MgProto and MgProtoMe; it also inhibited (by at least 50%) the light-induced accumulation of several LHCB transcripts (Oster et al. 1996). Moreover, inhibition of etiolated barley seedlings with amitrole, an inhibitor of carotenoid biosynthesis, also caused increased levels of ALA, MgProto and MgProtoMe, but lower steady state levels of LHCB and RBCS transcripts. The authors proposed that the elevated Mg-porphyrin levels reduced the transcriptional activities of these two nuclear gene families (La Rocca et al. 2001).
Signaling role of tetrapyrroles in green algae
Prior to studies with higher plants, the first suggestive evidence for a role of porphyrins as a plastid signaling factor was provided by analyses on LHC gene expression in Chlamydomonas reinhardtii. Defects in tetrapyrrole metabolism either due to mutations or induced by the application of inhibitors affected LHC gene regulation and these changes were assigned to alterations in the pool levels of tetrapyrrole intermediates (Johanningmeier and Howell 1984; Johanningmeier 1988). One limitation of these studies, though, was the absence of porphyrin measurements.
Analysis of the signaling pathway that mediates the light induction of nuclear chaperone genes of the HSP70 family in Chlamydomonas provided the first solid evidence for a role of tetrapyrroles in chloroplast-to-nucleus signaling (Kropat et al. 1997). The biological sense behind the induction of HSP70 genes by light is evidenced by enhanced protection of PSII and its faster recovery from damage inflicted by light-stress conditions. Thus, the protection and recovery of PSII from photoinhibition positively correlated with levels of the plastidic chaperone HSP70B (Schroda et al. 1999).
In the case of the nuclear chaperone genes, an involvement of the chloroplast was first suggested by the finding that in a mutant (brs-1), defective in the H-subunit of Mg-chelatase (Chekounova et al. 2001), the HSP70 genes no longer were light inducible (Kropat et al. 1997). In contrast, the induction of these genes by heat shock was still intact in this mutant. A mutant blocked in a later step of chlorophyll synthesis, i.e., the conversion of Pchld to Chld, showed no defect in the light regulation of the chaperone genes. From these results, it was concluded that either the enzyme (or one of its subunits) that catalyzes the synthesis of MgProto from Proto or intermediates of chlorophyll biosynthesis after Proto and before Chld are involved in signaling from chloroplast to the nuclear HSP70 genes. The question whether intermediates of chlorophyll biosynthesis played a role in this induction was answered by feeding various precursors to cells in the dark. The feeding of MgProto or its methyl ester (but not of Proto, Pchld or Chld) resulted in an induction of the HSP70 genes, suggesting that these two compounds specifically could substitute for the light signal (Kropat et al. 1997).
From these data, a model was derived (Beck 2001): The signal cascade for induction of the HSP70 genes is activated by light within the chloroplast or its envelope. Following light activation, MgProto and/or MgProtoMe, synthesized in the plastid, become(s) accessible on the cytoplasmic side of the chloroplast. In cytoplasm or nucleus, these tetrapyrroles may be recognized by factor(s) that either control expression of the nuclear HSP70 genes directly or stimulate a signaling cascade that enters the nucleus (model 1 in Fig. 1).
An analysis of the wavelength dependence of this induction revealed a major peak around 600 nm and a minor one around 450 nm. This suggested that a novel photoreceptor, possibly a constituent of the chloroplast, mediates this induction (Kropat and Beck 1998). However, it remains to be determined whether this photoreceptor controls the light-induced accumulation of Mg-tetrapyrroles, the release of MgProto/MgProtoMe into the cytosol, or both steps.
While direct evidence for a transport of MgProto/MgProtoMe out of the chloroplast is still lacking, there is evidence that light may play a dual role in this signaling pathway: (1) A shift of dark-adapted cultures into the light caused a transient increase in the pool levels of Proto, MgProto, and MgProtoMe (Kropat et al. 1999, 2000) which preceded the accumulation of HSP70 mRNA. Although a subcellular localization of these tetrapyrroles was not yet possible, a plastidic location of these compounds was assumed. Conditions that abolished the light-induced increase in porphyrin pool levels, (e.g., inhibitors of cytoplasmic protein synthesis or the use of gametic cells) also abolished the light induction of the HSP70 genes but addition of MgProto to such cultures restored induction of the HSP70 genes. Thus, a causal relationship between the light-induced increase in chlorophyll precursor levels and the induction of nuclear genes was deduced (Kropat et al. 2000). (2) Evidence for the involvement of additional light-requiring steps in the tetrapyrrole-mediated light induction of the HSP70 genes came from analyses of porphyrin pool levels in C. reinhardtii after adding Proto in the dark which is taken up by the cells and converted into MgProto and MgProtoMe to increase their intracellular pool sizes that are located, presumably, within the plastids (Kropat et al. 2000). These plastid tetrapyrroles apparently do not access cytosol and/or nucleus in the dark as they do not contribute to HSP70 gene induction. Thus, these Mg-porphyrins appear to be retained by the chloroplasts in dark-grown cells. Since adding MgProto directly to the cells in the dark resulted in HSP70 induction, it is proposed that Mg-porphyrins are exported out of the chloroplast actively or passively by a light-triggered process.
Recent studies revealed that heme may also be assigned a role as an organellar signal in Chlamydomonas. In this organism, a single HEMA gene, encoding glutamyl-tRNA reductase, the rate limiting enzyme in tetrapyrrole biosynthesis, is induced by heme and also by MgProto (Vasileuskaya et al. 2005). While the physiological role of heme as a signal from either plastids or mitochondria remains to be elucidated, it appears to be warranted to add it to the list of tetrapyrroles with a regulatory role at least in algae.
Genes that are targets of MgProto regulation
Which nuclear genes are regulated by tetrapyrroles? In C. reinhardtii out of a group of 20 light-inducible genes that were considered as candidates for a regulation by tetrapyrroles, four (HSP70A, HSP70B, HSP70E, HEMA) exhibited induction by MgProto (Vasileuskaya et al. 2004). Two of these genes (HSP70A, HSP70E) encode cytosolic chaperones while the HSP70B and HEMA gene products are plastid-localized. Proto had no effect on gene expression. A repressing effect of tetrapyrroles in Chlamydomonas was not observed for any of the genes analyzed. However, this does not exclude a long-term effect of MgProto in the downregulation of some genes. The situation may be different in cells with elevated mRNA levels, e.g., after light induction. Here, as suggested by the work of Johanningmeier and Howell (1984), MgProto or its monomethyl ester, may well be employed for reducing mRNA levels of, e.g., LHC genes, by turning down their rate of transcription and/or by destabilization of their mRNAs. Due to the high toxicity of MgProto even at low light intensities, this idea could not be tested by feeding this tetrapyrrole to cells in the light.
Using a more comprehensive microarray-based analysis, the expression of 8,200 nuclear Arabidopsis genes was investigated (Strand et al. 2003). The mRNA levels of wild type and the gun2 and gun5 mutants (see above) were compared during treatment with norflurazon. In the wild type, this treatment resulted in a repression of the LHCB gene, causally linked to an about 15-fold increase in MgProto levels. In mutants gun2 (defective in heme oxygenase) and gun5 (partially defective in the H-subunit of Mg-chelatase), norflurazon treatment also resulted in an increase in MgProto pool levels but the highest levels observed were substantially lower than those seen in the wild type after the same treatment (Strand et al. 2003). In the gun2 and gun5 mutants, in contrast to wild type, about 70 out of 182 genes repressed by norflurazon treatment were found not to be repressed (the genes registered exhibited a more than threefold change in expression upon addition of the herbicide). The majority (10 out of 11) of the LHC genes analyzed exhibited at least two-fold higher mRNA levels in the norflurazon-treated gun5 mutant as compared to wild type treated with the same compound. In the gun2 mutant, the mRNA increases observed were lower than those in the gun5 strain for all of these genes. In addition, tetrapyrrole biosynthetic genes GSA and one encoding a non-specified subunit of Mg-chelatase showed an upregulation of more than two-fold in the norflurazon-treated gun5 mutant as compared to norflurazon-treated wild type (Strand et al. 2003). Since in the gun5 mutant, the MgProto levels remained low, the non-repression of these genes may be viewed as a consequence of lower Mg-porphyrin levels (Strand et al. 2003). These results imply a prominent role for MgProto as a regulator of nuclear gene expression in higher plants. However, in these experiments, differences in photosensitization and thus ROS production between mutant and wild type after norflurazon treatment (see above) as a source of altered gene regulation could not be excluded. Indeed, the recent observation that Arabidopsis lines overexpressing PORA (encoding Pchld-oxido-reductase) and thus presumably contain lower levels of Mg-tetrapyrroles upon norflurazon treatment, exhibited only a partial release from repression of LHCB (McCormac and Terry 2004) points to a participation of multiple plastid signaling pathways in controlling a single gene.
Targets of tetrapyrrole signaling
The elucidation of target sequences for plastid signals (MgProto and others) has made progress recently. Light responsive promoters of genes that are also subject to control by a plastid signal—in most cases coding for photosynthetic machinery components—were compared and subjected to mutational analyses. A conclusion derived from these studies suggested that light and the plastidic signal control expression of nuclear genes via the same cis-acting elements (Bolle et al. 1996; Kusnetsov et al. 1996). In these studies, the plastidic signal involved in controlling nuclear gene expression was modulated by the addition of norflurazon that, in the light, leads to photooxidation of the thylakoid membrane. As shown recently for Arabidopsis, treatment with norflurazon also leads to the accumulation of MgProto (Strand et al. 2003).
Subsequently it was shown that regulation of nuclear genes by light and a plastid factor, activated by norflurazon treatment, employs complex cis elements formed by aggregation of cognate sequences of different transcription factors (Terzaghi and Cashmore 1995). A modular array of two motifs: an I-box and a G-box were recently shown to be sufficient for the regulation of a minimal promoter by light and a norflurazon treatment-derived plastidal signal (Martínez-Hernández et al. 2002).
New insights into promoter elements that are responsive to the norflurazon-induced plastidal signal came from the functional analysis of a G-box motif with an ACGT core that is specifically bound by a factor termed CUF1. In the promoter of the LHCB1 gene of Arabidopsis, this element contributes to high level expression but is not required for light or circadian regulation (Anderson et al. 1994). The LHCB1 promoter, mutated in the G-box and cloned in front of a reporter gene, was found not to be repressed by norflurazon treatment. In contrast, transgenic lines with the LHCB1 wild-type promoter were repressed by norflurazon in a wild type, but not in a gun5 mutant background. Since the gun5 mutant in the presence of norflurazon accumulates less MgProto, it was deduced that the CUF-1 binding site is the target of a MgProto-mediated signal (Strand et al. 2003).
Analysis of the MgProto-inducible HSP70A promoter region of C. reinhardtii revealed the existence of two promoters and between their start sites, a tandem of cis-regulatory elements. These elements with the motif GCGAC-N11-TATACATA were shown to confer inducibility by MgProto not only to both HSP70A promoters but also to heterologous promoters. The orientation and distance-independent function of these cis-regulatory elements qualifies them as enhancers that mediate the response of nuclear genes to chloroplast-derived MgProto (von Gromoff, Schroda, Oster and Beck, unpublished).
While the search for transcription factors that mediate MgProto-activated gene expression is on, the heme activated transcription of nuclear genes in yeast may provide a model. Here, transcription factor Hap1 activates gene expression after interaction with mitochondria-derived heme (Forsburg and Guarente 1989). In the non-DNA binding form, Hap1 is associated with molecular chaperones HSP90 and Ydj1 forming a higher order complex. Heme disrupts this complex and permits Hap1 to bind to DNA with high affinity, thereby activating transcription (Hon et al. 1999).
Transport of tetrapyrroles
With the documented role for MgProto (and MgProtoMe) in chloroplast-to-nucleus signaling an open question still remaining unresolved is how this signal is perceived in cytosol/nucleus. It was proposed that MgProto exits the chloroplast in order to modulate the activity and/or nuclear translocation of regulatory proteins (Kropat et al. 1997, 2000; Strand et al. 2003). A release of Protogen (for the transport to mitochondria) and of phytochromobilin (the chromophore of the cytosolic/nuclear photoreceptor phytochrome) from the chloroplast may be deduced from the fact that these molecules are synthesized exclusively in plastids, but have their function in cytosol/nucleus. The efflux of heme, Protogen and Proto from isolated chloroplasts has been observed (Thomas and Weinstein 1990; Jacobs and Jacobs 1993). The routes for their transport out of the chloroplasts are still unknown. However, transporters for heme have been identified in eubacteria that are human pathogens (Stojiljkovic and Hantke 1994; Drazek et al. 2000; Skaar et al. 2004). In Arabidopsis mitochondria peripheral-type benzodiazepine receptors were demonstrated to transport Proto (Lindemann et al. 2004). Mutant analyses in Arabidopsis lead to the discovery of a plastid-localized ABC-like transporter that appears to be involved in the distribution of Proto (Møller et al. 2001). A transporter for Mg-porphyrin, though, remains to be discovered and the exit of MgProto from chloroplasts has not yet been demonstrated experimentally. Active transport and facilitated diffusion, triggered by the light signal, both seem feasible. In this process, some data point to a role for the Mg-chelatase subunit CHL H that binds Proto and releases MgProto (Walker and Willows 1997), and/or the GUN4 protein shown to bind Proto and MgProto (Larkin et al. 2003). Significant proportions of these two proteins were found attached to the inner envelope of plastids, in the case of CHL H preferentially at elevated Mg2+ concentrations (5 μM) (Nakayama et al. 1998; Larkin et al. 2003). It is therefore tempting to speculate about their involvement in controlling the release of MgProto from the chloroplast.
Plants with knock-down constructs for Mg-porphyrin biosynthesis genes reveal additional complexity
While the data presented above point to a direct regulatory role of Mg-porphyrins in the communication of the chloroplast with the nucleus, studies with tobacco plants that harbor transgenes in antisense or sense orientation for Mg-chelatase subunits CHL H and CHL I as well as for MgProto monomethyl transferase CHL M indicate a more complex role of tetrapyrrole biosynthesis and intermediates in signaling. Tobacco plants with reduced CHL H and CHL I proteins exhibited lower Mg-chelatase activity and reduced steady state levels of chlorophyll precursors MgProto and MgProtoMe when compared to wild type (Papenbrock et al. 2000a, b). Reduction in CHL M caused a decrease in MgProtoMe and an increase in MgProto (Alawady and Grimm 2005). In all transformants, reduced levels of Proto, the Mg-chelatase substrate, were observed that correlated with diminished activities in ALA synthesis in all three types of modified plants. This diminished ALA synthesizing capacity was attributed to lower transcript levels of nuclear genes HEMA, GSA, and ALAD encoding glutamyl-tRNA reductase, glutamate-1-semialdehyde aminotransferase and 5-aminolevulinic acid dehydratase, respectively (Papenbrock et al. 2000a; Alawady and Grimm 2005). Recently, also a semidominant CHLI mutant allele of Arabidopsis was shown to result in a reduced synthesis of ALA and Proto, corroborating previous results with transgenic knock-down strains (B. Grimm, personal communication).
Transgenic lines overexpressing CHL M offer additional information: pools of Proto, MgProto, and MgProtoMe as well as the Chl concentration in the antisense line were about the same as in wild type. Nonetheless, transcript levels of HEMA, GSA, and LHCB were elevated (Alawady and Grimm 2005). Clearly these results show a lack of correlation between the steady state levels of MgProto or MgProtoMe pool levels and the expression of genes of tetrapyrrole biosynthesis. This lack of correlation between Mg-porphyrin pool levels and nuclear gene expression in transgenic lines is in contrast to data in Arabidopsis where, upon norflurazon treatment, elevated MgProto levels were linked to reduced LHCB gene expression (Strand et al. 2003).
Here, differences in experimental setups and possibly in the organisms used may play an important role. For transgenic tobacco lines with altered Mg-chelatase or MgProto methyltransferase, leaves from regenerated plants were assayed (Papenbrock et al. 2000a). In the case of the gun mutants, seedlings bleached by treatment with norflurazon for 5 days were employed (Mochizuki et al. 2001). In the transgenic lines, the changes in gene expression observed may reflect the long term acclimation of the regenerated plants to a severe defect in tetrapyrrole metabolism. In contrast, the norflurazon-treated Arabidopsis seedlings were exposed to the damaging effect of light for a rather short period; the changes assayed may thus reflect a short time response. Also the developmental state of a plant appears to affect chloroplast to nucleus signaling. While in Arabidopsis seedlings, norflurazon treatment caused an inhibition of LHCB and HEMA1 expression, treatment of mature plants resulted in an inhibition of LHCB only. The HEMA1 promoter in plants no longer was responsive to the loss of a functional chloroplast (McCormac and Terry 2004).
Two alternative models integrate the present knowledge on the role of tetrapyrrole biosynthesis and biosynthetic intermediates in the regulation of nuclear genes (Beck and Grimm 2005). Model 1 is based on the observation of regulatory responses in the nucleus upon adding Mg-porphyrins: the induction of HSP70 genes and HEMA in Chlamydomonas and the repression of LHCB genes in higher plants (Fig. 1). These findings can be explained by a direct interaction of MgProto and/or MgProtoMe with regulatory factors present in cytoplasm/nucleus. The communication from plastids to cytosol/nucleus is ensured by the transport of Mg-porphyrins through the plastid envelopes. This concept is supported by an experiment in which the feeding of Proto in the dark resulted in an accumulation of Mg-porphyrins, but not in the induction of the HSP70 genes, because the Mg-porphyrins presumably were held within the chloroplast (Kropat et al. 2000).
According to model 2, the Mg-porphyrin levels are sensed at the sites of their synthesis by an enzyme complex consisting of Mg-chelatase, MgProto monomethyl transferase and other putative MgProto-binding proteins (Fig. 1). This complex possibly mediates transmembrane signaling from the stroma side of the inner plastid envelope to the cytoplasm. A crucial role may be assigned to alterations in MgProto/MgProtoMe steady state levels that, via the plastidic protein complex, are communicated to the nucleus.
Some data are consistent with a role of CHL H and/or GUN4 in the signal transfer according to models 1 and 2. CHL H has been found in the inner envelope of chloroplasts in the presence of 5 μM Mg2+, which is in the range of plastidal Mg2+ concentration, although the enzyme is also localized in the stroma at lower Mg2+ concentrations (Gibson et al. 1996; Leegood et al. 1985). The interaction with the inner envelope may activate signaling on the cytoplasmic side by yet unknown mechanisms.
In summary, while a role for tetrapyrroles in inter-organellar signaling is clearly emerging, the data also suggest that the signaling network triggered by these compounds is of complex nature.
Global changes in nuclear gene expression observed in mutants defective in photosynthesis or signaling
The employment of DNA arrays with 2,661 and 631 gene probes for nuclear chloroplast genes and genes encoding non-chloroplast proteins of Arabidopsis, respectively, permitted a comprehensive analysis of regulatory consequences of mutations that affect the chloroplast (Richly et al. 2003). Of particular interest in the context of chloroplast-to-nucleus communication are mutants altered in tetrapyrrole metabolism (gun5, flu) and those defective in photosynthesis, i.e., PSI, the cytb6/f complex, or plastidic ATPase. When compared to wild type, the gun5 mutant exhibited an upregulation of about 1,600 genes and a downregulation of about 100 genes. Approximately 300 of the upregulated genes encode non-chloroplast proteins. These experiments, in contrast to those reported by Strand et al. (2003) (see above), were performed in the absence of norflurazon. This indicates that a defect in Mg-chelatase by itself may affect the regulation of many nuclear genes. When assayed in the dark and compared to wild type, the flu mutant that under these conditions accumulates Pchld (Meskauskiene et al. 2001), showed an upregulation of about 1,900 genes, 25% of which encode non-chloroplast proteins, and a downregulation of about 100 genes. In the light, where Pchld levels are strongly decreased, the same mutant exhibited a downregulation of about 850 genes, one quarter being non-plastid genes. Under these conditions an upregulation of about 50 genes was observed (Richly et al. 2003).
Similar complex patterns of deregulation were also observed in mutants defective in the cytb6/f, PSI, or ATPase complexes (Maiwald et al. 2003; Richly et al. 2003; Ihnatowicz et al. 2004). These data may offer a first glimpse into a regulatory network that integrates information about perturbations within the plastids with nuclear gene expression. However, the situation appears to be even more complex since mutants defective in the same photosynthetic complex may show opposite regulatory responses. An atpd mutant showed an upregulation of about 1,400 genes; about 100 genes were down regulated. In an atpc mutant, defective in the same protein complex, about 1,750 genes were downregulated and 100 genes showed an upregulation (Richly et al. 2003). The analysis of individual mutants with knock-out alleles in genes PSAE, PSAD,PSAN, PSAO, (encoding components of PSI), as well as PETC (encoding the Rieske protein of the cytochrome b6/f complex) and ATPD (encoding the δ subunit of the chloroplast ATP synthase) revealed two groups with similar regulatory patterns in nuclear gene expression: the petC-2, psae1-1, and psad1-1 mutants and the atpd-1, psan-1, and psao-1 mutants (Maiwald et al. 2003). A defined pattern of deregulation observed thus could not be assigned to defects in a distinct component of the photosynthetic apparatus. Rather, different types of plastid signaling pathways appear to be triggered, presumably as a consequence of the physiological effects of individual mutations. In addition, single genes may be subject to regulatory input from more than one plastid signal (Yabuta et al. 2004).
Conclusions
The present state of knowledge about chloroplast-to-nucleus signaling is still rather limited. For several pathways, summarized in Fig. 1, genetic and physiological evidence has accumulated, but the molecular characterization of individual components has only just begun. Neither the route by which the plastid signal passes the organelle’s envelope nor the downstream signaling molecules in cytosol/nucleus have yet been identified for any of these pathways. Apparently, it is the interaction of various signaling pathways that make these types of studies very complex. The recent discovery that perturbations in photosynthesis affect the expression of a large number of nuclear genes in a highly complex manner, may be viewed as evidence for the general importance of the chloroplast in determining nuclear output in plants. Nevertheless, these data are difficult to reconcile with simple one-dimensional signaling pathways. Rather, a highly complex network of interacting signaling pathways appears to link the chloroplast to nuclear gene expression.
Abbreviations
- ALA:
-
5-aminolevulinic acid
- CHL H:
-
H subunit of Mg-chelatase
- CHL I:
-
I subunit of Mg-chelatase
- Chl:
-
Chlorophyll
- Chld:
-
Chlorophyllide
- DBMIB:
-
2,5-di-bromo-3-methyl-6-isopropyl-p-benzoquinone
- DCMU:
-
3-(3,4-dichlorophenyl)-1,1-dimethyl urea
- gun :
-
Genome uncoupled
- LHC :
-
Gene for light-harvesting chlorophyll a/b binding protein
- MgProto:
-
Mg protoporphyrin IX
- MgProtoMe:
-
Mg protoporphyrin monomethyl ester
- Pchld:
-
Protochlorophyllide
- Proto:
-
Protoporphyrin IX
- Protogen:
-
Protoporphyrinogen
- PS:
-
Photosystem
- RBCS :
-
Gene for small subunit of ribulose bisphosphate carboxylase/oxygenase
- ROS:
-
Reactive oxygen species
References
Abdallah F, Salamini F, Leister D (2000) A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci 5:141–142
Adamska I (1995) Regulation of early light-inducible protein gene expression by blue and red light in etiolated seedlings involves nuclear and plastid factors. Plant Physiol 107:1167–1175
Ahlert D, Ruf S, Bock R (2003) Plastid protein synthesis is required for plant development in tobacco. Proc Natl Acad Sci USA 100:15730–15735
Alawady AE, Grimm B (2005) Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis. Plant J 41:282–290
Anderson SL, Teakle GR, Martino-Catt SJ, Kay SA (1994) Circadian clock- and phytochrome-regulated transcription is conferred by a 78 bp cis-acting domain of the Arabidopsis CAB2 promoter. Plant J 6:457–470
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Bajracharya D, Bergfield R, Hatzfeld W-D, Klein S, Schopfer P (1987) Regulatory involvement of plastids in the development of peroxisomal enzymes in the cotyledons of mustard (Sinapis alba L.) seedlings. J Plant Physiol 126:421–436
Beale SI (1999) Enzymes of chlorophyll biosynthesis. Photosynth Res 60:43–73
Beck CF (2001) Signalling pathways in chloroplast-to-nucleus communication. Protist 152:175–182
Beck CF, Grimm B (2005) Involvement of tetrapyrroles in cellular regulation. In: Grimm B, Porra RJ, Rüdiger W, Scheer H (eds) Biochemistry, biophysics and biological functions of Chlorophylls. Springer, The Netherlands (in press)
Boldt R, Börner T, Schnarrenberger C (1992) Repression of the plastidic isoenzymes of aldolase, 3-phosphoglycerate kinase, and triosephosphate isomerase in the barley mutant “albostrians”. Plant Physiol 99:895–900
Boldt R, Koshuchowa S, Gross W, Börner T, Schnarrenberger C (1997) Decrease in glycolate pathway enzyme activities in plastids and peroxisomes of the albostrians mutant of barley (Hordeum vulgare L.). Plant Sci 124:33–40
Bolle C, Kusnetsov VV, Herrmann RG, Oelmüller R (1996) The spinach AtpC and AtpD genes contain elements for light-regulated, plastid-dependent and organ-specific expression in the vicinity of the transcription start sites. Plant J 9:21–30
Börner T (1981) Enzymes of plastid ribosome-deficient mutants. Ferredoxin-NADP+ reductase. Biochem Physiol Pflanzen 176:737–743
Börner T (1986) Chloroplast control of nuclear gene function. Endocyt Cell Res 3:265–274
Börner T, Mendel RR, Schiemann J (1986) Nitrate reductase is not accumulated in chloroplast-ribosome-deficient mutants of higher plants. Planta 169:202–207
Bradbeer JW, Börner T (1978) Activities of glyceraldehyde-phosphate dehydrogenase (NADP+) and phosphoribulokinase in two barley mutants deficient in chloroplast ribosomes. In: Akoyunoglou G et al (eds) Chloroplast development. North-Holland Biomedical Press, Elsevier, pp 727–732
Bradbeer JW, Atkinson YE, Börner T, Hagemann R (1979) Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesised RNA. Nature 279:816–817
Bray EA, Bailey-Serres J, Weretilnyk E (2000) Responses to abiotic stresses. In: Buchanan BB, Gruissem W, Jones RL (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, pp 1158–1203
Chekounova E, Voronetskaja V, Papenbrock J, Grimm B, Beck CF (2001) Characterization of Chlamydomonas mutants defective in the H-subunit of Mg-chelatase. Mol Gen Genomics 266:363–373
Drazek ES, Hammack CA Sr, Schmitt MP (2000) Corynebacterium diphteriae genes required for acquisition of iron from haemin and haemoglobin are homologous to ABC haemin transporters. Mol Microbiol 36:68–84
Durnford DG, Falkowski PG (1997) Chloroplast redox regulation of nuclear gene transcription during photoacclimation. Photosynth Res 53:229–241
Emanuel C, Weihe A, Graner A, Hess WR, Börner T (2004) Chloroplast development affects expression of phage-type RNA polymerases in barley leaves. Plant J 38:460–472
Escoubas JM, Lomas M, La Roche J, Falkowski PG (1995) Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA 92:10237–10241
Fey V, Wagner R, Bräutigam K, Wirtz M, Hell R, Dietzmann A, Leister D, Oelmüller R, Pfannschmidt T (2005) Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem 280:5318–5328
Forsburg SL, Guarente L (1989) Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu Rev Cell Biol 5:153–180
Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR (2003) Control of ascorbate peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J 33:691–705
Gibson LCD, Marrison JL, Leech RM, Jensen PE, Bassham DC, Gibson M, Hunter CN (1996) A putative Mg chelatase subunit from Arabidopsis thaliana cv C24. Plant Physiol 111:61–71
Goldschmidt-Clermont M (1998) Coordination of nuclear and chloroplast gene expression in plant cells. Int Rev Cytol 177:115–180
Gray JC (2003) Chloroplast-to-nucleus signalling: a role for Mg-protoporphyrin. Trends Genet 19:526–529
Gray JC, Sornarajah R, Zabron AA, Duckett CM, Khan MS (1995) Chloroplast control of nuclear gene expression. In: Mathis P (ed) Photosynthesis: from light to biosphere, vol III. Kluwer Acad Publishers, The Netherlands, pp 543–550
Gray JC, Sullivan JA, Wang JH, Jerome CA, MacLean D (2003) Coordination of plastid and nuclear gene expression. Philos Trans R Soc Lond B Biol Sci 358:135–144
Hagemann R, Börner T (1978) Plastid ribosome-deficient mutants of higher plants as a tool in studying chloroplast biogenesis. In: Akoyunoglou G et al (eds) Chloroplast development. North-Holland Biomedical Press, Elsevier, pp 709–720
Hammond-Kosack K, Jones JDG (2000) Responses to plant pathogens. In: Buchanan BB, Gruissem W, Jones RL (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, pp 1102–1156
Hedtke B, Wagner I, Börner T, Hess WR (1999) Interorganellar crosstalk in higher plants: impaired chloroplast development affects mitochondrial gene and transcript levels. Plant J 19:635–644
Hess WR, Schendel R, Börner T, Rüdiger W (1991) Reduction of mRNA level for two nuclear encoded light regulated genes in the barley mutant albostrians is not correlated with phytochrome content and activity. J Plant Physiol 138:292–298
Hess WR, Müller A, Nagy F, Börner T (1994) Ribosome-deficient plastids affect transcription of light-induced nuclear genes: genetic evidence for a plastid-derived signal. Mol Gen Genet 242:305–312
Hon T, Hach A, Tamalis D, Zhu Y, Zhang L (1999) The yeast heme-responsive transcriptional activator Hap1 is a preexisting dimer in the absence of heme. J Biol Chem 274:22770–22774
Ihnatowicz A, Pesaresi P, Varotto C, Richly E, Schneider A, Jahns P, Salamini F, Leister D (2004) Mutants for photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J 37:839–852
Jacobs JM, Jacobs NJ (1993) Porphyrin accumulation and export by isolated barley (Hordeum vulgare) plastids. Plant Physiol 101:1181–1187
Johanningmeier U (1988) Possible control of transcript levels by chlorophyll precursors in Chlamydomonas. Eur J Biochem 177:417–424
Johanningmeier U, Howell SH (1984) Regulation of light-harvesting chlorophyll-binding protein mRNA accumulation in Chlamydomonas reinhardtii. J Biol Chem 259:13541–13549
Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM (1997) Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9:627–640
Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P (1999) Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284:654–657
Kimura M, Manabe K, Abe T, Yoshida S, Matsui M, Yamamoto YY (2003) Analysis of hydrogen peroxide-independent expression of the high-light-inducible ELIP2 gene with the aid of the ELIP2 promoter-luciferase fusion. Photochem Photobiol 77:668–674
Krieger-Liszkay A (2004) Singlet oxygen production in photosynthesis. J Exp Bot 56:337–346
Kropat J, Beck CF (1998) Characterization of photoreceptor and signalling pathway for light induction of the Chlamydomonas heat-shock gene HSP70A. Photochem Photobiol 68:414–419
Kropat J, Oster U, Rüdiger W, Beck CF (1997) Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc Natl Acad Sci USA 94:14168–14172
Kropat J, Pöpperl G, Rüdiger W, Beck CF (1999) Identification of Mg-protoporphyrin IX as a chloroplast signal that mediates the expression of nuclear genes. In: Wagner E, Normann J, Greppin H, Hackstein JHP, Hermann RG, Kowallik KV, Schenk HEA, Seckbach J (eds) From symbiosis to eukaryotism. Endocytobiol VII. University of Geneva, Geneva, pp 341–348
Kropat J, Oster U, Rüdiger W, Beck CF (2000) Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J 24:523–531
Kusnetsov V, Bolle C, Lubberstedt T, Sopory S, Herrmann RG, Oelmüller R (1996) Evidence that the plastid signal and light operate via the same cis-acting elements in the promoters of nuclear genes for plastid proteins. Mol Gen Genet 252:631–639
La Rocca N, Rascio N, Oster U, Rüdiger W (2001) Amitrole treatment of etiolated barley seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta 213:101–108
Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299:902–906
Leegood RC, Walker DA, Foyer CH (1985) Regulation of the Benson-Calvin cycle. In: Barber J, Baker NR (eds) Photosynthetic mechanisms and the environment. Elsevier Science Publishers, Amsterdam, pp 199–258
Leon P, Arroyo A, Mackenzie S (1998) Nuclear control of plastid and mitochondrial development in higher plants. Annu Rev Plant Physiol Plant Mol Biol 49:453–480
Lindemann P, Koch A, Degenhardt B, Hause G, Grimm B, Papadopoulos V (2004) A novel Arabidopsis thaliana protein is a functional peripheral-type benzodiazepine receptor. Plant Cell Physiol 45:723–733
Møller SG, Kunkel T, Chua N-H (2001) A plastidic ABC protein involved in intercompartmental communication of light signalling. Genes Dev 15:90–103
Maiwald D, Dietzmann A, Jahns P, Pesaresi P, Joliot P, Joliot A, Levin JZ, Salamini F, Leister D (2003) Knock-out of the genes coding for the Rieske protein and the ATP-synthase δ-subunit of Arabidopsis. Effects on photosynthesis, thylakoid protein composition, and nuclear chloroplast gene expression. Plant Physiol 133:191–202
Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA 99:12246–12251
Martínez-Hernández A, López-Ochoa L, Argüello-Astorga G, Herrera-Estrella L (2002) Functional properties and regulatory complexity of a minimal RCBS light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol 128:1223–1233
Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14:2659–2679
Maxwell DP, Laudenbach DE, Huner NPA (1995) Redox regulation of light-harvesting complex II and cab mRNA abundance in Dunaliella salina. Plant Physiol 109:787–795
McCormac AC, Terry MJ (2004) The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid-signalling pathways during de-etiolation. Plant J 40:672–685
Meskauskiene R, Apel K (2002) Interaction of FLU, a negative regulator of tetrapyrrole biosynthesis, with the glutamyl-tRNA reductase requires the tetratricopeptide repeat domain of FLU. FEBS Lett 532:27–30
Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001). FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98:12826–12831
Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN 5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98:2053–2058
Mullineaux P, Karpinski S (2002) Signal transduction in response to excess light: getting out of the chloroplast. Curr Opin Plant Biol 5:43–48
Nakayama M, Masuda T, Bando T, Yamagata H, Ohta H, Takamiya K (1998) Cloning and expression of the soybean chlH gene encoding a subunit of Mg-Chelatase and localization of the Mg2+ concentration-dependent ChlH protein within the chloroplast. Plant Cell Physiol 39:275–284
Oelmüller R (1989) Photooxidative destruction of chloroplasts and its effect on nuclear gene expression and extraplastidic enzyme levels. Photochem Photobiol 49:229–239
Oelmüller R, Mohr H (1986a) Photooxidative destruction or chloroplasts and its consequences for expression of nuclear genes. Planta 167:106–113
Oelmüller R, Levitan I, Bergfeld R, Rajasekhar VK, Mohr H (1986b) Expression of nuclear genes is affected by treatments acting on plastids. Planta 168:482–492
Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, Sassa S, Hayashi N, Yamamoto M, Shibehara S, Fujita H, Igarashi K (2001) Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J 20:2835–2843
op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Göbel C, Feussner I, Nater M, Apel K (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15:2320–2332
Oster U, Brunner H, Rüdiger W (1996) The greening process in cress seedlings. V. Possible interference of chlorophyll precursors, accumulated after thujaplicin treatment, with light-regulated expression of Lhc genes. J Photochem Photobiol B 36:255–261
Papenbrock J, Grimm B (2001) Regulatory network of tetrapyrrole biosynthesis—studies for intracellular signalling involved in metabolic and developmental control of plastids. Planta 213:667–681
Papenbrock J, Mock H-P, Tanaka R, Kruse E, Grimm B (2000a) Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol 122:1161–1169
Papenbrock J, Pfündel E, Mock H-P, Grimm B (2000b) Decreased and increased expression of the subunit CHL I diminishes Mg-chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J 22:155–164
Petracek ME, Dickey LF, Thompson WF (1997) Light-regulated changes in abundance and polyribosome association of ferrodoxin are dependent on photosynthesis. Plant Cell 9:2291–2300
Petracek ME, Dickey LF, Nguyen TT, Gatz G, Sowinski DA, Allen GC, Thompson WF (1998) Ferredoxin-1 mRNA is destabilized by changes in photosynthetic electron transport. Proc Natl Acad Sci USA 95:9009–9013
Pfannschmidt T (2003) Chloroplast redox signals: how photosynthesis controls its own genes. Trends Plant Sci 1:33–41
Pfannschmidt T, Nilsson A, Allen JF (1999) Photosynthetic control of chloroplast gene expression. Nature 397:625–628
Pfannschmidt T, Schütz K, Brost M, Oelmüller R (2001) A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J Biol Chem 276:36125–36130
Pfannschmidt T, Schütz K, Fey V, Sherameti I, Oelmüller R (2003) Chloroplast redox control of nuclear gene expression—a new class of plastid signals in interorganellar communication. Antiox Redox Signal 5:95–101
Pursiheimo S, Mulo P, Rintamäki E, Aro E-M (2001) Coregulation of light-harvesting complex II phosphorylation and Lhcb accumulation in winter rye. Plant J 26:317–327
Reiss T, Bergfeld R, Link G, Thien W, Mohr H (1983) Photooxidative destruction of chloroplasts and its consequences to cytosolic enzyme levels and plant development. Planta 159:518–528
Richly E, Dietzmann A, Biehl A, Kurth J, Laloi C, Apel K, Salamini F, Leister D (2003) Covariations in the nuclear chloroplast transcriptome reveal a regulatory master-switch. EMBO reports 4:491–498
Rodermel S (2001) Pathway of plastid-to-nucleus signalling. Trends Plant Sci 6:471–478
Rujan T, Martin W (2001) How many genes in Arabidopsis came from cyanobacteria? An estimate from 386 protein phylogenies. Trends Genet 17:113–120
Schroda M, Vallon O, Wollman F-A, Beck CF (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11:1165–1178
Sherameti I, Sopory SK, Trebicka A, Pfannschmidt T, Oelmüller R (2002) Photosynthetic electron transport determines nitrate reductase gene expression and activity in higher plants. J Biol Chem 277:46594–46600
Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O (2004) Iron-source preference of staphylococcus aureus infections. Science 305:1626–1628
Stojiljkovic I, Hantke K (1994) Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol 13:719–732
Strand A (2004) Plastid-to-nucleus signalling. Curr Opin Plant Biol 7:621–625
Strand A, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421:79–83
Sullivan JA, Gray JC (1999) Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant Cell 11:901–910
Sullivan JA, Gray JC (2002) Multiple plastid signals regulate the expression of the pea plastocyanin gene in pea and transgenic tobacco plants. Plant J 32:763–774
Surpin M, Larkin RM, Chory J (2002) Signal transduction between the chloroplast and the nucleus. Plant Cell 14 (Suppl):S327–S338
Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74:787–799
Taylor WC (1989) Regulatory interactions between nuclear and plastid genomes. Annu Rev Plant Physiol Plant Mol Biol 40:211–233
Terry MJ, Kendrick RE (1999) Feedback inhibition of chlorophylI synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol 119:143–152
Terzaghi WB, Cashmore AR (1995) Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46:445–474
Thomas J, Weinstein JD (1990) Measurement of heme efflux and heme content in isolated developing chloroplasts. Plant Physiol 94:1414–1423
Vasileuskaya Z, Oster U, Beck CF (2004) Involvement of tetrapyrroles in inter-organellar signaling in plants and algae. Photosynth Res 82:289–299
Vasileuskaya Z, Oster U, Beck CF (2005) Identification of Mg-protoporphyrinIX and heme as plastid/mitochondrial factors that control HEMA expression in Chlamydomonas. Eukaryotic Cell 4 (in press)
Vothknecht UC, Kannangara CG, von Wettstein D (1996) Expression of catalytically active barley glutamyl tRNAGlu reductase in Escherichia coli as a fusion protein with glutathione S-transferase. Proc Natl Acad Sci USA 93:9287–9291
Wagner D, Przybyla D, op den Camp R, Kim C, Landgraf F, Lee KP, Würsch M, Laloi C, Nater M, Hideg E, Apel K (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306:1183–1185
Walker CJ, Willows RD (1997) Mechanism and regulation of Mg-chelatase. Biochem J 327:321–333
Weinstein JD, Beale SI (1985) Enzymatic conversion of glutamate to delta-aminolevulinate in soluble extracts of the unicellular green alga, Chlorella vulgaris. Arch Biochem Biophys 237:454–464
Yabuta Y, Maruta T, Yoshimura K, Ishikawa T, Shigeoka S (2004) Two distinct redox signaling pathways for cytosolic APX induction under photooxidative stress. Plant Cell Physiol 45:1586–1594
Acknowledgements
The author wishes to thank Bernhard Grimm, Wolfgang Hess, Anja Liszkay-Krieger, Thomas Pfannschmidt and Michael Schroda for constructive comments on the manuscript. He also wishes to thank Ning Shao and Zinaida Vasileuskaya for drawing of the figures. This project was supported by a grant of the DFG (BE903/13-1).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Beck, C.F. Signaling pathways from the chloroplast to the nucleus. Planta 222, 743–756 (2005). https://doi.org/10.1007/s00425-005-0021-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-005-0021-2