Abstract
The anatomical uniqueness of the frontal sinus morphology has been widely used for comparative forensic identification using various techniques, mostly including 2D X-rays or one fixed slice of an axial computed tomography (CT) scan image. However, computer-aided 3D automatic graphical comparison techniques can provide accurate comparisons between two 3D models that allow users to comply with even the strictest deviation standards, avoiding error-prone identification of frontal sinuses with similar morphologies. The study proposes the use of a computer-aided comparative paradigm based on the 3D-3D frontal sinus model superimposition process and further assesses the anatomical uniqueness of frontal sinuses using a large Chinese Han sample. Three hundred thirty-six patients older than 20 years with two multi-slice CT scans were collected. Frontal sinus 3D models were semi-automatically segmented through Dolphin Imaging software. Automatic pairwise comparisons of 336 matched pairs from the same person and 340 mismatched pairs from different individuals with an analysis of average root mean square (RMS) point-to-point distance were performed using Geomagic Studio Qualify software. RMS ranged between 0.005 and 1.032 (mean RMS 0.390 ± 0.25 mm) in the group of matches and between 1.107 and 19.363 (mean RMS 4.49 ± 2.69 mm) in the group of mismatches. On average, the RMS value was over ten-fold greater in mismatches than in matches. Statistically significant differences in RMS between the group of matches and mismatches were assessed using the Mann–Whitney U test (p < 0.05). This study supports the value of the frontal sinus with a 3D computer-aided superimposition method for human identification with large samples when DNA, fingerprints, and dental materials are not accessible.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Comparative frontal sinus identification using antemortem (AM) and postmortem (PM) radiological imaging data is one of the prominent research areas in forensic science [1,2,3,4,5,6]. The frontal sinus is a reliable structure given its well-proven anatomical uniqueness among populations. Moreover, there is a considerable amount of literature on side-by-side comparisons using regional morphologic and metric measurement features depending on one or several 2D frontal sinus images derived from X-rays or computed tomography (CT) scans [7,8,9,10,11,12,13]. However, minor variations in head position and different imaging modalities can alter the resulting images, adding difficulty to the process of comparison and reducing the reliability of the identification results, especially using few fixed sectional images.
Multi-slice CT is widely used in clinical practice given its reduced invasiveness and high spatial and temporal resolutions, which have made three-dimensional (3D) frontal sinus visualization possible [14]. Reconstructed visualizations provide information about frontal sinus pneumatization patterns and an accurate assessment of the spatial relationship between the frontal sinus and the ethmoid sinus. Furthermore, CT images are increasingly used in forensic science, particularly as a routine preprogram before forensic autopsy in some countries [2]. The advantage of excellent visualization of the intact frontal sinuses compared with the partial information presented by 2D X-rays and multiple 2D axial CT images makes 3D frontal sinus visualization a good candidate for future forensic identification. In addition, every angle of the whole 3D frontal sinus model can be easily observed with random rotation, and the compared features were no longer subtle or regional anatomical structures but were 3D features. Therefore, the influence on available 3D features of minor variations in head position and different imaging modalities is less marked than the influence of the 2D regional features.
In fact, a growing body of literature has evaluated the value of 3D frontal sinus models as antemortem (AM) and postmortem (PM) comparative materials for human identification. A series of studies have analyzed the use of 3D frontal sinus cavities/volume for sex estimation. For example, in 2018, Choi et al. suggested that the frontal sinus cavity outline exhibits sexual dimorphism in 3D cone beam CT images and can be used for sex determination by developing a multiple logistic regression based on several significant variables, such as basal/ellipse fit, basal/width, and volume, with an accuracy of 80% of 130 CT scans [15]. In 2019, Čechová et al. applied the form and shape of the external surface of the frontal bone and the 3D frontal sinuses (surface area and volume) for sex estimation by developing a support vector machine model, suggesting an accuracy of 84.46% for 103 cranial CT images [16]. In 2019, Wanzeler et al. combined adult paranasal sinus volumes (total volume of the maxillary sinus, the frontal sinus, and the sphenoid sinus) with foramen magnum measurements to determine the sex, achieving a high accuracy of 100% for 200 CT scans [17].
In addition, some studies have been published on further morphologic and metric features of the human frontal sinuses using the intact 3D digital model. Kim et al. in 2013, Gibelli et al. in 2019, and Zhao et al. in 2020 presented a series of morphologic and metric classifications for the frontal sinus [18,19,20]. Furthermore, some studies have discovered the usefulness of entire frontal sinus 3D model superimposition to establish identification, suggesting high identification accuracy with the root mean square (RMS) of the mean point-to-point distance between the two models. For example, in 2015, Beaini et al. first applied the 3D-3D superimposition method to the comparison of 3D frontal sinus models using MeshLab software (www.meshlab.sourceforge.net, Visual Computing Lab-ISTI CNR) based on 20 paired samples and CloudCompare software (www.danielgm.net/cc) for volumetric comparison with an absolute distance scale indicating high coincidence when concentrated next to zero [21]. In 2019, Gibelli et al. compared the geometric distance information of the 3D model without focusing on specific regional metrics to achieve a positive match [22]. Compared with Beaini et al.’s study, Gibelli et al. analyzed not only the group of matches but also the group of mismatches, identifying statistically significant differences in the average RMS point-to-point distance between the group of matches (ranging from 0.07 to 0.96 mm, mean RMS 0.35 ± 0.23 mm) and the group of mismatches (ranging from 0.96 to 10.29 mm, mean RMS 2.59 ± 1.79 mm).
However, the samples of these 3D superimposition studies are small, ranging from 20 to 40 matched subjects. To the best of our knowledge, large samples have not yet been investigated for the frontal sinus. This study aimed to explore an innovative procedure for the assessment of frontal sinus individuality based on a 3D-3D superimposition procedure with a database of 672 head CT scans collected from 336 individuals, 336 matched pairs, and 340 mismatched pairs, applying Geomagic Quality, a standard technique tool, for the comparison of frontal sinus 3D models. The results may provide a new point of view for the assessment of individual variants of the frontal sinus.
Materials and methods
Acquisition of the 3D model of the frontal sinus
The present study was performed with the approval of the ethics committee of the West China Hospital of Sichuan University (KS2020001). Informed consent was obtained from all patients. A retrospective collection of 336 people who underwent head CT scans twice at the West China Hospital of Sichuan University between March 2014 and January 2020 was conducted. All participants were from the Chinese Han population and did not show any deformity or disease in the cranial region.
Table 1 depicts the demographic information of the participants. The subjects were between 20 and 63 years old with a mean age of 38.34 years for men and 43.57 years for women. The time between the two CT examinations was between 1 and 53 months in the entire sample.
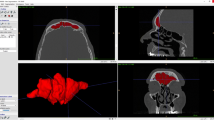
All CT scans were performed using a second-generation dual-source scanner (Somatom Definition Flash; Siemens, Erlangen, Germany) with the following acquisition parameters: kV, 120; mAs, 110; collimation, 1 mm; tube rotation, 0.3 s; and reconstruction thickness, 1 mm. All CT scans were for their normal clinical diagnosis. Imaging data were processed using Dolphin Imaging (version 11.5; Chatsworth, CA, USA) to extract the 3D model of the frontal sinus. A semi-automatic approach was applied to segment the 3D frontal sinus models. One or more seeds were manually placed in the voids on the image that are part of the patient’s frontal sinus. Starting at these seed points, Dolphin selects adjacent empty areas, manfully adding more seed points to permeate the entire volume, and then calculating volume and rendering the frontal sinus (Fig. 1).
Alignment and 3D model visual comparison procedure
The performed comparisons were as follows: 336 pairwise comparisons of the 3D models from the two CT scans from the same individual (in STL format) and 340 pairwise comparisons of the 3D models from two different individuals selected from the mismatched pair data with a relatively similar volume among all samples by random combination. These comparisons were performed using Geomagic Studio 2013 to obtain the point cloud of the 3D model of the frontal sinus.
The point cloud data (wrap) were then imported into the second software Geomagic Qualify 2013 (3D Systems Company, America), which is a standard tool for technical comparisons of 3D models (computer aided design systems). Comparison analysis between models was performed using an automatic algorithm implemented in Geomagic Qualify software. For the comparison, we randomly set one of the paired frontal sinuses as the reference model and the other as the test model. Then, the differences in the datasets of the reference and test groups were compared and analyzed.
For a correct implementation of the comparative process, it was necessary to overlap individual models such that they would occupy the exact same position in the 3D space. This so-called registration process was performed for all models in two stages using Geomagic Quality software. Registration was performed using the “best-fit alignment” option in the Geomagic Control X software. The precision of the registration was set to at least 0.3 mm (tolerance type: “3D Deviation”) with a maximum of 100,000 polygons for surface representation. The corresponding polygons from the selected reference areas were automatically superimposed.
The distances between the corresponding areas of the paired frontal sinuses were compared to obtain color-coded maps. The yellow-to-red fields indicate that the definitive casts were larger than the reference model, and the turquoise-to-dark blue fields indicate that the definitive casts were smaller than the reference model. The 3D deviation analysis has a tolerance range (green) of ± 0.50 mm with a maximum of 2 mm. Parameters of minimum distance, maximum distance, average distance (minimum and maximum), standard deviation, and RMS (root mean square) between the surfaces of the 3D models were automatically reported by Geomagic Quality software. All the values in this range indicate the matching percentage between the two specular 3D models.
Statistical analysis
The average point-to-point distance and RMS values were calculated for all the superimpositions. Kolmogorov–Smirnov analysis was used to verify the normality of the distribution of average point-to-point distances and RMS values (p < 0.05). The Mann–Whitney test was used to assess significant differences in RMS values according to group (matches and mismatches) and sex. In both cases, a significance level of 5% was set. Statistical analyses were performed using SPSS (IBM SPSS Statistics for Windows, Version 22.0, IBM Corp).
Results
When frontal sinus pairs belonging to the same individual were compared, the RMS value was 0.390 on average, ranging between 0.005 and 1.032. The average error value was 0.222, ranging between 0.002 and 0.766. On average, the standard deviation was 0.291, ranging between 0.005 and 0.796. Figure 2 shows the color-coded maps of six cases of registration and superimposition of two 3D models belonging to the same individual. The concordant areas are colored green.
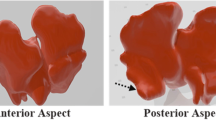
When frontal sinus pairs belonging to different individuals were compared, the RMS value was 4.49 on average, ranging between 1.107 and 19.363. The average error value was 3.260, ranging between 0.900 and 15.877. On average, the standard deviation was 1.322, ranging between 0.8488 and 2.251. Figure 3 shows the color-coded maps of six cases of registration and superimposition of two 3D models belonging to the same individual. The most mismatched areas are colored blue, yellow, and red.
Three examples of registration and superimposition of two 3D models that belong to different individuals (mismatches). a, c, and e are references; b, d, and f are tests. The first row of each example is anterior view images, and the second row is posterior views. a–b, c–d, and e–f are chromatic representations of the RMS point-to-point distance between the two models
For both matched pairs and mismatched pairs, the frequency distribution of point-to-point deviation errors did not fit a normal distribution (Kolmogorov–Smirnov, p > 0.05). Statistically significant differences of RMS between the group of matches and mismatches were assessed through Mann–Whitney U test (p < 0.05).
Discussion
All human bones are three-dimensional, and the analysis of their 3D anatomical data provides a direct increase in comparative accuracy over the traditional 2D method [23]. An increasing number of studies have analyzed the personal identification value of 3D anatomical structures using a quantitative assessment by a point-to-point distance approach derived from 3D model superimposition. Examples of assessed structures are the frontal sinuses [21, 22], the palatal rugae [24], the sphenoid sinuses [25], and the lumbar vertebrae [26]. However, the sample scale of these two studies on the frontal sinus was 20 individuals and 30 individuals, respectively. The aim of this study was to test this method for forensic personal identification in a large-scale sample of 336 individuals, by introducing a 3D-3D computer-aided inspection software (Geomagic Qualify) for developing standard tool individual identification based on technical comparisons of 3D models.
The 3D model superimposition software allows automatic graphical comparisons and automatic alignment between the AM and the PM 3D models by providing valid statistical data, which reduces the required level of expertise for the identification of a person by comparing the frontal sinus morphology compared to a 2D analysis. Deviation levels between two frontal sinuses are displayed graphically using customized colors. Forensics can view the entire range of deviations for a detailed analysis. This study successfully demonstrated the usefulness of another 3D tool, Geomagic Qualify, for human identification.
In addition to the above advantages of 3D superimposition software, the greatest advantage of Geomagic Qualify is that it can generate HTML reports automatically with a single click, providing multiple, user-defined, and annotated views, numeric details, VRML objects, and notes and conclusions. The RMS error, average error, max upper deviation, max lower deviation, average deviation, standard deviation, number of points, deviation distribution histogram and deviation distribution table, standard deviation histogram and deviation distribution table, and six views of customized maps of the 3D frontal sinus model were recorded in this study for each comparison. Inspections can be performed with one click of the mouse and shared with colleagues via automatically generated, web-ready HTML reports. These reports can be very useful as supportive materials to ensure the admissibility of forensic conclusions in court (see the supplementary materials: one report of matched pair report and one report of mismatched pair report). Regarding automatic alignment, Geomagic Qualify provides three different alignment opinions for different types of models: RPS alignment for 3D models with holes, best-fit alignment for 3D models with irregular shapes, and feature-based alignment for regular models. Thus, for frontal sinus samples, we chose the best-fit alignment to obtain the best match between the entire surfaces of both models.
Other recent studies have revealed that the RMS values between mismatch and match groups are significantly different. For example, in Gibelli et al.’s study [22] on 3D-3D frontal sinus superimposition, the mean RMS was 0.35 ± 0.23 mm for the match group and 2.59 ± 1.79 mm for the mismatch group. On average, the RMS value was almost seven times smaller in matches than in mismatches. In Capplella et al.’s [25] study on 3D-3D sphenoid sinus superimposition, the mean RMS was 0.22 ± 0.11 mm in the match group and 2.16 ± 0.57 mm in the mismatch group. On average, the RMS value was almost ten times smaller in matches compared with mismatches. In our case, even though we used different 3D segmented software and 3D-3D superimposition software, our results are consistent with those previous findings: the mean RMS value in matched pairs was 0.390 (range, 0.007–1.032), and the mean RMS value in mismatched pairs was 0.451 (range, 1.107–19.363). On average, the RMS value was approximately ten-fold reduced in matches than in mismatches. Notably, both Gibelli et al. [22] and Capplella et al. [25] studied small samples as their matched groups: 30 matched pairs of frontal sinuses and 40 matched pairs of sphenoid sinuses, respectively. Our study assessed greater than 300 matched pairs, which represents a much larger sample, and our results thus reinforce the anatomical uniqueness of the frontal sinus and provide more evidence supporting the reliability of the 3D-3D superimposition method for forensic personal identification.
In addition to bony cavities (frontal sinus and sphenoid sinus), Decker et al. [26] reported the usefulness of lumbar models for positive identification based on their individuality through 3D-3D superimposition. In their study, when 0.5 mm with at least a 90% match was considered a positive identification, the comparison results exhibited a 100% match rate. These researchers selected a 1-mm range, explaining that most current clinical CT scanners cannot reliably detect objects smaller than 0.6 mm because they have an in-plane spatial resolution of 0.3 mm and a slice thickness of approximately 0.6 mm. Our study and Gibelli et al. [22] reported that the RMS errors range from 0.007 to 1.032 and from 0.07 to 0.96, respectively. Thus, we conclude that most of the distance error produced in the point-to-point comparison of matched pairs is caused by the inherent CT scan and reconstruction technology.
Although 3D modeling is becoming increasingly important for assessing the uniqueness of the frontal sinus, there are some limitations related to the general aspects of 3D model segmentation and the specific application of 3D frontal sinus superimposition for human identification.
First, the reliability of the superimposition result is closely related to the 3D segmentation and reconstruction of the frontal sinus if it can ideally depict the frontal sinus. There are two concerning dimensions for the accurate segmentation of the 3D frontal sinus. One dimension is whether the software itself can accurately segment the frontal sinus. There is no currently available evidence regarding which technique or software is most reliable to efficiently segment the airway [27]. The study of Hakan Ela et al. [28] showed that the reliability of three automatic programs for upper airway segmentation, including Dolphin3D (version 11, Dolphin Imaging & Management Solutions, Chatsworth, Calif), InVivoDental (version 4.0.70, Anatomage, San Jose, CA), and OnDemand3D (version 1.0.1.8407, CyberMed, Seoul, Korea), was high but with poor accuracy, suggesting that systematic errors were needed. They also suggested that the volume calculated by Dolphin is probably lower than it should be because it does not allow control of the threshold for the area identified and occasionally fails to render some parts of the airway. However, their study was based on the upper airway, which is affected by head posture and breathing stage. The frontal sinus cavity is surrounded by bone with constant volume, and the thin-slice CT scan has strong power to determine boundaries between frontal sinus cavities with a relatively low density and the bone structure with the highest density. Therefore, it is not reasonable to directly compare the frontal sinus to the airway. The second dimension is that different data acquisition procedures, such as different scanning protocols or different CT machines, could affect the segmentation accuracy of the same subject. The study of Kulczyk et al. [29] has showed that the 3D model of canine teeth obtained from cone beam CT (CBCT) with high resolution presents a better quality than that obtained from CBCT with standard resolution, and micro-CT exhibits the highest quality. It seems unreasonable to expect consistency among different CT scanning protocols for the same images. Studies reviewing segmentation scanning protocols for the frontal sinus are severely lacking. More works on testing the reliability of 3D frontal sinus segmentation techniques by comparing different softwares and different scanning protocols should be investigated.
Second, in our study, we used the semi-automatic segmentation procedure provided by Dolphin software, which is based on regional expansion. This approach to segmentation examines neighboring pixels of initial seed points and determines whether the pixel neighbors should be added to the regions. Seed point selection is based on the airway CT threshold criterion. The regions are grown from these seed points to adjacent points depending on a region membership criterion. Given that the regions are grown on the basis of the CT threshold, the image information itself is important. Thus, this superimposition method was not available to identify unknown decedents with frontal sinusitis, which would definitely change the model cavity and result in the difference between AM and PM in frontal sinus superimposition. This methodology is also not applicable for people who have AM CT data of less than 18 years presented with the developing frontal sinus [30].
Finally, semi-automatic 3D segmentation of the frontal sinus can be very challenging, especially in the complex anatomy of the frontal sinus. Manually placing the seeds by sliding the CT sectional slices to separate the frontal sinus from its complex surrounding structures and precise reconstruction requires a high level of professional knowledge in imaging anatomy, and the whole procedure is very time-consuming.
In conclusion, this study provides more insight into the 3D assessment of the anatomical uniqueness of the frontal sinus and successfully applies well-developed automatic graphical comparison software for comparative forensic identification.
Data availability
The data are not public.
Code availability
There is no code for software.
References
Murphy M, Drage N, Carabott R, Adams C (2012) Accuracy and reliability of cone beam computed tomography of the jaws for comparative forensic identification: a preliminary study. J Forensic Sci 57(4):964–968. https://doi.org/10.1111/j.1556-4029.2012.02076.x
Dedouit F, Savall F, Mokrane FZ, Rousseau H, Crubézy E, Rougé D, Telmon N (2014) Virtual anthropology and forensic identification using multidetector CT. Br J Radiol 87(1036):20130468. https://doi.org/10.1259/bjr.20130468
Ognard J, Deloire L, Saccardy C, Burdin V, Salem DB (2020) Automated contour detection in spine radiographs and computed tomography reconstructions for forensic comparative identification. Forensic Sci Med Pathol 16(1):99–106. https://doi.org/10.1007/s12024-019-00189-0
Ali Z, Mourtzinos N, Ali BB, Fowler D (2020) A pilot study comparing postmortem and antemortemct for the identification of unknowns: could a forensic pathologist do it? J Forensic Sci 65(2):492–499. https://doi.org/10.1111/1556-4029.14199
Boer H, Blau S, Delabarde T, Hackman L (2019) The role of forensic anthropology in disaster victim identification (DVI): recent developments and future prospects. Forensic Sci Res 4(4):303–315. https://doi.org/10.1080/20961790.2018.1480460
Deloire L, Diallo I, Cadieu R, Auffret M, Alavi Z, Ognard J, Salem D (2019) Post-mortem X-ray computed tomography (PMCT) identification using ante-mortem CT-scan of the sphenoid sinus. J Neuroradiology 46(4):248–255. https://doi.org/10.1016/j.neurad.2018.08.003
Nikolova S, Toneva D, Georgiev I, Dandov A, Lazarov N (2018) Morphometric analysis of the frontal sinus: application of industrial digital radiography and virtual endocast. J Forensic Radiology Imag 12:31–39. https://doi.org/10.1016/j.jofri.2018.02.001
Xavier TA, Dias Terada ASS, Da Silva RHA (2015) Forensic application of the frontal and maxillary sinuses: a literature review. J Forensic Radiology Imag 3(2):105–110. https://doi.org/10.1016/j.jofri.2DD015.05.001
Campobasso CP, Dell’Erba AS, Belviso M, Di VG (2007) Craniofacial identification by comparison of antemortem and postmortem radiographs: two case reports dealing with burnt bodies. Am J Foren Med Path 28(2):182–186. https://doi.org/10.1097/PAF.0b013e31806195cb
Taniguchi M, Sakoda S, Kano T, Zhu BL, Maeda H (2003) Possible use of nasal septum and frontal sinus patterns to radiographic identification of unknown human remains. Osaka City Med J 49(1):31–38
Silva RFD, Prado FB, Caputo IGC, Devito KL, Botelho TDL, Eduardo DJ (2009) The forensic importance of frontal sinus radiographs. J Forensic Legal Med 16(1):18–23. https://doi.org/10.1016/j.jflm.2008.05.016
Tatlisumak E, Ovali GY, Aslan A, Asirdizer M, Zeyfeoglu Y, Tarhan S (2007) Identification of unknown bodies by using CT images of frontal sinus. Forensic Sci Int 166(1):42–48. https://doi.org/10.1016/j.forsciint.2006.03.023
Brun CN, Christensen AM, Kravarski M, Gorincour G, Schweitzer W, Thali MJ, Gascho D, Hatch GM, Ruder TD (2017) Comparative radiologic identification with standardized single CT images of the paranasal sinuses—evaluation of inter-rater reliability. Forensic Sci Int 280:81–86. https://doi.org/10.1016/j.forsciint.2017.08.029
Kushwah A, Bhalse R, Pande S (2015) CT evaluation of diseases of paranasal sinuses & histopathological studies. Int J Med Res Rev 3: 1306–1310. https://doi.org/10.17511/ijmrr.2015.i11.237.
Choi IG, Duailibi-Neto EF, Beaini TL, Da Silva RL, Chilvarquer I (2018) The frontal sinus cavity exhibits sexual dimorphism in 3D cone-beam ct images and can be used for sex determination. J Forensic Sci 63(3):692–698. https://doi.org/10.1111/1556-4029.13601
Čechová M, Dupej J, Brůžek J, Bejdová Š, Horák M, Velemínská J (2019) Sex estimation using external morphology of the frontal bone and frontal sinuses in a contemporary Czech population. Int J Legal Med 133(4):1285–1294. https://doi.org/10.1007/s00414-019-02063-8
Wanzeler AMV, Alves-Júnior SM, Ayres L, Da Costa Prestes MC, Gomes JT, Tuji FM (2019) Sex estimation using paranasal sinus discriminant analysis: a new approach via cone beam computerized tomography volume analysis. Int J Legal Med 133(6):1977–1984. https://doi.org/10.1007/s00414-019-02100-6
Kim DI, Lee UY, Park SO, Kwak DS, Han SH (2013) Identification Using Frontal Sinus By Three- Dimensional Reconstruction From Computed Tomography. J Forensic Sci 58(1):5–12. https://doi.org/10.1111/j.1556-4029.2012.02185
Gibelli D, Cellina M, Gibelli S, Oliva AG, Termine G, Sforza C (2020) Are coding systems of frontal sinuses anatomically reliable? A study of correlation among morphological and metrical features. Int J Legal Med 134(5):1897–1903. https://doi.org/10.1007/s00414-020-02293-1
Zhao H, Li Y, Xue H, Deng ZH, Liang WB, Zhang L (2020) Morphological analysis of three-dimensionally reconstructed frontal sinuses from Chinese Han population using computed tomography. Int J Legal Med. https://doi.org/10.1007/s00414-020-02443-5
Beaini TL, Duailibi-Neto EF, Chilvarquer I, Melani RFH (2015) Human identification through frontal sinus 3D superimposition: pilot study with cone beam computer tomography. J Forensic Legal Med 36:63–69. https://doi.org/10.1016/j.jflm.2015.09.003
Gibelli D, Cellina M, Cappella A, Gibelli S, Panzeri MM, Oliva AG, Termine G, Ang De, elis D, Cattaneo C, Sforza C, (2019) An innovative 3D–3D superimposition for assessing anatomical uniqueness of frontal sinuses through segmentation on CT scans. Int J Legal Med 133(4):1159–1165. https://doi.org/10.1007/s00414-018-1895-4
Brough A, Rutty G, Villa C, Colman K, Dedouit F, Decker S (2019) The benefits of medical imaging and 3D modelling to the field of forensic anthropology positional statement of the members of the forensic anthropology working group of the International Society of Forensic Radiology and Imaging. J Forensic Radiology Ima 18:18–19. https://doi.org/10.1016/j.jofri.2019.07.003
Gibelli D, De Angelis D, Pucciarelli V, Riboli F, Ferrario VF, Dolci C, Sforza C, Cattaneo C (2018) Application of 3D models of palatal rugae to personal identification: hints at identification from 3D–3D superimposition techniques. Int J Legal Med 132:1241–1245. https://doi.org/10.1007/s00414-017-1744-x
Cappella A, Gibelli D, Cellina M, Mazzarelli D, Oliva AG, De Angelis D, Sforza C, Cattaneo C (2019) Three-dimensional analysis of sphenoid sinus uniqueness for assessing personal identification: a novel method based on 3D-3D superimposition. Int J Legal Med 133(6):18 95–1901. https://doi.org/10.1007/s00414-019-02139-5
Decker SJ, Ford JM (2019) Forensic personal identification utilizing part-to-part comparison of CT-derived 3D lumbar models. Forensic Sci Int 280:21–29. https://doi.org/10.1016/j.forsciint.2018.10.018
Alsufyani NA, Flores-Mir C, Major PW (2012) Three-dimensional segmentation of the upper airway using cone beam CT: a systematic review. Dentomaxillofac Radiol 41(4):276. https://doi.org/10.1259/dmfr/79433138
El H, Palomo JM (2010) Measuring the airway in 3 dimensions: a reliability and accuracy study. Am J Orthod Dentofac 137.4-supp-S: S50.e1-S50.e9. https://doi.org/10.1016/j.ajodo.2009.11.010
Kulczyk T, Rychlik M, Lorkiewicz-Muszyńska D, Abreu-Głowacka M, Czajka-Jakubowska A, Przystańska A (2019) Computed tomography versus optical scanning: a comparison of different methods of 3D data acquisition for tooth replication. Bio Med Res Int. https://doi.org/10.1155/2019/4985121
Shah RK, Dhingra JK, Carter BL, Rebeiz EE (2010) Paranasal sinus development: a radiographic study. Laryngoscope 113(2):205–209. https://doi.org/10.1097/00005537-200302000-00002
Funding
This work was supported by Post-doctoral Research and Development Fund of Sichuan University (20826041D4013) and the 68th China Postdoctoral Science Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval
The present study was performed with the approval of the ethics committee of the West China Hospital of Sichuan University. Informed consent was obtained from all the patients.
Consent to participate
The present study was performed with the approval of the ethics committee of the West China Hospital of Sichuan University. Informed consent was obtained from all the patients.
Consent for publication
All the authors are consent for the publication.
Conflicts of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Xu, C., Yu, D. et al. Computer-aided superimposition of the frontal sinus via 3D reconstruction for comparative forensic identification. Int J Legal Med 135, 1993–2001 (2021). https://doi.org/10.1007/s00414-021-02585-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-021-02585-0