Abstract
All hitherto identified aromatic compounds accumulating in leaves of Arabidopsis thaliana (L.) Heynh. upon infection with virulent or avirulent strains of Pseudomonas syringae pathovar tomato (Pst) were indolic metabolites. We now report the strong accumulation of a novel type of natural product, 3′-O-β-d-ribofuranosyl adenosine (3′RA), exclusively during compatible interactions. In contrast to the various indolic metabolites, 3′RA was undetectable in incompatible interactions of A. thaliana leaves with an avirulent Pst strain, as well as in uninfected control leaves. A similar, strong induction of 3′RA was observed in compatible but, again, not in incompatible interactions of Pst with its natural host, Lycopersicon esculentum. The strength of the effect and its confinement to compatible interactions suggests that it may be applicable as a diagnostic tool.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
We have recently initiated investigations on the chemical nature, induction modes, and — as a long-term goal — defense-related functions of aromatic secondary metabolites in Pst-infected A. thaliana leaves (Hagemeier et al. 2001). The results obtained so far indicated that all major induced compounds were indolic substances, with the notable exception of one distinct type of metabolite that was provisionally designated as compound X. This compound represented one of the most prominent peaks on HPLC chromatograms of soluble extracts from compatible Pst/A. thaliana interactions, but was absent in uninfected controls and remained undetectable in incompatible Pst/A. thaliana interactions (Hagemeier et al. 2001).
The strong accumulation of compound X exclusively during compatible Pst/A. thaliana interactions, as well as its apparent, non-indolic nature (as deduced from the UV spectrum), prompted us to further investigate its chemical structure, its mode of accumulation, and its occurrence or non-occurrence in a different pathosystem, i.e., during the interaction of Pst with its natural host, L. esculentum.
Materials and methods
Plant material
Arabidopsis thaliana (L.) Heynh. ecotype Col-0 (Lehle Seeds, Round Rock, USA), and Lycopersicon esculentum Mill. cv. Moneymaker (Bruno Nebelung Kipenkerl-Pflanzenzüchtung, Everswinkel, Germany) plants were grown under 16-h light periods at 400 μmol photons m−2 s−1 for 5 weeks at 22 °C and for 8 weeks at 26 °C, respectively.
Bacterial infections
Pseudomonas syringae pathovar tomato, strain DC3000 or strain DC3000, carrying either the avirulence gene avrRpm1 (for infections of A. thaliana) or the avirulence gene avrPto (for infections of L. esculentum), were grown and infiltrated into A. thaliana or L. esculentum leaves as described by Hagemeier et al. (2001). Leaf samples (approx. 0.2 g) were collected 48 h post inoculation, frozen in liquid N2 and stored at −80 °C.
Extraction procedure and HPLC analysis
After addition of 50% aqueous methanol (v/v; 0.6 ml each), the tissue was homogenized using zirconia beads (1 mm; Roth, Karlsruhe, Germany) in a Mini-Beadbeater-8 (Biospec Products, Bartlesville, USA) and centrifuged for 15 min at 20,000 g. The residues were re-extracted in 0.6 ml methanol and supernatants were combined where appropriate. The solvent was removed at 30 °C using a Speed-Vac (Eppendorf, Hamburg, Germany) and the residue was re-dissolved in 80% aqueous methanol (v/v; 2 μl mg−1 initial FW). Extracts (20 μl) were subjected to HPLC on a Nucleosil C-18 column (EC 250/4, 120-5; Macherey & Nagel, Düren, Germany) using 0.1% trifluoroacetic acid as solvent A and 98% acetonitrile/0.1% trifluoroacetic acid as solvent B at a flow rate of 1 ml min−1 at 24 °C (gradient of solvent A: 100% at 0 min, 94% at 3 min, 80% at 13 min, 76% at 20 min, 20% at 33 min) and a Photodiode Array Detector 540 at 254 nm as part of the Bio-Tek System (Solvent Delivery System 522, Autosampler 565, Jet-Stream plus, Degasy DG 1210, software CHROMA 2000; Bio-Tek, Neufahrn, Germany). For preparative HPLC, a Nucleosil C-18 SP 250/10 120-5 column and the respective part of the gradient were used under otherwise identical conditions.
Nuclear magnetic resonance (NMR) spectroscopy
1H, 1H-1H COSY (correlation spectroscopy), NOESY (nuclear overhouser effect spectroscopy), HMBC (heteronuclear multiple bond correlation), and HMQC (heteronuclear multiple quantum coherence) spectra were recorded on an Avance DRX 500 NMR spectrometer (Bruker, Karlsruhe, Germany) using an inverse detection microprobe head (2.5 mm). Methanol-d 4 was used as a solvent and trimethylsilane (TMS) as internal standard. NMR data of compound X:
-
1H NMR (500 MHz): δ 8.40 (s, H-8), 8.21 (s, H-2), 6.00 (d, J=4.3 Hz, H-1′), 5.05 (s, H-1″), 4.70 (dd, J=5.1, 4.3 Hz, H-2′), 4.43 (dd, J=5.1, 5.1 Hz, H-3′), 4.37 (dd, J=7.2, 4.5 Hz, H-3″), 4.21 (ddd, J=5.1, 2.8, 2.3 Hz, H-4′), 4.04 (d, J=4.5 Hz, H-2″), 3.98 (ddd, J=7.2, 3.1, 2.3 Hz, H-4″), 3.92 (dd, J=12.5, 2.3 Hz, H-5′a), 3.82 (dd, J=12.5, 2.3 Hz, H-5″a), 3.77 (dd, J=12.5, 2.8 Hz, H-5′b), 3.67 (dd, J=12.5, 3.1 Hz, H-5″b).
-
13C NMR (125 MHz): δ 153.3 (C-2), 150.0 (C-4), 142.0 (C-8), 109.4 (C-1″), 91.2 (C-1′), 85.0 (C-4″), 84.9 (C-4′), 78.9 (C-3′), 76.8 (C-2″), 75.6 (C-2′), 70.8 (C-3″), 62.4 (C-5′), 61.8 (C-5″).
Mass spectrometry
Samples were analyzed by ESI–MS using a Hewlett-Packard (Avondale, PA, USA) HP 1100 HPLC coupled to a Micromass Quattro II (Waters, Micromass, Manchester, UK) tandem quadrupole mass spectrometer (geometry quadrupole–hexapole–quadrupole) equipped with an electrospray (ESI) source. The capillary and cone voltages in ESI mode were 3.3 kV and 18 V, respectively. Nitrogen was used for nebulization (15 l h−1) and as drying gas (250 l h−1 250 °C). Source and capillary were heated at 80 °C and 250 °C, respectively. The mass spectrometer was operated in conventional scanning mode using the first quadrupole. Positive-ion full-scan mass spectra were recorded over the range from m/z 50–450 in a scan time of 1.5 s. Fixed precursor ion (MS/MS) spectra (a daughter ion scan) were recorded by setting the first quadrupole to transmit the parent ion of interest and scanning the product ions obtained after collision of parent ions in the hexapole gas cell using the second quadrupole analyzer. Argon was used for collision-induced dissociations (CID) at 1.5×10−3 mbar and the collision energy was varied from 16 to 25 eV for fragmentation. Synthetic standard was dissolved in methanol and introduced into the spectrometer using a Rheodyne valve. Ca. 5–10 μl of each sample was injected into the mobile phase [50:50 (v/v) acetonitrile:water, flow of 0.05 ml min−1]. Separation of compounds was achieved on a reverse phase column (5 μm C18 phase, 100 mm long, 4.1 mm i.d.; Supelco, Bellefonte, USA) equipped with a precolumn (Supelco). Solvent system and gradient program were used as indicated above. The flow was maintained at 1.0 ml min−1 at a column temperature of 30 °C. The UV detector was set at 254 nm.
Results
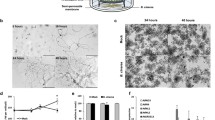
The data shown in Fig. 1 confirm and extend our previous, preliminary results on the induction of compound X (Hagemeier et al. 2001). More precisely, they (i) demonstrate again the highly predominant or even exclusive accumulation of X in the compatible Pst/A. thaliana interaction (Fig. 1a–c), (ii) indicate that the induced increase commences around 12 h post inoculation and reaches a plateau around 48 h (Fig. 1d), and (iii) reveal its chemical identity as 3′-O-β-d-ribofuranosyl adenosine (Fig. 1e).
HPLC analysis, induction kinetics and chemical structure of compound X. a Mock-infected Arabidopsis thaliana leaf. b Compatible Pst/A. thaliana interaction. c Incompatible Pst/A. thaliana interaction. d Time course of X accumulation during the compatible Pst/A. thaliana interaction. Bars indicate deviations from mean values obtained from three independent determinations. e Chemical structure of X, as derived from MS and NMR analysis of the HPLC-purified compound
The chemical identity was established by MS and NMR analysis. Mass spectra were obtained using the electrospray ionization source (ESI). A quasi-molecular ion at m/z 400, accompanied by the m/z 422 sodium ion adduct, indicated that compound X contains an odd number of nitrogen atoms. A series of product ion spectra of X gave the following results. Fragmentation of m/z 400 yielded, even at higher collision energy, a rather simple mass spectrum that was dominated by m/z 268 and 136 ions, both of which presumably contained an odd number of nitrogen atoms. The repeated difference by 132 Da was in accordance with two sequential losses of furanoside moieties (C5H8O4; see below). Fragmentation of the aglycone ion, m/z 136, at a collision energy of 25 eV yielded the following product ion spectrum [m/z (%)]: 136 (26), 119 (100), 109 (5), 94 (12), 92 (32), 67 (12). These data indicate a dominant loss of 17 Da (presumably NH3) and two losses of 27 Da (presumably HCN).
The 1H NMR spectrum exhibited singlets of two isolated protons at δ 8.40 and 8.21 and signals of two pentose units. Striking similarities with the 1H NMR spectrum of commercially available adenosine and with literature data on 2′- and 5′-β-ribosyl adenosine (Markiewicz et al. 1998; Mikhailov et al. 1998) suggested that compound X was a ribosyl adenosine. The 1H-1H COSY and 2D heteronuclear correlation spectra (HMBC and HMQC) enabled the assignment of all 1H signals, of the carbon atoms of the two ribosyl units, and of most of the carbon atoms of the adenine heterocyclus. HMBC connectivities of H-1′ (δ 6.00) with C-4 (δ 150.0) and C-8 (δ 142.0) and a strong NOE between H-1′ and H-8 (δ 8.40) verified the attachment of the carbohydrate unit to N-9. The nature of the interglycosidic linkage was deduced from an HMBC correlation between H-1″ (δ 5.05) and C-3 (δ 78.9). Owing to the glycosylation shift (Agrawal 1992), this carbon resonance appeared approximately 8–9 ppm downfield from that observed for adenosine, for 2′- and 5′-β-ribosyl adenosines, and for C-3″ of the second ribosyl unit of compound X. These data reveal X to be 3′-O-β-d-ribofuranosyl adenosine.
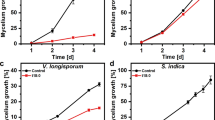
Both findings, the selective accumulation of X in compatible interactions and its chemical identity as an adenosine derivative, were novel and unexpected. To our knowledge, there has been no previous report on the natural occurrence of 3′RA, nor on a compound whose induction is so strictly confined to compatible plant/pathogen interactions. This two-fold novelty prompted us to extend the analysis to a different pathosystem and test whether 3′RA is also induced in interactions of Pst with its host plant, L. esculentum. The results (Fig. 2a–c) demonstrate that, indeed, 3′RA accumulates strongly in the compatible Pst/L. esculentum interaction but, again, not in the incompatible interaction, and not in the mock-infected control. At 48 h post inoculation, when the accumulation during the compatible interaction (Fig. 2b) had reached about half the level (approx. 0.4 nmol mg−1) that had been observed at the same time point for the compatible Pst/A. thaliana interaction (approx. 0.8 nmol mg−1; Fig. 1b), little or no 3′RA was detectable during the corresponding incompatible interaction in this case either.
Discussion
One of the most intriguing, unresolved questions in plant pathology concerns the role of those numerous soluble and wall-bound secondary metabolites that accumulate rapidly, often to high levels, around pathogen infection sites. While the induction of wall-bound compounds appears to be largely confined to a narrow range of similarly substituted benzoic and cinnamic acids and aldehydes that participate in the formation of a physical barrier (Hahlbrock et al. 2003), the numerous concomitantly accumulating, soluble metabolites usually represent a rich, species-specific bouquet of compounds from a great diversity of biosynthetic origins and with largely unknown functions (Mansfield 2000; Dixon 2001). Some of them possess antibiotic activity, such as the indolic phytoalexin camalexin (Rogers et al. 1996) and various glucosinolate breakdown products in A. thaliana (Tierens et al. 2001), or several terpenoid (Tjamos and Smith 1974) and polyacetylenic (de Wit and Kodde 1981) phytoalexins in L. esculentum. However, mechanistically defined or functionally decisive roles of these compounds in pathogen defense have hardly been demonstrated, save for a few, exceptional cases in both A. thaliana (Thomma et al. 1999) and L. esculentum (Suleman et al. 1996).
In nearly all pathosystems investigated so far, there was no qualitative, but usually a considerable quantitative, difference in the induction modes of these compounds between compatible and incompatible plant/pathogen interactions. For example, camalexin was induced in both types of Pst/A. thaliana interaction, but accumulated more rapidly and to much higher levels in incompatible than in compatible interactions; and none of the various co-induced indolic compounds accumulated exclusively in one of the two types of interaction, although the induction strengths and kinetics differed in all cases (Hagemeier et al. 2001). Similar observations had been made with the phytoalexins of L. esculentum (de Wit and Flach 1979), and numerous other such examples have been reported. At least during the initial stages of infection, the induction usually proceeds more rapidly and more strongly in incompatible than in compatible interactions. Hence, the mode of 3′RA induction, as observed in this study, is exceptional in both respects: 3′RA accumulated either exclusively or at least with a very high preference in only one of the two types of interaction, and in contrast to expectation, this was not the incompatible but the compatible interaction.
Equally surprising was the induction of an adenosine derivative among otherwise exclusively indolic metabolites in A. thaliana. It remains open at this stage of analysis whether 3′RA is a bacterial or a plant product, or a combination of both, particularly because it has not been observed previously as a naturally occurring compound in any type of organism. In any case, its highly predominant or even exclusive accumulation during compatible interactions of Pst with two widely different plant species makes it an interesting target for further studies on both its possible role in the infection process and its potential as a diagnostic tool for the molecular definition of particular Pst/plant interaction types, especially in phenotypically doubtful cases. One obvious possibility would be that the accumulation of 3′RA in the compatible interaction is associated with a virulence mechanism of the pathogen, or conversely, that its absence in the incompatible interaction either indicates the lack of virulence of the pathogen or results from a resistance response of the plant. If true, such a functional connection would remain valid even if 3′RA were an artificial product generated during the extraction procedure, and the original, accumulating metabolite were not 3′RA itself but a structurally related compound — a possibility that cannot be excluded.
Irrespective of the precise function and metabolic origin of 3′RA, its solitary, rapid and strong induction among otherwise metabolically widely distinct compounds, at least in A. thaliana, is another exceptional phenomenon. Whether the indolics in A. thaliana (Hagemeier et al. 2001), the terpenoids (Tjamos and Smith 1974) or polyacetylenes (de Wit and Kodde 1981) in L. esculentum, or yet other classes of compounds in other plant species or families, they are always induced as a rich bouquet of structural variants, apparently in sharp contrast to 3′RA. Furthermore, all of them fall into the large group of secondary metabolites, whereas the classification of 3′RA is doubtful in this regard. Various ribofuranosyl nucleosides, but not 3′RA, have been described as minor tRNA components (Keith et al. 1990; Glasser et al. 1991; Luyten et al. 2000) and as the prosthetic group of citrate lyase (Oppenheimer et al. 1979), and numerous bacterial nucleoside antibiotics have been isolated and structurally identified (Isono 1991). However, we are not aware of the occurrence of an adenosine derivative with structural similarity to 3′RA in plants or in plant pathogens. Moreover, the obvious lack of co-induced, structurally related compounds in Pst-infected A. thaliana leaves seems to indicate a different kind of metabolic connection than that, for example, of the various indole derivatives, thus further substantiating the special role of 3′RA. Again, these considerations would hold true regardless of whether 3′RA is of plant or of bacterial origin.
Abbreviations
- Pst:
-
Pseudomonas syringae pathovar tomato
- 3′RA:
-
3′-O-β-d-ribofuranosyl adenosine
References
Agrawal PK (1992) NMR spectroscopy in the structural elucidation of oligosaccharides and glycosides. Phytochemistry 31:3307–3330
De Wit PJGM, Flach W (1979) Differential accumulation of phytoalexins in tomato leaves but not in fruits after inoculation with virulent and avirulent races of Cladosporium fulvum. Physiol Plant Pathol 15:257–267
De Wit PJGM, Kodde E (1981) Induction of polyacetylenic phytoalexins in Lycopersicon esculentum after inoculation with Cladosporium fulvum (syn Fulvia fulva). Physiol Plant Pathol 18:143–148
Dixon RA (2001) Natural products and disease resistance. Nature 411:843–847
Glasser AL, Degres J, Heitzler J, Gehrke CW, Keith G (1991) O-Ribosyl-phosphate purine as a constant modified nucleotide located at position 64 in cytoplasmic initiator tRNAsMet of yeasts. Nucleic Acids Res 19:5199–5203
Hagemeier J, Schneider B, Oldham NJ, Hahlbrock K (2001) Accumulation of soluble and wall-bound indolic metabolites in Arabidopsis thaliana leaves infected with virulent or avirulent Pseudomonas syringae pathovar tomato strains. Proc Natl Acad Sci USA 98:753–758
Hahlbrock K, Bednarek P, Ciolkowski I, Hamberger B, Heise A, Liedgens H, Logemann L, Nürnberger T, Schmelzer E, Somssich IE, Tan JW (2003) Non-self recognition, transcriptional reprogramming, and secondary metabolite accumulation during plant/pathogen interactions. Proc Natl Acad Sci USA. DOI 10.1073/pnas.0831246100
Isono K (1991) Current progress in nucleoside antibiotics. Pharmacol Ther 52:269–286
Keith G, Glasser AL, Degres J, Kuo KC, Gehrke CW (1990) Identification and structural characterization of O-β-ribosyl-(1″→2′)-adenosine-5″-phosphate in yeast methionine initiator tRNA. Nucleic Acids Res 18:5989–5993
Luyten I, Esnouf RM, Mikhailov SN, Efimtseva EV, Michiels P, Heus HA, Hilbers CW, Herdewijn P (2000) Solution structure of a RNA decamer duplex containing 9-[2-O-(β-d-ribofuranosyl)-β-d-ribofuranosyl]adenine, a special residue in lower eucariotic initiator tRNAs. Helv Chim Acta 83:1278–1289
Mansfield JW (2000) Antimicrobial compounds and resistance. The role of phytoalexins and phytoanticipins. In: Slusarenko A, Fraser RSS, van Loon LC (eds) Mechanisms of resistance to plant diseases. Kluwer, Dordrecht, pp 325–370
Markiewicz WT, Niewcyk A, Gdaniec Z, Adamiak DA, Dauter Z, Rypniewski W, Chmielewski M (1998) Studies on synthesis and structure of O-β-d-ribofuranosyl(1″→2′)ribonucleosides and oligonucleotides. Nucleosides Nucleotides 17:411–424
Mikhailov SN, Rodionov AA, Efimtseva EV, Ermolinsky MV, Fomitcheva NS, Padyukova NS, Rothenbacher K, Lescrinier E, Herdiwijn P (1998) Formation of trisaccharide nucleosides during disaccharide nucleoside synthesis. Eur J Org Chem 10:2193–2199
Oppenheimer NJ, Singh M, Sweeley CC, Sung SJ, Srere PA (1979) The configuration and location of the ribosidic linkage in the prosthetic group of citrate lyase (Klebsiella aerogenes). J Biol Chem 254:1000–1002
Rogers EE, Glazebrook J, Ausubel FM (1996) Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis–pathogen interactions. Mol Plant Microbe Interact 9:748–757
Suleman P, Tohamy AM, Saleh AA, Madkour MA, Straney DC (1996) Variation in sensitivity to tomatine and rishitin among isolates of Fusarium oxysporum f. sp. Lycopersici, and strain not pathogenic on tomato. Physiol Plant Pathol 48:131–144
Thomma BPHJ, Nelissen I, Eggermont K, Broekaert WF (1999) Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J 19:163–171
Tierens KFMJ, Thomma BPH, Brouwer M, Schmidt J, Kistner K, Porzel A, Mauch-Mani B, Cammue BPA, Broekaert WF (2001) Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol 125:1688–1699
Tjamos EC, Smith IM (1974) The role of phytoalexins in the resistance of tomato to Verticillium wilt. Physiol Plant Pathol 4:249–259
Acknowledgements
We thank Drs. Jeff Dangl (Capel Hill, NC, USA) and Gregory B. Martin (Cornell University, Ithaca, NY, USA) for kind gifts of P. syringae DC3000 avrRpm1 and avrPto, respectively, and Dr. Ales Svatos (MPI for Chemical Ecology, Jena, Germany) for valuable help with data interpretation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bednarek, P., Winter, J., Hamberger, B. et al. Induction of 3′-O-β-d-ribofuranosyl adenosine during compatible, but not during incompatible, interactions of Arabidopsis thaliana or Lycopersicon esculentum with Pseudomonas syringae pathovar tomato. Planta 218, 668–672 (2004). https://doi.org/10.1007/s00425-003-1146-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-003-1146-9