Abstract
DNA and the machinery for gene expression have been discovered in chloroplasts during the 1960s. It was soon evident that the chloroplast genome is small, that many genes for chloroplast-localized proteins must reside in the nucleus, and that the expression of the genes in both cellular compartments must be coordinated. In the 1970s, the first evidence for plastid signals controlling nuclear gene expression was provided for plastid ribosome-deficient mutants. This review describes the discovery and the first studies on plastid-to-nucleus signaling. Today, many retrograde signals are known, which coordinate plastid and nuclear gene expression during the development of the organelle and in response to environmental changes. The nucleus receives information about the flux through the heme branch of the tetrapyrrole pathway, the expression of plastid genes, the metabolite stage in the organelle, and the efficiency of the photosynthetic electron flow. Singlet oxygen generated during light stress and breakdown products of carotenoids initiate signaling events in the organelle which alter nuclear gene expression. Operational signals permanently coordinate gene expression in both organelles. The biosynthesis of phytohormones like jasmonic, salicylic, and abscisic acids or cytokinins starts in the plastids, and these hormones became crucial players in coordinating plastid and nuclear gene expression under stress. Methylerythritol cyclodiphosphate, a biochemical intermediate of the methylerythritol phosphate pathway, alters the chromatin structure in the nucleus which in turn affects the expression of a particular subset of stress-inducible genes. Dual targeted proteins with plastid and nuclear locations participate in the interorganellar communication. We discuss our current knowledge about retrograde signaling and address open questions.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Jasmonic acid
- Photosynthesis-associated nuclear genes
- Plastids
- Redox
- Salicylic acid
- Signaling
- Singlet oxygen
- Tetrapyrroles
1 Discovery of Plastid Retrograde Signals and Early Steps in Their Function
In the 1970s, it became clear that many genes for plastid proteins must be located in the nucleus, because the genetic information in the organelle is too small for the huge amount of functions that chloroplasts, etioplasts, leucoplasts, amyloplast, or chromoplasts fulfill in their different cellular environments (Kirk and Tilney-Bassett 1967; Kirk 1971; Börner et al. 1973; Bogorad 1975; Taylor 1989). More than 3000 different proteins were identified in plastids, and it is estimated that more than 95% of them are encoded by nuclear genes (Leister 2005, 2016; Tiller and Bock 2014). The plastome of higher plants contains only approximately 100 genes for photosynthesis, fatty acid biosynthesis, components of the import machinery, ribosomal proteins, and RNA polymerase subunits as well as rRNAs and tRNAs. Thus, the expression of the genes in both compartments have to be coordinated (Brunkard and Burch-Smith 2018; Van Dingenen et al. 2016; Greiner and Bock 2013). An obvious idea was that the expression of these genes in the nucleus are only expressed when the gene products are required in the organelles, therefore the nucleus should be informed about the stage of the plastids in a particular organ, tissue, or cell. The first hints for the existence of such a control mechanism came from mutants defective in plastid development (Börner 2017). Plastid-ribosome-deficient mutants do not only lack the plastid-encoded components of multiprotein complexes (such as the ribulose-1,5-bisphosphate carboxylase, the photosynthesis complexes, or the 70S ribosomes of the plastids) but also the nuclear-encoded partners. Further analyses of these mutants, as well as plants which were chemically or physically treated to inhibit plastid gene expression or development showed that the absence of the nuclear-encoded proteins of these multiprotein complexes is caused by the absence or reduction of their expression. Tom Börner (2017) recently summarized early steps of the discovery of plastid retrograde signals and focused on the genetic evidence based on mutants with lesions in plastid functions. The historical overview also described the contribution of the researchers in this field and their interaction across the iron curtain. We only summarize a few additional historical aspects which were not in the main focus of Börner’s review.
With the knowledge that the small subunit of ribulose-1,5-bisphosphate carboxylase is nuclear- and the large subunit plastid-encoded, early research focused on the identification of the mechanisms of how the expression of the genes in the two genetic compartments is coordinated (Bradbeer et al. 1979; Criddle et al. 1970; Givan and Criddle 1972; Chan and Wildman 1972; Blair and Ellis 1973; Ellis 1975, 1977; Börner et al. 1972, 1973, 1974, 1976; Hagemann and Börner 1987; Reichenbächer et al. 1978). Finally, mRNA measurements for RBCS transcript levels (for the small subunit of ribulose-1,5-bisphosphate carboxylase) in mutants impaired in plastid functions let to the hypothesis that the expression of the nuclear RBCS genes is controlled by signals from the plastids (Mayfield and Taylor 1984, 1987; Oelmüller and Mohr 1986; Harpster et al. 1984; Batschauer et al. 1986; Oelmüller et al. 1986a, b; Burgess and Taylor 1988; Giuliano and Scolnik 1988). The studies were extended to other nuclear-encoded genes for plastid proteins, with a main focus on genes for light-harvesting chlorophyll-a/b-binding proteins (LHCPs) (Mayfield and Taylor 1984; Oelmüller et al. 1986b; Oelmüller and Schuster 1987; Johanningmeier and Howell 1984). Physiological experiments initially demonstrated that LHCP expression is far more sensitive to photooxidative damage of the plastids than RBCS gene expression, and comparable differences were observed when plastids recovered from photodamage (Schuster et al. 1988). It appeared that more than one signal might be involved in the interorganellar cross talk and that there might be specificity for individual genes in their response to the information deriving from the plastids. Intermediates of chlorophyll biosynthesis have been postulated as signaling molecules mediating plastid-to-nucleus signaling, with the main focus on LHCP expression (Johanningmeier and Howell 1984; Kropat et al. 1997). Furthermore, also etioplasts are able to inform the nucleus about the stage of the organelle, as shown with inhibitor studies in etiolated mustard seedlings (Oelmüller et al. 1986b).
Tom Börner (2017) already described the interesting observation that also the activity and expression of nitrate reductase, an enzyme located in the cytoplasm, is decreased in leaves with impaired plastids, suggesting that the organelle also controls non-plastidal enzymes which require functional plastids (Börner et al. 1986; Oelmüller et al. 1988; Mohr et al. 1992; Hess et al. 1994; Oelmüller 1989; Oelmüller and Briggs 1990; Sherameti et al. 2002b). Nitrate reductase activity and expression is induced by nitrate and light, and both stimuli are only active when functional plastids are present. Besides effects in the cytoplasm (Reiss et al. 1983), also peroxisomal enzyme activities are controlled by the state of the plastids (Bajracharya et al. 1987). How the interorganellar signaling could occur, how specific such a signal has to operate, and which are the targets of plastid-derived signals in the nucleus/cytoplasm or peroxisomes were a matter of intensive discussion. The original studies were performed with plants in which chloroplast development was severely impaired by either mutation (Börner 2017; Bradbeer and Börner 1978; Hagemann and Börner 1987; Bradbeer et al. 1979), chemical (Oelmüller 1989) or heat (Feierabend 1977; Feierabend and Schrader-Reichhardt 1976; Feierabend and Mikus 1977) treatments. It was difficult to imagine that these badly damaged organelles, often without any detectable organelle structure, repress nuclear gene expression highly specifically, and that only one signaling molecule is responsible for the altered gene expression in the nucleus. Therefore, the discussions about the nature of the information flow from the organelle to the nucleus ranged from organellar cross talk with information exchange at many levels and multiple actors to highly specific plastid-derived signals which control the expression of individual genes in the nucleus. Quite early, it became obvious that the regulatory scenario must be somehow coupled to light signaling, since all known plastid-responsive genes were also light regulated (cf. Lepistö and Rintamäki 2012; Lepistö et al. 2012). However, at that time, we were only at the beginning to understand which signaling molecules mediate light responsiveness, and nobody could envision at that time that light-, hormone-, and other signaling processes share common signaling compounds, cross talk to each other and integrate the information from internal and external sources (e.g., Gollan et al. 2015).
During the discovery of plastid retrograde signaling, a similar process was already discussed intensively for mitochondria, based on studies with petite mutants from yeast. These mutants were impaired in mitochondrial functions and had severe alterations in the nuclear/cytoplasm cross talk, including altered expression of nuclear genes. The petite mutants were already discovered in the 1950 (summarized in Bernardi 1979) in yeast, and besides mitochondrial retrograde signals which control nuclear gene expression, also many other processes in the cytoplasm were affected. The available information about these mutants stimulated the discussion about a comparable role of plastids for nuclear gene expression and plastid-related enzymes located in the cytosol. Even now, plant researchers can still learn from the cross talk between the mitochondria and the nucleus/cytoplasm, in particular with regard to signaling components which transfer the information from the plastids to the nucleus and integrate organelle information with those from other sources. Butow and Avadhani (2004) described “mitochondrial retrograde signaling as a pathway of communication from mitochondria to the nucleus that influences many cellular and organismal activities under both normal and pathophysiological conditions. In yeast it is used as a sensor of mitochondrial dysfunction that initiates readjustments of carbohydrate and nitrogen metabolism. In both yeast and animal cells, retrograde signaling is linked to TOR signaling, but the precise connections are unclear. In mammalian cells, mitochondrial dysfunction sets off signaling cascades through altered Ca2+ dynamics, which activate factors such as NFκB, NFAT, and ATF. Retrograde signaling also induces invasive behavior in otherwise nontumorigenic cells implying a role in tumor progression.” This short description by Butow and Avadhani (2004) also highlights that much more has to be discovered for plastid retrograde signaling even now (cf. Pesaresi et al. 2006, 2007).
Initially, the expression levels of nuclear genes for plastid proteins were detected by their translatability in vitro, Northern analyses, and run-on transcription assays. In particular, the experiments by Batschauer et al. (1986) demonstrated that the plastid-derived signals must control transcriptional events in the nucleus. This implies the involvement of nuclear-localized transcription factors and responsive cis-regulatory elements in the promoters of the responding genes as targets of the signals from the plastids. Since light-responsive cis-regulatory elements in the promoter regions of light-inducible genes were studied in many laboratories at that time, one research direction focused on the identification of plastid-responsive elements in the promoters of genes for plastid proteins. The overall results of these studies uncovered that light-responsive and plastid-responsive elements were either identical or at least overlapping. Apparently, signals from the plastids and those from light converge before regulation the expression of their target genes in the nucleus (Kusnetsov et al. 1996). For example, Bolle et al. (1996a) showed that the spinach AtpC and AtpD genes for two of the three nuclear-encoded proteins of the plastid ATP synthase contain elements for light-regulated, plastid-dependent, and organ-specific expressions in the vicinity of the transcription start sites. Bolle et al. (1996b) also demonstrated that intron sequences are involved in the plastid- and light-dependent expression of the spinach PsaD gene. A number of quite short additional promoter sequence of genes for thylakoid proteins were identified to be involved in plastid-dependent expression (Kusnetsov et al. 1996, 1999; Oelmüller et al. 1993; Lübberstedt et al. 1994; Bolle et al. 1994); however a common plastid-responsive element which is present in the promoter region of more than one gene for plastid proteins could not be identified (Oelmüller et al. 1993; Bolle et al. 1996a, b). Overall, it appears that quite different target sequences are coupled to the signals from the plastids and that light signals and plastid-derived signals merge before controlling nuclear gene expression (Bolle et al. 1994; Kusnetsov et al. 1996). Finally, Kusnetsov et al. (1999) showed that the assembly of the CAAT-box-binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the plastids. Apparently, also hormone signals target the same or similar cis-elements as plastid signals.
While these studies support transcriptional control by retrograde signals from the plastids, Sherameti et al. (2002a) investigated polyribosome loading of spinach mRNA species. They found that in light-grown, but not in dark-grown, spinach seedlings, the mRNAs for the nuclear-encoded photosystem (PS) I subunits D, F, and L are associated with polyribosomes, and this association is prevented by the application of 3-(3′,4′-dichlorophenyl)-1,1′-dimethyl urea (DCMU), an inhibitor of the photosynthetic electron transport. To identify the cis-elements which are responsible for this regulation, they generated a series of chimeric PsaD constructs and tested them in transgenic tobacco. The spinach PsaD 5′-untranslated region is sufficient to confer light- and photosynthesis-dependent polyribosome association onto a reporter gene, while the tobacco PsaD 5′-untranslated region directs constitutive polyribosome association. These results suggest that signals from photosynthetic electron flow control also posttranslational events. Thus, retrograde signals may be involved in quite different steps of nuclear gene expression, from transcription in the nucleus to the efficiency of the translation of specific mRNAs in the cytoplasm. Since the main focus on the research was directed toward transcriptional control, and the nature of the signals from the plastids, posttranscriptional events controlling the translatability and stability of specific RNA species were only considered much later.
A breakthrough in the research on retrograde signaling came with the identification of the gun (genomes uncoupled) mutants in Joanne Chory’s laboratory (Susek et al. 1993). They used Arabidopsis plants with an LHCP reporter gene construct and screened for mutants which express the nuclear gene in seedlings in which plastids were destroyed by photooxidative damage due to inhibition of carotenoid biosynthesis with Norflurazon, an inhibitor that blocks carotenoid biosynthesis and thus leads to photooxidative destruction of the plastids. The herbicide treatment results in the downregulation of LHCP gene expression, and the mutants thus uncouple the expression from the state of the plastids. Ultimately, six GUN genes were identified, five of them are related to tetrapyrrole biosynthesis. This showed that at least one retrograde pathway is based on Mg-ProtoporphyrinIX, the first intermediate in the chlorophyll branch of the tetrapyrrole biosynthetic pathway (Nott et al. 2006; Pogson et al. 2008; Woodson et al. 2011). The sixth protein, GUN1, is a chloroplast-localized PPR protein (Nott et al. 2006, cf. below).
2 Nature of the Plastid Signal
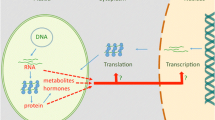
Quite early after the discovery of chloroplast retrograde signaling, four different starting points in the organelle have been postulated: components of the tetrapyrrole biosynthesis, products deriving from chloroplast gene expression, chloroplast redox homeostasis, and photosynthesis-derived reactive oxygen species (ROS). Later, after the discovery that the whole scenario is more complex than anticipated that far, the retrograde signals were classified as those exerting biogenic control during early chloroplast development in seedlings which leads to the transition from etioplast to chloroplast development, and operational signals that inform the nucleus about the state of the mature and functional organelle (e.g., in Brunkard and Burch-Smith 2018; Kleine and Leister 2016; de Souza et al. 2017). This includes the efficiency of photosynthetic electron transport but also metabolite requirements of the cell from the plastids or compounds such as hormone precursors and secondary metabolites including volatiles to respond to stress or pathogens. Ultimately, with the identification of specific metabolites as retrograde signals, such as methylerythritol cyclodiphosphate (MEcPP) (Xiao et al. 2012), the role of plastidal control on phytohormone synthesis and signaling for biotic stress responses became an important facet in the cross talk scenario. The nature of the plastid signals and the cross talk with nucleus in regulating the expression of genes is depicted in Fig. 18.1.
Schematic diagram depicting the retrograde signaling pathways originating from chloroplasts. Plastid-to-nucleus retrograde signaling can be classified into two processes: “biogenic signals” that are relayed to the nucleus during early chloroplast development and “operational signals” that inform the nucleus about the state of the mature and functional organelle. Sensing and processing of plastid signals are mediated by diverse pathways, some of which appear to be interconnected through proteins, metabolites, and/or ROS. The pathways include various components of plastid genes expression, tetrapyrrole synthesis, redox state of photosynthetic electron transport, and chloroplast metabolite stage as well as different kinds of reactive oxygen species and hormones (green arrowheads). Several regulatory proteins have been found to be involved in signal transduction (orange). The signals cause transcriptional responses and may influence chromatin modeling and/or post-transcriptional processes in the nucleus and cytoplasm. Targets of nuclear gene regulation (gray boxes) frequently include transcription factors (orange)
2.1 GUN1, a Biogenic Control Signal
Functional plastid gene expression (PGE) is crucial to initiate the expression of Photosynthesis Associated Nuclear Genes (PhANG) during early chloroplast development (Koussevitzky et al. 2007). In this process, perturbation of plastid gene expression triggers retrograde signals that control nuclear gene expression. Evidence for this type of regulation comes from studies with inhibitors of plastid translation and transcription (Oelmüller et al. 1986a, b; Gray et al. 1995; Woodson et al. 2013). The inhibitory effect can be attributed to a decline in protein synthesis rate in plastids or a blockade in chloroplast development. Genetic analysis placed GUN1 in the PGE pathway as an important factor (Koussevitzky et al. 2007). GUN1 is a pentatricopeptide repeat protein (PPR) that was originally identified in a screen with other gun mutants which were involved in the tetrapyrrole biosynthesis (Susek et al. 1993). However, GUN1 is not involved in this pathway and operates differently. It has been shown that only in gun1 mutants, mRNA levels of the photosynthesis-related genes LHCB1 and RBCS are altered in the presence of lincomycin, whereas these genes are sensitive to the treatment in the gun2, gun3, gun4, and gun5 mutants (Susek et al. 1993; Mochizuki et al. 2001; Larkin et al. 2003).
Based on its evolutionary relationships with other members of the PPR family, a role in nucleic acid recognition can be assigned (Lurin et al. 2004; Barkan and Small 2014), although the experimental evidence of such conclusion remains scarce (Koussevitzky et al. 2007; Tadini et al. 2016). Recent studies found now that it interacts with multiple proteins, likely in a transient manner. Among the interacting partners are those involved in plastid transcription, translation, and protein homeostasis as well as tetrapyrrole biosynthesis enzymes (Tadini et al. 2016). According to this work, GUN1 appears to modulate the formation of protein complexes in the chloroplast. The authors further suggested that retrograde signaling might be linked to GUN1-dependent formation of protein complexes (Tadini et al. 2016; Colombo et al. 2016).
The GUN1 protein was associated with signals which are based on perturbations of plastid translation and transcription, as well as oxidative stress induced by carotenoid deficiency. The current model proposes that GUN1 integrates several signals originating from chloroplasts (e.g., signals related to the tetrapyrrole biosynthesis pathway, PGE-triggered retrograde signals, signals derived from the photosynthetic electron transport chain) and subsequently controls downstream nuclear gene expression (Koussevitzky et al. 2007; Woodson et al. 2011; Kindgren et al. 2012; Pfalz et al. 2012; Hernández-Verdeja and Strand 2018; Colombo et al. 2016). However, the exact mechanism of signal transduction by GUN1 and downstream components has not yet been fully understood. Recent work suggested that plastid-derived signals upon stress induction direct the plant homeodomain transcription factor PTM from the chloroplast outer envelope membrane into the nucleus, where it regulates PhANG expression. Furthermore, genetic analysis provided a molecular link to GUN1-mediated responses (Sun et al. 2011), although some controversy remains (Page et al. 2017). Downstream, the nucleus-localized transcription factors ABA INSENSITIVE 4 (ABI4) and Golden 2-Like1/2 (GLK1/GLK2) appear to be major determinants for transcription (Brunkard and Burch-Smith 2018). GUN1 activates ABI4, an ERF/AP2 transcription factor which negatively regulates the expression of PhANGs (Koussevitzky et al. 2007). GUN1 also represses glk1/2 transcription, which positively regulate expression of PhANGs and promote photomorphogenesis by antagonizing PHYTOCHROME-INTERACTING FACTORs (PIFs) (Waters et al. 2009; Martin et al. 2016). PIFs promote skotomorphogenetic development in dark-grown seedlings. Based on recent genetic information, activities of ABI4 and GLK1/2 represent two independent GUN1-mediated signaling events, in which phytochrome and retrograde signals converge antagonistically to control nuclear transcription during dark-to-light transition (Martin et al. 2016).
2.2 Redox, an Operational Signal
Imbalanced energy distribution between PSII and PSI generates redox signals within the plastoquinone (PQ) pool that controls both nuclear and plastid gene expression (Pfannschmidt et al. 1999; Fey et al. 2005; Dietzel et al. 2015). Likewise, it has been shown that redox states of acceptor or donor components of the PSI induce changes in the expression of nuclear genes for plastid proteins (Baier et al. 2004; Piippo et al. 2006; Barajas-López et al. 2013). Tuning gene expression to fluctuating light condition is necessary to maintain the efficiency of photosynthesis and metabolism and allows plants to survive unfavorable conditions. In this perspective, plants have developed mechanisms for both short- and long-term regulatory adaptations. A rapid reaction, a so-called short-term response, is state transition for balancing light energy distribution between the PSs by lateral movement of the LHCII antenna (Bellafiore et al. 2005; Bonardi et al. 2005). It takes place in a range of seconds or a few minutes. The details of molecular processes during short-term adaptation have been reviewed elsewhere (Dietzel et al. 2008; Rochaix 2013a, b). Longer term acclimation responses, which proceed at a slower tempo, are related to cellular strategies keeping PS stoichiometry adjusted to external light variations. This includes complex sensing and signaling pathways which regulate gene expression. Here, we focus on the role of redox signals from photosynthesis in regulation of nuclear gene expression. For details of the redox-regulatory mechanism controlling plastid gene expression see reviews by Barajas-López et al. (2013) and Dietzel et al. (2008).
Light acclimation and the molecular mechanism underlying this process have been an intense focus in recent years (Karpiński et al. 2013). Early evidence that redox-signals emanating from the photosynthetic electron transport chain regulate nuclear gene expression (e.g., genes associated with photosynthesis) was first demonstrated in the green algae Dunaliella tertiolecta (Escoubas et al. 1995; Maxwell et al. 1995). Escoubas et al. (1995) showed that light intensity alters the transcriptional activity of LHCB genes during photoacclimation and concluded that the changes in gene expression are associated with changes of the redox state of the PQ pool, as LHCB expression levels increased or decreased upon application of the selective chemical photosynthesis inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethyl urea (DCMU) or 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB), respectively. A redox-regulatory mechanism on the expression of nuclear genes by the redox state of the PQ pool was also found in higher plants. In Arabidopsis, for example, an increase in transcript abundance of two cytosolic ascorbate peroxidase genes (APX1 and APX2) was measured in response to high light and DCMU treatments (Karpinski et al. 1997). Subsequent studies revealed a link between cytosolic defense mechanism and the redox state of the PQ pool by which H2O2 might act as a systemic signal molecule (Karpinski et al. 1999). In the following decade, a few single nuclear genes related to photosynthesis have been identified to be regulated in response to light intensity as well as light quality by photosynthetic redox signals (Petracek et al. 1998; Pursiheimo et al. 2001; Eguchi et al. 2002; Pfannschmidt et al. 2001). These signals effect nuclear gene expression on almost all levels, including the regulation of transcription, stability, and translational efficiency (Pfannschmidt et al. 2003). The application of array-based technologies combined with physiological and genetic analyses have facilitated discovery of redox-responsive genes through comparison of the expression profiles of Arabidopsis plants exposed to wavelengths that preferentially excited either PSII or PSI (Fey et al. 2005; Piippo et al. 2006; Bräutigam et al. 2009; Pesaresi et al. 2009). Besides transcriptional control of photosynthesis-related genes, light quality shifts also effected the expression of genes involved in regulation, signal transduction, gene expression, stress responses, transport, and metabolism. According to the observed dynamics of transcriptional changes, redox signals rapidly (within 30 min to 2 h) alter the transcriptome pattern, with significant temporal changes during the period of 48 h light acclimation (Bräutigam et al. 2009). Related efforts by Dietzel et al. (2015) exhibited a set of early regulated genes. They fell into functional groups with defined processes including genes for the mitochondrial electron transport chain, tetrapyrrole biosynthesis, photosynthesis, and lipid metabolism. The light shift experiments showed expression profiles that were clearly different from those in plants exposed to high light treatments (Jung et al. 2013). In summary, these studies emphasize that the mechanism triggering the changes in expression of nuclear genes involves diverse redox signals emanating from the photosynthetic electron transport chain (Barajas-López et al. 2013; Hernández-Verdeja and Strand 2018). The existence of different sets of regulatory genes suggest a complex relationship between sensing, signaling, gene expression, and adaptation to the environment and may reflect a high degree of variability in light acclimation capabilities.
Efforts in understanding the transduction pathway of signals in response to the redox state of the photosynthetic machinery have combined multiple genetic and physiological analyses, but an answer still remains elusive. In this context, a phosphorylation-mediated signal cascade has been suggested. Among the components to be discovered, the STN7 kinase, which induces state transition to ensure balanced excitation within the photosynthetic system (Bellafiore et al. 2005; Bonardi et al. 2005), has been proposed to transduce signals due to its redox-sensitive kinase activity (Pesaresi et al. 2009, 2011; Bräutigam et al. 2009). However, studies by Tikkanen et al. (2012) have shown that the genetic disruption of stn7 in Arabidopsis does not fully inactivate the redox signaling pathway, indicating that STN7 is not essential for this process. In this work, STN7 was proposed to exert its signaling effect by maintaining the steady-state phosphorylation of the light-harvesting II proteins and the redox balance in the thylakoid membrane, thereby controlling chloroplast ROS homeostasis. In turn, alterations in redox homeostasis trigger signals that regulate the entire cellular network, probably by modification of hormone-mediated pathways (Tikkanen et al. 2012).
2.3 Metabolite Stage of Cell in Retrograde Signaling
Besides highly specific signaling molecules (cf. below) which potentially leave the organelle and control nuclear gene expression, changes in metabolite concentrations or intermediates of biochemical pathways are likely to be involved in the interorganellar cross talk (Estavillo et al. 2013; Brunkard and Burch-Smith 2018). The metabolite state in the cell or in a subcellular compartment permanently changes and is redirected according to the requirements of the organism. These changes result in the alteration of expression of the genes which are involved in the redirection of the metabolite pathways. Metabolite changes in the organelle, caused by, for instance, changes in light conditions, externally applied abiotic or biotic stresses or nutrient shortages, pathogen attack, and also developmental processes which result in a specific metabolite requirement at a particular place, time and organ, or circadian rhythm, cause appropriate changes in the metabolite profiles outside of the plastid in the cytoplasm, and consequently altered expression of responsive nuclear genes (Kleine and Leister 2016). Therefore, it is reasonable to assume that the nucleus is permanently informed about metabolite alterations in the organelle, either directly or indirectly due to metabolite adjustments between the plastid and cytoplasmic compartments, and adjusts its gene expression profile according to the metabolomic situation. This is particularly striking since many essential metabolites required for cellular functions and plant development are synthesized in the plastids and are exported into the cytoplasm. Obviously, metabolite concentrations represent an additional source of retrograde signaling during plant growth and upon responses to stress (Chi et al. 2013, 2015). Metabolite fluxes with plastidal involvement have been reviewed repeatedly and include carbon (Demmig-Adams et al. 2017; Tamoi and Shigeoka 2015), sulfur (Przybyla-Toscano et al. 2018; Eisenhut et al. 2015; Hanke and Mulo 2013; Tripathy et al. 2010; Hawkesford and De Kok 2006), nitrogen (Otori et al. 2017; Dörmann et al. 2014), and phosphorous (Karlsson et al. 2015; Rausch and Bucher 2002). Recently, de Souza et al. (2017) summarized the cross talk of multiple signaling events from mitochondria and plastids to coordinate nuclear gene expression and proposed that retrograde signals act as integrators of interorganellar communication and orchestrators of plant development. Interorganellar communication signals mediate reallocation of metabolic resources and energy currencies to balance growth and development against adaptive responses. Kleine and Leister (2016) highlight genetic screens which have already been performed and should be extended in the future to identify additional components in the cross talk. Metabolite profiling combined with bioinformatic tools is also a promising approach to identify novel players which are directly involved in retrograde signaling. Overall, it is reasonable to assume that changes in metabolite concentrations integrate information from the plastids, peroxisomes, mitochondria, the cytosol, as well as extracellular regions to regulate the activity of already existing signaling pathways and molecules to adjust nuclear gene expression.
Metabolite transporters in the plastid envelope membrane play a crucial role in the connection of plastidal and cytoplasmic metabolite pools. One would expect that they are of prokaryotic origin; however, the story appears to be more complex (cf. Weber and Linka 2011). A connection between the organellar metabolism and the host cell was probably an important issue after establishment of the symbiosis, and it must have been established early in evolution. The plastidic phosphate translocators were the first transporters identified in the plastid envelope. The discovery of triose phosphate/phosphate translocator, glucose 6-phosphate/phosphate translocator, xylulose 5-phosphate/phosphate translocators, and phosphoenolpyruvate/phosphate transporter highlights the important role of phosphate homeostasis between organelles and cytoplasm. Nucleotide carriers facilitate exchange of this essential metabolite across the organellar membrane. ADP/glucose, folate, S-adenoylmethionine, ATP and NAD carriers, dicarboxylate, glycolate and glycerate, maltose and glucose, as well as amino acid transporters are well known. Some of them are members of the mitochondrial carrier family and were redirected to the plastid envelop in the evolution. The function and evolution of these transporters are summarized by Weber and Linka (2011). This also highlights the importance of the metabolite exchange between the plastids, cytoplasm, and other cellular subcompartments, which consequently affects the expression of metabolite-related genes in the nucleus (Eisenhut et al. 2015; Mehrshahi et al. 2014; Linka and Theodoulou 2013; Flügge et al. 2011; Linka and Weber 2010; Weber and Fischer 2007; Hawkesford and De Kok 2006; Weber 2004). Thus, plastid metabolite levels might have an indirect effect on nuclear gene expression.
3 Specific Plastid Metabolites Control Specific Sets of Nuclear Genes
3.1 Tetrapyrroles
The role of more specific metabolites located in the plastids for the expression of nuclear genes has been investigated intensively. As mentioned above, five gun (gun2–6) mutants affect the branch point in the tetrapyrrole pathway (Susek et al. 1993; Larkin et al. 2003; Strand et al. 2003; Mochizuki et al. 2001, 2008; Moulin et al. 2008; Woodson et al. 2011; Thomas and Weinstein 1990). Protoporphyrin IX is chelated with iron by the ferrochelatase 1 or 2. The Fe-containing heme either remains in the plastids or further metabolizes to phytochromobilin, which is exported and associated with the apoprotein of phytochromes in the cytoplasm. The gun2 and gun3 mutants are affected in the conversion of heme to phytochromobilin. Alternatively, protoporphyrin IX is chelated with magnesium for chlorophyll biosynthesis. The gun4 and gun5 mutations prevent the insertion of magnesium. GUN5 is the H subunit of Mg-chelatase, and GUN4 binds the substrate of the Mg-chelatase reaction and activates the enzyme. Independent evidence of the involvement of chlorophyll precursors in the retrograde signaling came from the analysis of LHCP gene expression in Chlamydomonas (Johanningmeier and Howell 1984; Kropat et al. 1997, 2000). Whether one of the intermediates of the pathway triggers retrograde signaling and if so which of them is involved in it remains an open question. In the heme branch of the tetrapyrrole biosynthesis, the plastid ferrochelatase 1 synthesizes heme which results in the stimulation of nuclear gene expression. gun6 overexpresses the plastid-localized ferrochelatase 1, stimulates the flux through the heme branch of the tetrapyrrole pathway and the expression of the responsive genes in the nucleus. Therefore, it has been postulated that heme is a positively acting retrograde signal for nuclear genes (Woodson et al. 2011). Heme is also known to be released from the organelle (Thomas and Weinstein 1990), which further supports the idea. Finally, algae like Chlamydomonas synthesize billin, which might have a similar signaling function (discussed in Duanmu et al. 2013). In contrast, Mg-protoporphyrin IX represses the responding genes in the nucleus. Whether Mg-protoporphyrin IX acts as negative signal (Strand et al. 2003) or heme as positive signal (Woodson et al. 2011), or both metabolites are involved, remain an open question. Currently, it appears more likely that the flux through the two branches of the pathway might activate so far unknown signaling compounds in the plastids, which trigger retrograde signaling.
3.2 Singlet Oxygen (1O2) and Carotenoids
It is long known that reactive oxygen species (ROS) trigger nuclear gene expression (Galvez-Valdivieso and Mullineaux 2010), whereas the responding genes depend largely on the amount of location of ROS in and around the cell: low ROS levels have often signaling functions whereas high ROS levels are lethal. In photosynthetically highly active chloroplasts, singlet oxygen is produced in huge amounts, which is associated with the damage at the thylakoid membrane and altered gene expression in the nucleus (e.g., Kim and Apel 2013a; Ramel et al. 2012; Laloi et al. 2006). Originally proposed as retrograde signal, the short half-life of singlet oxygen suggests that it is unable to leave the organelle; however, it reacts with numerous compounds in its direct environment including carotenoids which have ROS-quenching functions (Ramel et al. 2012; 2013a). One of the carotenoid oxidation products is β-cyclocitral (β-CC), a volatile, which induces massive alteration of nuclear gene expression when applied to leaves in physiologically relevant concentrations (Ramel et al. 2012). The list includes 1O2-responsive genes (Ramel et al. 2012, 2013b), genes involved in light-stress acclimation (Lv et al. 2015), but also ISOCHORISMATE SYNTHASE 1 (ICS1), which synthesizes salicylic acid (SA) in the organelle. Elevated SA levels in the cell stimulate nuclear localization of NONEXPRESSOR OF PATHOGENESIS-RELATED GENE 1 (NPR1) which in turn activates SA-responsive genes (Lv et al. 2015). We are only at the beginning to understand how the abiotic and biotic stress acclimation responses are linked (cf. Maruta et al. 2012, 2016; Padmanabhan and Dinesh-Kumar 2010) and what is the exact role of events in the plastid that affect the expression of the genes in the nucleus. Nevertheless, as lipid-soluble volatile β-CC appears to be an ideal candidate for retrograde signaling, β-CC is not the only or the most important singlet oxygen-derived signaling compound. Apocarotenoids as enzymatic cleavage products of carotenoids may also have signaling functions (Auldridge et al. 2006; Avendaño-Vázquez et al. 2014). However, there must be additional pathways involved in the cross talk between the two organelles which become activated after singlet oxygen generation. Klaus Apel’s group demonstrated that the nuclear-encoded and plastid-localized EXECUTER1 and EXECUTER2 (Lee et al. 2007) are required for the activation of an independent plastid-localized signaling pathway by singlet oxygen, and the target genes in the nucleus differ from those responding to β-CC (Lee et al. 2007; Ramel et al. 2012). Single oxygen plays a crucial role in programmed cell death (PCD). Green leaves initiate PCD to restrict pathogen growth and distribution, a process that is stimulated by or even dependent on light perceived by photosynthesis. The fluorescent (flu) mutants show these lesions in the absence of any pathogen in light, but not in the dark. They accumulate excess protochlorophyllide in the dark, which are photosensitizing agents after transfer of the plants from the dark to light where they synthesize the toxic single oxygen leading to PCD phenotypes (Meskauskiene et al. 2001; op den Camp et al. 2003; Kim and Apel 2013a, b). EXECUTER1 and EXECUTER2 are required for the transduction of the single oxygen signal to the nucleus to initiate the PCD responses (Wagner et al. 2004). EXCECUTER1 is degraded in the flu mutants by the FtsH2 protease (Wang et al. 2016; Dogra et al. 2017). Obviously, high EXECUTOR1 levels are necessary for retrograde signaling from the plastids to the nucleus (Wang et al. 2016; Dogra et al. 2017) and are crucial for the survival of a cell.
3.3 3′-Phosphoadenosine 5′-Phosphate
3′-Phosphoadenosine 5′-phosphate (PAP) is proposed as a retrograde-active metabolite and accumulates, under stress conditions such as drought or high light, in plastids (Estavillo et al. 2011). The plastid- and mitochondria-localized enzymes SAL1 dephosphorylate PAP to AMP (Klein and Papenbrock 2004; Wilson et al. 2009) and a mutant of the plastid SAL1 protein accumulate high levels of PAP, similar to exposure of wild-type plants to stress (Rossel et al. 2006; Estavillo et al. 2011). In contrast, constitutively high levels of SAL1 in either the nucleus or the plastids result in lower PAP levels, even when the enzyme is expressed in the other compartment, suggesting that the metabolite can travel in the cell. Based on these and additional studies, it was proposed that accumulation of PAP stimulates the expression of nuclear-encoded stress genes, in particular those for antioxidant enzymes, including ascorbate peroxidase 2 (APX2), which was used for an initial mutant screen (Rossel et al. 2006). Targeting of SAL1 to either the nucleus or chloroplasts decreased the PAP levels and consequently APX2 expression (Estavillo et al. 2011). Since PAP appears to move between the plastid and cytoplasm, probably by a specific transporter (Gigolashvili et al. 2012), it fulfills a major criteria as retrograde signal. PAP is also produced during sulfonation reactions, whereby sulfate is transferred from PAPS to different metabolic substrates (Klein and Papenbrock 2004), and PAP is released during this reaction. However, quite interesting is the observation that PAP binds irreversibly to yeast 5′-3′ exoribonucleases and inhibits their activities (van Dijk et al. 2011). It appears that also in plants, PAP can alter RNA metabolism and thus acts posttranscriptionally. Although there is no doubt that PAP fulfills all criteria to transfer stress information from the plastids to the nuclear/cytoplasmic compartment, there might be many more such metabolites with similar functions.
4 Methylerythritol Cyclodiphosphate (MEcPP) as Defense-Related Retrograde Signal
MEcPP is a biochemical intermediate of the methylerythritol phosphate (MEP) pathway for the isoprenoid synthesis in chloroplasts (Vranova et al. 2013; Banerjee and Sharkey 2014). Not surprisingly, inhibition of this pathway leads to severe lesions in growth and development. The stress-inducible metabolite was identified as a plastid retrograde signal, which alters the chromatin structure in the nucleus that in turn affects the expression of a particular subset of stress-inducible genes (Xiao et al. 2012, 2013). Expression of the hydroperoxide lyase (HPL) and isochorismate synthase1 (ICS1) genes is altered in isolated mutants, and this results in increased SA levels, a phytohormone which confers resistance against biotrophic pathogens such as Pseudomonas syringae (Xiao et al. 2012). The authors showed that SA accumulation and the induction of the HPL gene are caused by the plastidal metabolite MEcPP and are not due to a general stress response due to the manipulation of the MEP pathway in the mutants (Xiao et al. 2012). MEcPP application also regulates HPL expression directly, confirming that the metabolite is active and plays a role as stress sensor in plastids. MEcPP is also present in bacteria and accumulates upon exposure to oxidative stress (Ostrovsky et al. 1992, 1998), suggesting a conserved mechanism of its occurrence and action during abiotic stresses (Walley et al. 2015; Xiao et al. 2012, 2013). Interestingly, MEcPP can disrupt histone H1-like protein interaction with DNA, which suggests that the metabolite remodels the chromatin structure to allow expression of stress-related genes (Grieshaber et al. 2004, 2006). MEcPP is probably the most direct evidence for the existence of metabolites in the plastid that control transcription in the nucleus. Besides functional conservation in evolution, it also differs from tetrapyrrole signaling, for which changes in flux rates play an important role for signal initiation. However, how MEcPP travels from the organelle to the nucleus is not known yet. Furthermore, MEcPP also highlights the important role of the plastid for biotic stress responses, in which SA and jasmonic acid (JA) are crucial phytohormones (Nomura et al. 2012; cf. below).
5 Dual Targeted Proteins in Plastids and Nucleus: Function as Transmitters or Integrators of Information?
Retrograde signal transduction is initiated by signaling molecules that are produced in and exported from plastids and then enter the nucleus to regulate the expression of appropriate genes. Signal transduction from plastids (and/or mitochondria) to the nucleus may also occur through the movement of proteins (Krause et al. 2012), such as transcription factors like PTM (for PHD type transcription factor with transmembrane domains), a chloroplast envelope-bound plant homeodomain transcription factor with transmembrane domains (Sun et al. 2011), PEND, a plastid envelope DNA-binding protein (Terasawa and Sato 2009), or WHIRLY1 (WHY1; Miao et al. 2013; Ren et al. 2017; Desveaux et al. 2004; Foyer et al. 2014; Isemer et al. 2012), a protein with specific functions in both organelles. Distinct retrograde signals may converge at PTM in the plastids, which then transmit common signals to the nucleus (Sun et al. 2011). In the nucleus, PTM promotes ABI4 transcription upon high light treatments. ABI4 was proposed to be involved in the integration of three plastids as well as mitochondrial retrograde signals (Koussevitzky et al. 2007). Retrograde signaling via members of the AP2/EREBP transcription factor gene family plays a role in the connection of metabolic, hormonal, and environmental signals during stress acclimation (Dietz et al. 2010). These examples demonstrate that signal information can also be transferred from plastids to the nucleus by traveling proteins. How this occurs is a matter of discussion. They might participate in signal integration in the plastids before transfer of the information to the nucleus (Koussevitzky et al. 2007). Others are part of signaling pathways or respond to them which are activated by different stimuli from outside of the plastids. This allows them to integrate information from plastids with those from other extraplastidic sources. Some of the proteins like WHY1 have defined functions in each of the organelle (Desveaux et al. 2004; Miao et al. 2013; Foyer et al. 2014; Isemer et al. 2012; Ren et al. 2017). As mentioned above, dual targeted proteins are often transcription factors or regulators of gene expression when they are in the nucleus. Since more and more dual targeted proteins with quite different functions are described (cf. Krause and Krupinska 2009; Nevarez et al. 2017; Mazzoleni et al. 2015; Gile et al. 2015; Langner et al. 2014; Ge et al. 2014; Berglund et al. 2009; Rokov-Plavec et al. 2008; Millar et al. 2006), it appears that there is a need for intensive investigations, including the import of nuclear-encoded proteins into the organelle (Inaba 2010; Inaba et al. 2011).
A well-studied example for a dual-targeted protein is WHY1. Like other members of the WHIRLY protein family, they perform numerous cellular functions in both locations (Krause et al. 2005; Grabowski et al. 2008; Miao et al. 2013; Ren et al. 2017; Foyer et al. 2014). These proteins were first discovered as nuclear transcriptional activators binding an elicitor response element in the promoter regions of pathogenesis-related genes in potato and Arabidopsis (Desveaux et al. 2000, 2004). They bind to various DNA sequences, including telomeres (Yoo et al. 2007), a distal element upstream of a kinesin gene (Xiong et al. 2009), the promoter region of the early senescence marker gene WRKY53 in a development-dependent manner in Arabidopsis (Miao et al. 2013), and the promoter region of the senescence-associated gene HvS40 which was induced during natural and stress-related senescence in barley (Krupinska et al. 2013). In plastids, WHY1 is present in the transcriptional active chromosome (TAC, Pfalz et al. 2006) and nucleoid preparations although it can be purified away from the transcriptional activity (Melonek et al. 2010) and binds to both single-stranded DNA and RNA with a role in intron splicing in maize chloroplasts (e.g., Prikryl et al. 2008). In barley chloroplasts, WHY1 also was found to be associated with intron-containing RNAs (Melonek et al. 2010). Moreover, the Brission group demonstrated that WHY proteins in organelles function as antirecombinant proteins favoring accurate DNA repair to maintain organellar genome stability (Cappadocia et al. 2010, 2012; Lepage et al. 2013). These results suggest that WHY proteins might function differently depending on their intracellular localization and/or the developmental stage of the plant (Ren et al. 2017). Recently, the Miao group constructed “compartmental mutants” of WHY1 that differentially accumulate different isoforms of the WHY1 protein in plastids (pWHY1) or nuclei (nWHY1) of Arabidopsis. Based on these mutants, the group identified differentially expressed nuclear genes in plants with constitutive and inducible pWHY1 or nWHY1 versions. The results shine new light on the role of WHY1 in integrating metabolic, hormonal, and environmental signals in retrograde signaling. In particular, the group demonstrates that WHY1-mediated retrograde signals involve ROS (H2O2)- and SA-dependent compounds and are integrated into known signaling events. The quite strong phenotypes of the compartmentalized WHY1 mutants generated in the Miao lab in response to external signals will be important tools to unravel the function of the dual targeted protein in the interorganellar cross talk.
6 The Role of Plastids in Stress Response: Importance for Retrograde Signaling?
Biogenic control signals inform the nucleus about developmental changes of the organelles, such as the development of chloroplasts from etioplasts or proplastids. Operational signals, such as redox signals inform the nucleus about the events that occur in functional plastids/chloroplasts such as the efficiency of the photosynthetic electron transport. Dramatic changes in nuclear gene expression occur also when the plants are exposed to stress (Fernández and Strand 2008). Abiotic stresses such as drought are counteracted by the synthesis of the phytohormone abscisic acid, biotic stresses involve SA and JA. Other plastid-related hormones such as cytokinins also participate in defense responses (Chan et al. 2010, 2016). Since the synthesis of these hormones starts in the plastids (SA is also synthesized in the cytoplasm), and is strongly stimulated upon stress, the organelle plays the essential role in the response of the cell to stress. Furthermore, SA accumulates in response to the retrograde signaling metabolite MEcPP and in response to the plastid-localized isoform of WHY1, connecting phytohormones to other retrograde signaling. Finally, MEcPP is a regulator of SA and JA cross talk (Lemos et al. 2016). Since these phytohormones strongly activate defense genes in the nucleus upon stress or pathogen attack, phytohormones also play a crucial role in retrograde signaling.
6.1 Salicylic Acid
Salicylic acid (SA) in plants is synthesized via two biosynthetic pathways: the plastid-localized isochorismate synthase (ICS) and the cytosolic phenylalanine ammonia lyase (PAL) pathways. Both pathways use chorismate as precursor, which is synthesized via the shikimate pathway in plastids (Poulsen and Verpoorte 1991; Schmid and Amrhein 1995). The plastid-localized isochorismate pathway is the main source of SA upon exposure of the plant to abiotic stress or pathogen attacks (Vlot et al. 2009; Dempsey et al. 2011). Furthermore, SA is the main defense hormone upon attack of plant by biotrophic pathogens, while necrotrophic pathogens activate the JA defense pathway. SA is also involved in a number of developmental processes (Martínez et al. 2004; Morris et al. 2000; Zhang et al. 2013; Abreu and Munné-Bosch 2009; Seguel et al. 2018) in which not only chloroplasts but also other types of plastids participate. The plastid-localized enzyme ICS1 (Strawn et al. 2007) converts chorismate to isochorismate which is subsequently converted to SA by a so-far unknown organellar enzyme. The SA biosynthesis is negatively regulated by an autoinhibitory feedback loop operating around ICS1. Export of SA from the chloroplast to the cytoplasm is mediated by the multidrug and toxin-extrusion transporter ENHANCED DISEASE SUSCEPTIBILITY5 (EDS5) in the chloroplast envelope. Interestingly, analysis of the eds5 mutant in Arabidopsis has demonstrated that SA is trapped in the chloroplast of the mutant and inhibits its own accumulation by the autoinhibitory feedback mechanism which couples SA export to its synthesis (Serrano et al. 2013; Yamasaki et al. 2013).
The cross talk between plastids and cytoplasm is a result of the evolution of the two pathways. In Arabidopsis, the basal SA level is produced via the PAL pathway (Huang et al. 2010), whereas under pathogen attack or abiotic stress, the vast majority of the SA is synthesized by the isochorismate pathway in the plastids (Wildermuth et al. 2001; Garcion et al. 2008). This appears to be species specific, since in soybean, both pathways contribute equally to the SA production upon pathogen attack (Shine et al. 2016). Arabidopsis and soybean contain two genes for the key enzyme of the plastid ICS pathway. In other species, different ICS isoforms are produced by alternative splicing of a single ICS gene (Macaulay et al. 2017). Apparently, the plastid-localized pathway for SA is highly sophisticated and an evolutionary result of intensive cross talk between the two organelles.
6.2 Jasmonic Acid
It is long known that Jasmonic acid (JA) precursors and, in particular, the JA precursor 12-oxo-phytodienoic acid (OPDA) are synthesized in plastids. Jasmonates are derived from the α-linolenic acid (18:3) or 7(Z)-, 10(Z)-, and 13(Z)-hexadecatrienoic acid (16:3). A lipoxygenase catalyzes the addition of molecular oxygen to α-linolenic acid which initiates JA biosynthesis by providing the substrate for the formation of an allene oxide by the allene oxide synthase (AOS), which is further converted to OPDA. The reactions until OPDA formation take place in plastids, while the subsequent steps in the JA biosynthesis occur in peroxisomes. In the plastids, OPDA can also be esterified to lipids. JA is converted to jasmonoyl-isoleucine (JA-Ile) in the cytoplasm, and after binding to its receptor, JAR1 activates specific defense genes in the nucleus (Huang et al. 2017; Zhang et al. 2017; Han 2017; Wasternack and Song 2017). Thus, besides being integrated into a complex hormone network, jasmonate also functions as retrograde signals in concert with other signals and plastid metabolites.
Lemos et al. (2016) showed that the plastidial retrograde signal methyl erythritol cyclopyrophosphate is a regulator of SA and JA cross talk. Wang et al. (2018) identified two ABA-responsive plastid-localized lipases which are involved in JA biosynthesis (cf. Mach 2018). Farmer and Mueller (2013), among others, proposed a link between jasmonate and ROS signaling. Thus, JA, SA, and ABA appear to be coupled to retrograde-active signals. Since not even the cross talk between the phytohormones is completely understood, it appears that their involvement in the cross talk between plastids and nucleus will become an interesting research field in the near future.
7 Concluding Remarks
Obviously, there is much more to be discovered in the interorganellar cross talk (cf. Godoy Herz et al. 2014). For instance, metabolites specifically responding to singlet oxygen in the organelle need to be identified. The redox signaling network is likely important for the distribution of information within the cell and entire organisms (Dietz 2016; Dietz et al. 2016) and does not only include redox signals from the photosynthetic electron transport but also other metabolic processes which are regulated by internal and environmental signals. The flux rate in the tetrapyrrole pathway needs to be translated into traveling metabolites or signals. Although much work has been performed to understand the role of light stress for retrograde signaling, there are many open questions to be addressed with novel tools (Szechyńska-Hebda and Karpiński 2013). For instance, little is known about processes balancing energy distribution and stress responses (Woodson 2016). Information transfer between organelles involves reversible phosphorylation events and Ca2+ signaling, and they have been barely investigated in this scenario (Chandok et al. 2001; Pesaresi et al. 2011; Guo et al. 2016). Whether proteins or peptides leave the organelle and inform the nucleus is also an open question. Finally, plastids play an essential role in phytohormone functions. They have a tremendous influence on gene expression profiles and developmental strategies (cf. Li et al. 2013; Serrano et al. 2016). Phytohormones determine the response of the plant to environmental signals and the decision of the plant to invest in either growth and productivity or defense. Not all concepts could be covered in this brief overview. For instance, Burch-Smith et al. (2011) proposed an organelle-nucleus cross talk via plasmodesmata. Signaling via Ca2+ levels coordinates many responses and integrates cell’s internal and external information (Guo et al. 2016; de Souza et al. 2017). The Ca2+ signaling network is well known to participate in mitochondrial retrograde signaling (cf. Butow and Avadhani 2004). Many volatiles and secondary metabolites are partially synthesized in plastids and have tremendous influences on nuclear gene expression. Considering the central role of plastids for all processes in the plant cell and entire plant, there are probably many more communication systems that will be discovered in the future. Finally, the cross talk between plastid- and mitochondria-derived signals has been little investigated (Van Aken and Pogson 2017).
Abbreviations
- ABI4:
-
abscisic acid insensitive 4
- β-CC:
-
β-cyclocitral
- GLK1/2:
-
golden 2-like 1/2
- GUN1/4/5:
-
genomes uncoupled 1/4/5
- EX1/EX2:
-
executer 1/2
- HDS1:
-
hydroxymethylbutenyl diphosphate synthase
- LHCB:
-
gene-encoding photosystem II chlorophyll a/b binding protein
- MEcPP:
-
methylerythritol cyclodiphosphate
- Mg-protop-IX:
-
Mg-protoporphyrin IX
- ΔPET:
-
impairment of photosynthetic electron transport chain
- PGE:
-
plastid gene expression
- PhANG:
-
photosynthesis-Associated Nuclear Genes
- PQ:
-
plastoquinone
- PRIN2:
-
plastid redox-insensitive 2
- PSI:
-
photosystem I
- ROS:
-
reactive oxygen species
- SAL1:
-
inositol polyphosphate 1-phosphatase
- TFs:
-
transcription factors
- STN7:
-
thylakoid protein kinase 7
- WHY1:
-
whirly 1
References
Abreu ME, Munné-Bosch S (2009) Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana. J Exp Bot 60:1261–1271
Auldridge ME, McCarty DR, Klee HJ (2006) Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol 9:315–321
Avendaño-Vázquez AO, Cordoba E, Llamas E, San Román C, Nisar N, De la Torre S, Ramos-Vega M, de la Luz Gutiérrez-Nava M, Cazzonelli CI, Pogson BJ, León P (2014) An uncharacterized apocarotenoid-derived signal generated in ζ-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. Plant Cell 26:2524–2537
Baier M, Stroher E, Dietz KJ (2004) The acceptor availability at photosystem I and ABA control nuclear expression of 2-Cys peroxiredoxin-A in Arabidopsis thaliana. Plant Cell Physiol 45:997–1006
Bajracharya D, Bergfield R, Hatzfeld W-D, Klein S, Schopfer P (1987) Regulatory involvement of plastids in the development of peroxisomal enzymes in the cotyledons of mustard (Sinapis alba L.) seedlings. J Plant Physiol 126:421–436
Banerjee A, Sharkey TD (2014) Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat Prod Rep 31:1043–1055
Barajas-López J de D, Blanco NE, Strand Å (2013) Plastid-to-nucleus communication, signals controlling the running of the plant cell. Biochim Biophys Acta 1833:425–437
Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65:415–442
Batschauer A, Mösinger E, Kreuz K, Dörr I, Apel K (1986) The implications of a plastid-derived factor in the transcriptional control of nuclear genes encoding the light-harvesting chlorophyll a/b protein. Eur J Biochem 154:625–634
Bellafiore S, Barneche F, Peltier G, Rochaix JD (2005) State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433:892–895
Berglund AK, Spånning E, Biverståhl H, Maddalo G, Tellgren-Roth C, Mäler L, Glaser E (2009) Dual targeting to mitochondria and chloroplasts: characterization of Thr-tRNA synthetase targeting peptide. Mol Plant 2:1298–1309
Bernardi G (1979) The petite mutation in yeast. Trends Biochem Sci 4:197–201
Blair GE, Ellis RJ (1973) Protein synthesis in chloroplasts. I. Light-driven synthesis of the large subunit of fraction I protein by isolated pea chloroplasts. Biochim Biophys Acta 319:223–234
Bogorad L (1975) Evolution of organelles and eukaryotic genomes. Science 188:891–898
Bolle C, Sopory S, Lübberstedt T, Klösgen RB, Herrmann RG, Oelmüller R (1994) The role of plastids in the expression of nuclear genes for thylakoid proteins studied with chimeric β-glucuronidase gene fusions. Plant Physiol 105:1355–1364
Bolle C, Kusnetsov VV, Herrmann RG, Oelmüller R (1996a) The spinach AtpC and AtpD genes contain elements for light-regulated, plastid-dependent and organ-specific expression in the vicinity of the transcription start sites. Plant J 9:21–30
Bolle C, Herrmann RG, Oelmüller R (1996b) Intron sequences are involved in the plastid- and light-dependent expression of the spinach PsaD gene. Plant J 10:919–924
Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D (2005) Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437:1179–1182
Börner T (2017) The discovery of plastid-to-nucleus retrograde signaling-a personal perspective. Protoplasma 254:1845–1855
Börner T, Knoth R, Herrmann F, Hagemann R (1972) Struktur und Funktion der genetischen Information in den Plastiden. V. Das Fehlen von ribosomaler RNS in den Plastiden der Plastommutante “Mrs. Parker” von Pelargonium zonale Ait. Theor Appl Genet 42:3–11
Börner T, Herrmann FH, Hagemann R (1973) Plastid ribosome deficient mutants in Pelargonium zonale. FEBS Lett 37:17–19
Börner T, Knoth R, Herrmann F, Hagemann R (1974) Struktur und Funktion der genetischen Information in den Plastiden. X. Das Fehlen von Fraktion-I-Protein in den weißen Plastiden einiger Sorten von Pelargonium zonale. Biochem Physiol Pflanz 165:429–432
Börner T, Schumann B, Hagemann R (1976) Biochemical studies on a plastid ribosome-deficient mutant of Hordeum vulgare. In: Bücher T, Neupert W, Sebald W, Werner S (eds) Genetics and biogenesis of chloroplasts and mitochondria. Elsevier/North-Holland Medical Press, Amsterdam, pp 41–48
Börner T, Mendel RR, Schiemann J (1986) Nitrate reductase is not accumulated in chloroplast-ribosome-deficient mutants of higher plants. Planta 169:202–207
Bradbeer JW, Börner T (1978) Activities of glyceraldehyde-dephosphate dehydrogenase (NADP+) and phosphoribulokinase in two barley mutants deficient in chloroplast ribosomes. In: Akoyunoglou G, Argyroudi-Akoyunoglou JG (eds) Chloroplast development. North Holland, Amsterdam, pp 727–732
Bradbeer JW, Atkinson YE, Börner T, Hagemann R (1979) Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesised RNA. Nature 279:816–817
Bräutigam K, Dietzel L, Kleine T, Ströher E, Wormuth D, Dietz KJ, Radke D, Wirtz M, Hell R, Dörmann P, Nunes-Nesi A, Schauer N, Fernie AR, Oliver SN, Geigenberger P, Leister D, Pfannschmidt T (2009) Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. Plant Cell 21:2715–2732
Brunkard JO, Burch-Smith TM (2018) Ties that bind: the integration of plastid signalling pathways in plant cell metabolism. Essays Biochem 62:95–107
Burch-Smith TM, Brunkard JO, Choi YG, Zambryski PC (2011) Organelle-nucleus cross-talk regulates plant intercellular communication via plasmodesmata. Proc Natl Acad Sci U S A 108:E1451–E1460
Burgess DG, Taylor WC (1988) Chloroplast photooxidation affects the transcription of a nuclear gene family. Mol Gen Genet 214:89–96
Butow RA, Avadhani NG (2004) Mitochondrial signaling: the retrograde response. Mol Cell 14:1–15
Cappadocia L, Maréchal A, Parent JS, Lepage E, Sygusch J, Brisson N (2010) Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 22:1849–1867
Cappadocia L, Parent JS, Zampini E, Lepage E, Sygusch J, Brisson N (2012) A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage. Nucleic Acids Res 40:258–269
Chan PH, Wildman SG (1972) Chloroplast DNA codes for the primary structure of the large subunit of fraction I protein. Biochim Biophys Acta 277:677–680
Chan KX, Crisp PA, Estavillo GM, Pogson BJ (2010) Chloroplast-to-nucleus communication: current knowledge, experimental strategies and relationship to drought stress signaling. Plant Signal Behav 5:1575–1582
Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol 67:25–53
Chandok MR, Sopory SK, Oelmüller R (2001) Cytoplasmic kinase and phosphatase activities can induce PsaF gene expression in the absence of functional plastids: evidence that phosphorylation/dephosphorylation events are involved in interorganellar crosstalk. Mol Gen Genet 264:819–826
Chi W, Sun X, Zhang L (2013) Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol 64:559–582
Chi W, Feng P, Ma J, Zhang L (2015) Metabolites and chloroplast retrograde signaling. Curr Opin Plant Biol 25:32–38
Colombo M, Tadini L, Peracchio C, Ferrari R, Pesaresi P (2016) GUN1, a Jack-of-all-trades in chloroplast protein homeostasis and signaling. Front Plant Sci 7:1427
Criddle RS, Dau B, Kleinkopf DE, Huffaker RC (1970) Differential synthesis of ribulose diphosphate subunits. Biochem Biophys Res Commun 41:621–627
de Souza A, Wang JZ, Dehesh K (2017) Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu Rev Plant Biol 68:85–108
Demmig-Adams B, Stewart JJ, Adams WW 3rd (2017) Environmental regulation of intrinsic photosynthetic capacity: an integrated view. Curr Opin Plant Biol 37:34–41
Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF (2011) Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9:e0156
Desveaux D, Després C, Joyeux A, Subramaniam R, Brisson N (2000) PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12:1477–1489
Desveaux D, Subramaniam R, Després C, Mess JN, Lévesque C, Fobert PR, Dangl JL, Brisson N (2004) A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev Cell 6:229–240
Dietz KJ (2016) Thiol-Based peroxidases and ascorbate peroxidases: why plants rely on multiple peroxidase systems in the photosynthesizing chloroplast? Mol Cells 39:20–25
Dietz KJ, Vogel MO, Viehhauser A (2010) AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245:3–14
Dietz KJ, Turkan I, Krieger-Liszkay A (2016) Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol 171:1541–1550
Dietzel L, Bräutigam K, Pfannschmidt T (2008) Photosynthetic acclimation: state transitions and adjustment of photosystem stoichiometry – functional relationships between short-term and long-term light quality acclimation in plants. FEBS J 275:1080–1088
Dietzel L, Gläßer C, Liebers M, Hiekel S, Courtois F, Czarnecki O, Schlicke H, Zubo Y, Börner T, Mayer K, Grimm B, Pfannschmidt T (2015) Identification of early nuclear target genes of plastidial redox signals that trigger the long-term response of Arabidopsis to light quality shifts. Mol Plant 8:1237–1252
Dogra V, Duan J, Lee KP, Lv S, Liu R, Kim C (2017) FtsH2-dependent proteolysis of EXECUTER1 Is essential in mediating singlet oxygen-triggered retrograde signaling in Arabidopsis thaliana. Front Plant Sci 8:1145
Dörmann P, Kim H, Ott T, Schulze-Lefert P, Trujillo M, Wewer V, Hückelhoven R (2014) Cell-autonomous defense, re-organization and trafficking of membranes in plant-microbe interactions. New Phytol 204:815–822
Duanmu D, Casero D, Dent RM, Gallaher S, Yang W, Rockwell NC, Martin SS, Pellegrini M, Niyogi KK, Merchant SS, Grossman AR, Lagarias JC (2013) Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc Natl Acad Sci U S A 110:3621–3626
Eguchi S, Takano H, Ono K, Takio S (2002) Photosynthetic electron transport regulates the stability of the transcript for the protochlorophyllide oxidoreductase gene in the liverwort, Marchantia paleacea var. diptera. Plant Cell Physiol 43:573–577
Eisenhut M, Hocken N, Weber AP (2015) Plastidial metabolite transporters integrate photorespiration with carbon, nitrogen, and sulfur metabolism. Cell Calcium 58:98–104
Ellis RJ (1975) Inhibition of plastid protein synthesis by lincomycin and 2-(4-methyl-2,6-dinitroanilino)-N-methylpropionamide. Phytochemistry 14:89–93
Ellis RJ (1977) Protein synthesis by isolated chloroplasts. Biochim Biophys Acta 463:285–315
Escoubas JM, Lomas M, LaRoche J, Falkowski PG (1995) Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci U S A 92:10237–11041
Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, Brearley C, Hell R, Marin E, Pogson BJ (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23:3992–4012
Estavillo GM, Chan KX, Phua SY, Pogson BJ (2013) Reconsidering the nature and mode of action of metabolite retrograde signals from the chloroplast. Front Plant Sci 3:300
Farmer EE, Mueller MJ (2013) ROS-mediated lipid peroxidation and RES-activated signaling. Annu Rev Plant Biol 64:429–450
Feierabend J (1977) Capacity for chlorophyll synthesis in heat-bleached 70S ribosome-deficient rye leaves. Planta 135:83–88
Feierabend J, Mikus M (1977) Occurrence of a high temperature sensitivity of chloroplast ribosome formation in several higher plants. Plant Physiol 59:863–867
Feierabend J, Schrader-Reichhardt U (1976) Biochemical differentiation of plastids and other organelles in rye leaves with a high-temperature-induced deficiency of plastid ribosomes. Planta 129:133–145
Fernández AP, Strand A (2008) Retrograde signaling and plant stress: plastid signals initiate cellular stress responses. Curr Opin Plant Biol 11:509–513
Fey V, Wagner R, Braütigam K, Wirtz M, Hell R, Dietzmann A, Leister D, Oelmüller R, Pfannschmidt T (2005) Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem 280:5318–5328
Flügge UI, Häusler RE, Ludewig F, Gierth M (2011) The role of transporters in supplying energy to plant plastids. J Exp Bot 62:2381–2392
Foyer CH, Karpinska B, Krupinska K (2014) The functions of WHIRLY1 and REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 in cross tolerance responses in plants: a hypothesis. Philos Trans R Soc Lond Ser B Biol Sci 369:20130226
Galvez-Valdivieso G, Mullineaux PM (2010) The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol Plant 138:430–439
Garcion C, Lohmann A, Lamodière E, Catinot J, Buchala A, Doermann P, Métraux JP (2008) Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol 147:1279–1287
Ge C, Spånning E, Glaser E, Wieslander A (2014) Import determinants of organelle-specific and dual targeting peptides of mitochondria and chloroplasts in Arabidopsis thaliana. Mol Plant 7:121–136
Gigolashvili T, Geier M, Ashykhmina N, Frerigmann H, Wulfert S, Krueger S, Mugford SG, Kopriva S, Haferkamp I, Flügge UI (2012) Much more than a thylakoid ADP/ATP carrier– enlightening a role of TAAC in plastidic phosphoadenosine 50-phosphosulfate (PAPS) supply to the cytosol. Plant Cell 24:4187–4204
Gile GH, Moog D, Slamovits CH, Maier UG, Archibald JM (2015) Dual organellar targeting of aminoacyl-tRNA synthetases in diatoms and cryptophytes. Genome Biol Evol 7:1728–1742
Giuliano G, Scolnik PA (1988) Transcription of two photosynthesis-associated nuclear gene families correlates with the presence of chloroplasts in leaves of the variegated tomato ghost mutant. Plant Physiol 86:7–9
Givan AL, Criddle RS (1972) Ribulose diphosphate carboxylase from Chlamydomonas reinhardi: purification, properties and mode of synthesis in the cell. Arch Biochem Biophys 149:153–154
Godoy Herz MA, Kornblihtt AR, Barta A, Kalyna M, Petrillo E (2014) Shedding light on the chloroplast as a remote control of nuclear gene expression. Plant Signal Behav 9:e976150
Gollan PJ, Tikkanen M, Aro EM (2015) Photosynthetic light reactions: integral to chloroplast retrograde signalling. Curr Opin Plant Biol 27:180–191
Grabowski E, Miao Y, Mulisch M, Krupinska K (2008) Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol 147:1800–1804
Gray JC, Sornarajah R, Zabron AA, Duckett CM, Khan MS (1995) Chloroplast control of nuclear gene expression. In: Mathis P (ed) Photosynthesis, from light to biosphere. Kluwer, Dordrecht, pp 543–550
Greiner S, Bock R (2013) Tuning a menage a trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. BioEssays 35:354–365
Grieshaber NA, Fischer ER, Mead DJ, Dooley CA, Hackstadt T (2004) Chlamydial histone-DNA interactions are disrupted by a metabolite in the methylerythritol phosphate pathway of isoprenoid biosynthesis. Proc Natl Acad Sci U S A 101:7451–7756
Grieshaber NA, Sager JB, Dooley CA, Hayes SF, Hackstadt T (2006) Regulation of the Chlamydia trachomatis histone H1-like protein Hc2 is IspE dependent and IhtA independent. J Bacteriol 188:5289–5292
Guo H, Feng P, Chi W, Sun X, Xu X, Li Y, Ren D, Lu C, Rochaix JD, Leister D, Zhang L (2016) Plastid-nucleus communication involves calcium-modulated MAPK signalling. Nat Commun 7:12173
Hagemann R, Börner T (1987) Plastid ribosome-deficient mutants of higher plants as a tool in studying chloroplast biogenesis. In: Akoyunoglou G, Argyroudi-Akoyunoglou JG (eds) Chloroplast development. North Holland, Amsterdam, pp 709–720
Han GZ (2017) Evolution of jasmonate biosynthesis and signaling mechanisms. J Exp Bot 68:1323–1331
Hanke G, Mulo P (2013) Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ 36:1071–1084
Harpster MH, Mayfield SP, Taylor WC (1984) Effects of pigment-deficient mutants on the accumulation of photosynthetic proteins in maize. Plant Mol Biol 3:258–263
Hawkesford MJ, De Kok LJ (2006) Managing sulphur metabolism in plants. Plant Cell Environ 29:382–395
Hernández-Verdeja T, Strand Å (2018) Retrograde signals navigate the path to chloroplast development. Plant Physiol 176:967–976
Hess WR, Müller A, Nagy F, Börner T (1994) Ribosome-deficient plastids affect transcription of light-induced nuclear genes: genetic evidence for a plastid-derived signal. Mol Gen Genet 242:305–312
Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153:1526–1538
Huang H, Liu B, Liu L, Song S (2017) Jasmonate action in plant growth and development. J Exp Bot 68:1349–1359
Inaba T (2010) Bilateral communication between plastid and the nucleus: plastid protein import and plastid-to-nucleus retrograde signaling. Biosci Biotechnol Biochem 74:471–476
Inaba T, Yazu F, Ito-Inaba Y, Kakizaki T, Nakayama K (2011) Retrograde signaling pathway from plastid to nucleus. Int Rev Cell Mol Biol 290:167–204
Isemer R, Mulisch M, Schafer A, Kirchner S, Koop HU, Krupinska K (2012) Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett 586:85–88
Johanningmeier U, Howell SH (1984) Regulation of light-harvesting chlorophyll-binding protein messenger-RNA accumulation in Chlamydomonas rheinhardi – possible involvement of chlorophyll synthesis precursors. J Biol Chem 259:3541–3549
Jung HS, Crisp PA, Estavillo GM, Cole B, Hong F, Mockler TC, Pogson BJ, Chory J (2013) Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc Natl Acad Sci U S A 110:1447–1479
Karlsson PM, Herdean A, Adolfsson L, Beebo A, Nziengui H, Irigoyen S, Ünnep R, Zsiros O, Nagy G, Garab G, Aronsson H, Versaw WK, Spetea C (2015) The Arabidopsis thylakoid transporter PHT4;1 influences phosphate availability for ATP synthesis and plant growth. Plant J 84:99–110
Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM (1997) Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9:627–640
Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P (1999) Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284:654–657
Karpiński S, Szechyńska-Hebda M, Wituszyńska W, Burdiak P (2013) Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ 36:736–744
Kim C, Apel K (2013a) Singlet oxygen-mediated signaling in plants: moving from flu to wild type reveals an increasing complexity. Photosynth Res 116:455–464
Kim C, Apel K (2013b) O2-mediated and EXECUTER-dependent retrograde plastid-to-nucleus signaling in norflurazon-treated seedlings of Arabidopsis thaliana. Mol Plant 6:1580–1591
Kindgren P, Kremnev D, Blanco NE, de Dios Barajas López J, Fernández AP, Tellgren-Roth C, Kleine T, Small I, Strand A (2012) The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J 70:279–291
Kirk JTO (1971) Chloroplast structure and biogenesis. Annu Rev Biochem 40:161–196
Kirk JTO, Tilney-Bassett RAE (1967) The plastids. Freeman, London
Klein M, Papenbrock J (2004) The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J Exp Bot 55:1809–1820
Kleine T, Leister D (2016) Retrograde signaling: organelles go networking. Biochim Biophys Acta 1857:1313–1325
Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316:715–719
Krause K, Krupinska K (2009) Nuclear regulators with a second home in organelles. Trends Plant Sci 14:194–199
Krause K, Kilbienski I, Mulisch M, Rödiger A, Schäfer A, Krupinska K (2005) DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett 579:3707–3712
Krause K, Oetke S, Krupinska K (2012) Dual targeting and retrograde translocation: regulators of plant nuclear gene expression can be sequestered by plastids. Int J Mol Sci 13:11085–11101
Kropat J, Oster U, Rüdiger W, Beck CF (1997) Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc Natl Acad Sci U S A 94:14168–14172
Kropat J, Oster U, Rüdiger W, Beck CF (2000) Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J 24:523–531
Krupinska K, Dähnhardt D, Fischer-Kilbienski I, Kucharewicz W, Scharrenberg C, Trösch M, Buck F (2013) Identification of WHIRLY1 as a factor binding to the promoter of the stress and senescence-associated gene HvS40. J Plant Growth Regul 33:91
Kusnetsov V, Bolle C, Lübberstedt T, Sopory S, Herrmann RG, Oelmüller R (1996) Evidence that the plastid signal and light operate via the same cis-acting elements in the promoters of nuclear genes for plastid proteins. Mol Gen Genet 252:631–639
Kusnetsov V, Landsberger M, Meurer J, Oelmüller R (1999) The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the plastids. J Biol Chem 274:36009–36014
Laloi C, Przybyla D, Apel K (2006) A genetic approach towards elucidating the biological activity of different reactive oxygen species in Arabidopsis thaliana. J Exp Bot 57:1719–1724
Langner U, Baudisch B, Klösgen RB (2014) Organelle import of proteins with dual targeting properties into mitochondria and chloroplasts takes place by the general import pathways. Plant Signal Behav 9:e29301
Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299:902–906
Lee KP, Kim C, Landgraf F, Apel K (2007) EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci U S A 104:10270–10275
Leister D (2005) Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene 354:110–116
Leister D (2016) Towards understanding the evolution and functional diversification of DNA-containing plant organelles. F1000 Res 5:330
Lemos M, Xiao Y, Bjornson M, Wang JZ, Hicks D, Ad S, Wang CQ, Yang P, Ma S, Dinesh-Kumar S, Dehesh K (2016) The plastidial retrograde signal methyl erythritol cyclopyrophosphate is a regulator of salicylic acid and jasmonic acid crosstalk. J Exp Bot 67:1557–1566
Lepage É, Zampini É, Brisson N (2013) Plastid genome instability leads to reactive oxygen species production and plastid-to-nucleus retrograde signaling in Arabidopsis. Plant Physiol 163:867–881
Lepistö A, Rintamäki E (2012) Coordination of plastid and light signaling pathways upon development of Arabidopsis leaves under various photoperiods. Mol Plant 5:799–816
Lepistö A, Toivola J, Nikkanen L, Rintamäki E (2012) Retrograde signaling from functionally heterogeneous plastids. Front Plant Sci 3:286
Li B, Kronzucker HJ, Shi W (2013) Molecular components of stress-responsive plastid retrograde signaling networks and their involvement in ammonium stress. Plant Signal Behav 8:e23107
Linka N, Theodoulou FL (2013) Metabolite transporters of the plant peroxisomal membrane: known and unknown. Subcell Biochem 69:169–194
Linka N, Weber AP (2010) Intracellular metabolite transporters in plants. Mol Plant 3:21–53
Lübberstedt T, Oelmüller R, Wanner G, Herrmann RG (1994) The role of plastids in the expression of nuclear genes for thylakoid proteins studied with chimeric β-glucuronidase gene fusions. Plant Physiol 105:1355–1364
Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, Lecharny A, Le Ret M, Martin-Magniette ML, Mireau H, Peeters N, Renou JP, Szurek B, Taconnat L, Small I (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16:2089–2103
Lv F, Zhou J, Zeng L, Xing D (2015) β-Cyclocitral upregulates salicylic acid signalling to enhance excess light acclimation in Arabidopsis. J Exp Bot 66:4719–4732
Macaulay KM, Heath GA, Ciulli A, Murphy AM, Abell C, Carr JP, Smith AG (2017) The biochemical properties of the two Arabidopsis thaliana isochorismate synthases. Biochem J 474:1579–1590
Mach J (2018) The lipase Link: abscisic acid induces PLASTID LIPASES, which produce Jasmonic acid precursors. Plant Cell 30(5):948–949
Martin G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7:11431
Martínez C, Pons E, Prats G, León J (2004) Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J 37:209–217
Maruta T, Noshi M, Tanouchi A, Tamoi M, Yabuta Y, Yoshimura K, Ishikawa T, Shigeoka S (2012) H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J Biol Chem 287:11717–11729
Maruta T, Sawa Y, Shigeoka S, Ishikawa T (2016) Diversity and evolution of ascorbate peroxidase functions in chloroplasts: more than just a classical antioxidant enzyme? Plant Cell Physiol 57:1377–1386
Maxwell DP, Laudenbach DE, Huner N (1995) Redox regulation of light-harvesting complex II and cab mRNA abundance in Dunaliella salina. Plant Physiol 109:787–795
Mayfield SP, Taylor WC (1984) Carotenoid-deficient maize seedlings fail to accumulate light-harvesting chlorophyll a/b binding protein (LHCP) mRNA. Eur J Biochem 144:79–84
Mayfield SP, Taylor WV (1987) Chloroplast photooxidation inhibits the expression of a set of nuclear genes. Mol Gen Genet 208:309–314
Mazzoleni M, Figuet S, Martin-Laffon J, Mininno M, Gilgen A, Leroux M, Brugière S, Tardif M, Alban C, Ravanel S (2015) Dual targeting of the protein methyltransferase PrmA contributes to both chloroplastic and mitochondrial ribosomal protein L11 methylation in Arabidopsis. Plant Cell Physiol 56:1697–1710
Mehrshahi P, Johnny C, DellaPenna D (2014) Redefining the metabolic continuity of chloroplasts and ER. Trends Plant Sci 19:501–507
Melonek J, Mulisch M, Schmitz-Linneweber C, Grabowski E, Hensel G, Krupinska K (2010) Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 232:471–481
Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A 98:12826–12831
Miao Y, Jiang J, Ren Y, Zhao Z (2013) The single-stranded DNA-binding protein WHIRLY1 represses WRKY53 expression and delays leaf senescence in a developmental stage-dependent manner in Arabidopsis. Plant Physiol 163:746–756
Millar AH, Whelan J, Small I (2006) Recent surprises in protein targeting to mitochondria and plastids. Curr Opin Plant Biol 9:610–615
Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci U S A 98:2053–2058
Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A (2008) The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci U S A 105:15184–15189
Mohr H, Neininger A, Seith B (1992) Control of nitrate reductase and nitrite reductase gene expression by light, nitrate and a plastidic factor. Bot Acta 105:81–89
Morris K, MacKerness SA, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23:677–685
Moulin M, McCormac AC, Terry MJ, Smith AG (2008) Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc Natl Acad Sci U S A 105:15178–15183
Nevarez PA, Qiu Y, Inoue H, Yoo CY, Benfey PN, Schnell DJ, Chen M (2017) Mechanism of dual targeting of the phytochrome signaling component HEMERA/pTAC12 to plastids and the nucleus. Plant Physiol 173:1953–1966
Nomura H, Komori T, Uemura S, Kanda Y, Shimotani K, Nakai K, Furuichi T, Takebayashi K, Sugimoto T, Sano S, Suwastika IN, Fukusaki E, Yoshioka H, Nakahira Y, Shiina T (2012) Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat Commun 3:926
Nott A, Jung HS, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57:739–759
Oelmüller R (1989) Photooxidative destruction of chloroplasts and its effect on nuclear gene expression and extraplastidic enzyme levels. Photochem Photobiol l49:229–239
Oelmüller R, Briggs WR (1990) Intact plastids are required for nitrate- and light-induced accumulation of nitrate reductase activity and mRNA in squash cotyledons. Plant Physiol 92:434–439
Oelmüller R, Mohr H (1986) Photooxidative destruction of chloroplasts and its consequences for expression of nuclear genes. Planta 167:106–113
Oelmüller R, Schuster C (1987) Inhibition and promotion by light of the accumulation of translatable mRNA of the light-harvesting chlorophyll a/b-binding protein of photosystem II. Planta 172:60–70
Oelmüller R, Dietrich G, Link G, Mohr H (1986a) Regulatory factors involved in gene expression (subunits of ribulose-1,5-bisphosphate carboxylase) in mustard (Sinapis alba L.) cotyledons. Planta 169:260–266
Oelmüller R, Levitan I, Bergfeld R, Rajasekhar VK, Mohr H (1986b) Expression of nuclear genes as affected by treatments acting on the plastids. Planta 168:482–492
Oelmüller R, Schuster C, Mohr H (1988) Physiological characterization of a plastidic signal required for nitrate-induced appearance of nitrate and nitrite reductases. Planta 174:75–83
Oelmüller R, Bolle C, Tyagi AK, Niekrawietz N, Breit S, Herrmann RG (1993) Characterization of the promoter from the single-copy gene encoding ferredoxin-NADP+-oxidoreductase from spinach. Mol Gen Genet 237:261–272
op den Camp R, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Göbel C, Feussner I, Nater M, Apel K (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15:2320–2332
Ostrovsky D, Kharatian E, Malarova I, Shipanova I, Sibeldina L, Shashkov A, Tantsirev G (1992) Synthesis of a new organic pyrophosphate in large quantities is induced in some bacteria by oxidative stress. Biofactors 3:261–264
Ostrovsky D, Diomina G, Lysak E, Matveeva E, Ogrel O, Trutko S (1998) Effect of oxidative stress on the biosynthesis of 2-C-methyl-d-erythritol-2,4-cyclopyrophosphate and isoprenoids by several bacterial strains. Arch Microbiol 171:69–72
Otori K, Tanabe N, Maruyama T, Sato S, Yanagisawa S, Tamoi M, Shigeoka S (2017) Enhanced photosynthetic capacity increases nitrogen metabolism through the coordinated regulation of carbon and nitrogen assimilation in Arabidopsis thaliana. J Plant Res 130:909–927
Padmanabhan MS, Dinesh-Kumar SP (2010) All hands on deck—the role of chloroplasts, endoplasmic reticulum and the nucleus in driving plant innate immunity. Mol Plant-Microbe Interact 23:1368–1380
Page MT, Kacprzak SM, Mochizuki N, Okamoto H, Smith AG, Terry MJ (2017) Seedlings lacking the PTM protein do not show a genomes uncoupled (gun) mutant phenotype. Plant Physiol 174:21–26
Pesaresi P, Masiero S, Eubel H, Braun HP, Bhushan S, Glaser E, Salamini F, Leister D (2006) Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell 18:970–991
Pesaresi P, Schneider A, Kleine T, Leister D (2007) Interorganellar communication. Curr Opin Plant Biol 10:600–606
Pesaresi P, Hertle A, Pribil M, Kleine T, Wagner R, Strissel H, Ihnatowicz A, Bonardi V, Scharfenberg M, Schneider A, Pfannschmidt P, Leister D (2009) Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21:2402–2423
Pesaresi P, Pribil M, Wunder T, Leister D (2011) Dynamics of reversible protein phosphorylation in thylakoids of flowering plants: the roles of STN7, STN8 and TAP38. Biochim Biophys Acta 1807:887–896
Petracek ME, Dickey LF, Nguyen TT, Gatz C, Sowinski DA, Allen GC, Thompson WF (1998) Ferredoxin-1 mRNA is destabilized by changes in photosynthetic electron transport. Proc Natl Acad Sci U S A 95:9009–9013
Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R (2006) pTAC2, -6 and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18:176–197
Pfalz J, Liebers M, Hirth M, Grübler B, Holtzegel U, Schröter Y, Dietzel L, Pfannschmidt T (2012) Environmental control of plant nuclear gene expression by chloroplast redox signals. Front Plant Sci 3:257
Pfannschmidt T, Nilsson A, Tullberg A, Link G, Allen JF (1999) Direct transcriptional control of the chloroplast genes psbA and psaAB adjusts photosynthesis to light energy distribution in plants. IUBMB Life 48:271–276
Pfannschmidt T, Schütze K, Brost M, Oelmüller R (2001) A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J Biol Chem 276:36125–36130
Pfannschmidt T, Schütze K, Fey V, Sherameti I, Oelmüller R (2003) Chloroplast redox control of nuclear gene expression – a new class of plastid signals in interorganellar communication. Antioxid Redox Signal 5:95–101
Piippo M, Allahverdiyeva Y, Paakkarinen V, Suoranta UM, Battchikova N, Aro EM (2006) Chloroplast-mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiol Genomics 25:142–152
Pogson BJ, Woo NS, Förster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13:602–609
Poulsen C, Verpoorte R (1991) Roles of chorismate mutase, isochorismate synthase and anthranilate synthase in plants. Phytochemistry 30:377–386
Prikryl J, Watkins KP, Friso G, Van Wijk KJ, Barkan A (2008) A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res 36:5152–5165
Przybyla-Toscano J, Roland M, Gaymard F, Couturier J, Rouhier N (2018) Roles and maturation of iron-sulfur proteins in plastids. J Biol Inorg Chem 23(4):545–566
Pursiheimo S, Mulo P, Rintamaki E, Aro EM (2001) Coregulation of light-harvesting complex II phosphorylation and lhcb mRNA accumulation in winter rye. Plant J 26:317–327
Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci U S A 109:5535–5540
Ramel F, Ksas B, Akkari E, Mialoundama AS, Monnet F, Krieger-Liszkay A, Ravanat JL, Mueller MJ, Bouvier F, Havaux M (2013a) Light-induced acclimation of the Arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell 25:1445–1462
Ramel F, Mialoundama AS, Havaux M (2013b) Nonenzymatic carotenoid oxidation and photooxidative stress signalling in plants. J Exp Bot 64:799–805
Rausch C, Bucher M (2002) Molecular mechanisms of phosphate transport in plants. Planta 216:23–37
Reichenbächer D, Börner T, Richter J (1978) Untersuchungen am Fraktion-I-Protein der Gerste mit Hilfe quantitativer Immunelektrophoresen. Biochem Physiol Pflanz 172:53–60
Reiss T, Bergfeld R, Link G, Thien W, Mohr H (1983) Photooxidative destruction of chloroplasts and its consequences for cytosolic enzyme levels and plant development. Planta 159:518–528
Ren Y, Li Y, Jiang Y, Wu B, Miao Y (2017) Phosphorylation of WHIRLY1 by CIPK14 shifts its localization and dual functions in Arabidopsis. Mol Plant 10:749–763
Rochaix JD (2013a) Plant science. Fine-tuning photosynthesis. Science 342:50–51
Rochaix JD (2013b) Redox regulation of thylakoid protein kinases and photosynthetic gene expression. Antioxid Redox Signal 18:2184–2201
Rokov-Plavec J, Dulic M, Duchêne AM, Weygand-Durasevic I (2008) Dual targeting of organellar seryl-tRNA synthetase to maize mitochondria and chloroplasts. Plant Cell Rep 27:1157–1168
Rossel JB, Walter PB, Hendrickson L, Chow WS, Poole A, Mullineaux PM, Poson BJ (2006) A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ 29:269–281
Schmid J, Amrhein N (1995) Molecular organization of the shikimate pathway in higher plants. Phytochemistry 39:737–749
Schuster C, Oelmüller R, Bergfeld R, Mohr H (1988) Recovery of plastids from photooxidative damage: significance of a plastidic factor. Planta 174:289–297
Seguel A, Jelenska J, Herrera-Vásquez A, Marr SK, Joyce MB, Gagesch KR, Shakoor N, Jiang S-C, Fonseca A, Wildermuth MC, Greenberg JT, Holuigue L (2018) PROHIBITIN3 forms complexes with ISOCHORISMATE SYNTHASE1 to regulate stress-induced salicylic acid biosynthesis in Arabidopsis. Plant Physiol 176:2515–2531
Serrano M, Wang B, Aryal B, Garcion C, Abou-Mansour E, Heck S, Geisler M, Mauch F, Nawrath C, Métraux JP (2013) Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol 162:1815–1821
Serrano I, Audran C, Rivas S (2016) Chloroplasts at work during plant innate immunity. J Exp Bot 67:3845–3854
Sherameti I, Nakamura M, Yamamoto YY, Pfannschmidt T, Obokata J, Oelmüller R (2002a) Polyribosome loading of spinach mRNAs for photosystem I subunits is controlled by photosynthetic electron transport. Plant J 32:631–639
Sherameti I, Sopory SK, Trebicka A, Pfannschmidt T, Oelmuller R (2002b) Photosynthetic electron transport determines nitrate reductase gene expression and activity in higher plants. J Biol Chem 277:46594–46600
Shine MB, Yang JW, El-Habbak M, Nagyabhyru P, Fu DQ, Navarre D, Ghabrial S, Kachroo P, Kachroo A (2016) Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol 212:627–636
Strand Å, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421:79–83
Strawn MA, Marr SK, Inoue K, Inada N, Zubieta C, Wildermuth MC (2007) Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J Biol Chem 282:5919–5933
Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang (2011) A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat Commun 2:477
Susek RE, Ausubel FM, Chory J (1993) Signal-transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene-expression from chloroplast development. Cell 74:787–799
Szechyńska-Hebda M, Karpiński S (2013) Light intensity-dependent retrograde signalling in higher plants. J Plant Physiol 170:1501–1516
Tadini L, Pesaresi P, Kleine T, Rossi F, Guljamow A, Sommer F, Mühlhaus T, Schroda M, Masiero S, Pribil M, Rothbart M, Hedtke B, Grimm B, Leister D (2016) GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol 170:1817–1830
Tamoi M, Shigeoka S (2015) Diversity of regulatory mechanisms of photosynthetic carbon metabolism in plants and algae. Biosci Biotechnol Biochem 79:870–876
Taylor WC (1989) Regulatory interactions between nuclear and plastid genomes. Annu Rev Plan Physiol Plant Mol Biol 40:211–233
Terasawa K, Sato N (2009) Plastid localization of the PEND protein is mediated by a noncanonical transit peptide. FEBS J 276:1709–1719
Thomas J, Weinstein JD (1990) Measurement of heme efflux and heme content in isolated developing chloroplasts. Plant Physiol 94:1414–1423
Tikkanen M, Gollan PJ, Suorsa M, Kangasjärvi S, Aro EM (2012) STN7 operates in retrograde signaling through controlling redox balance in the electron transfer chain. Front Plant Sci 3:277
Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7:1105–1120
Tripathy BC, Sherameti I, Oelmüller R (2010) Siroheme: an essential component for life on earth. Plant Signal Behav 5:14–20
Van Aken O, Pogson BJ (2017) Convergence of mitochondrial and chloroplastic ANAC017/PAP-dependent retrograde signalling pathways and suppression of programmed cell death. Cell Death Differ 24:955–960
van Dijk EL, Chen CL, D’Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Né P, Loeillet S (2011) XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475:114–117
Van Dingenen J, Blomme J, Gonzalez N, Inze D (2016) Plants grow with a little help from their organelle friends. J Exp Bot 67:6267–6281
Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47:177–206
Vranova E, Coman D, Gruissem W (2013) Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 64:665–700
Wagner D, Przybyla D, op den Camp R, Kim C, Landgraf F, Lee KP, Würsch M, Laloi C, Nater M, Hideg E, Apel K (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306:1183–1185
Walley JW, Xiao Y, Wang JZ, Baidoo EE, Keasling JD, Shen Z, Briggs SP, Dehesh K (2015) Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc Natl Acad Sci U S A 112:6212–6217
Wang L, Kim C, Xu X, Piskurewicz U, Dogra V, Singh S, Mahler H, Apel K (2016) Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc Natl Acad Sci U S A 113:E3792–E3800
Wang K, Guo Q, Froehlich JE, Hersh HL, Zienkiewicz A, Howe GA, Benning C (2018) Two abscisic acid responsive plastid lipase genes involved in jasmonic acid biosynthesis in Arabidopsis thaliana. Plant Cell 29:1678–1696
Wasternack C, Song S (2017) Jasmonates: biosynthesis, metabolism and signaling by proteins activating and repressing transcription. J Exp Bot 68:1303–1321
Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21:1109–1128
Weber AP (2004) Solute transporters as connecting elements between cytosol and plastid stroma. Curr Opin Plant Biol 7:247–253
Weber AP, Fischer K (2007) Making the connections – the crucial role of metabolite transporters at the interface between chloroplast and cytosol. FEBS Lett 581:2215–2222
Weber AP, Linka N (2011) Connecting the plastid: transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu Rev Plant Biol 62:53–77
Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414:562–565
Wilson PB, Estavillo GM, Field KJ, Pornsiriwong W, Carroll AJ, Howell KA, Woo NS, Lake JA, Smith SM, Harvey Millar A, von Caemmerer S, Pogson BJ (2009) The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J 58:299–317
Woodson JD (2016) Chloroplast quality control – balancing energy production and stress. New Phytol 212:36–41
Woodson JD, Perez-Ruiz JM, Chory J (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21:897–903
Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73:1–13
Xiao Y, Savchenko T, Baidoo EE, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K (2012) Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149:1525–1535
Xiao Y, Wang J, Dehesh K (2013) Review of stress specific organelles-to-nucleus metabolic signal molecules in plants. Plant Sci 212:102–107
Xiong JY, Lai CX, Qu Z, Yang XY, Qin XH, Liu GQ (2009) Recruitment of AtWHY1 and AtWHY3 by a distal element upstream of the kinesin gene AtKP1 to mediate transcriptional repression. Plant Mol Biol 71:437–449
Yamasaki K, Motomura Y, Yagi Y, Nomura H, Kikuchi S, Nakai M, Shiina T (2013) Chloroplast envelope localization of EDS5, an essential factor for salicylic acid biosynthesis in Arabidopsis thaliana. Plant Signal Behav 8:e23603
Yoo HH, Kwon C, Lee MM, Chung IK (2007) Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J 49:442–451
Zhang K, Halitschke R, Yin C, Liu CJ, Gan SS (2013) Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci U S A 110:14807–14812
Zhang L, Zhang F, Melotto M, Yao J, He SY (2017) Jasmonate signaling and manipulation by pathogens and insects. J Exp Bot 68:1371–1385
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Pfalz, J., Oelmüller, R. (2019). Plastid Retrograde Signals: More to Discover. In: Sopory, S. (eds) Sensory Biology of Plants. Springer, Singapore. https://doi.org/10.1007/978-981-13-8922-1_18
Download citation
DOI: https://doi.org/10.1007/978-981-13-8922-1_18
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-8921-4
Online ISBN: 978-981-13-8922-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)