Abstract
Most supernumerary (B) chromosomes are parasitic elements carrying out an evolutionary arms race with the standard (A) chromosomes. A variety of weapons for attack and defense have evolved in both contending elements, the most conspicuous being B chromosome drive and A chromosome drive suppression. Here, we show for the first time that most microspermatids formed during spermiogenesis in two grasshopper species contain expulsed B chromosomes. By using DNA probes for B-specific satellite DNAs in Eumigus monticola and Eyprepocnemis plorans, and also 18S rDNA in the latter species, we were able to count the number of B chromosomes in standard spermatids submitted to fluorescence in situ hybridization, as well as visualizing B chromosomes inside most microspermatids. In E. plorans, the presence of B-carrying microspermatids in 1B males was associated with a significant decrease in the proportion of B-carrying standard spermatids. The fact that this decrease was apparent in elongating spermatids but not in round ones demonstrates that meiosis yields 1:1 proportions of 0B and 1B spermatids and hence that B elimination takes place post-meiotically, i.e., during spermiogenesis, implying a 5–25% decrease in B transmission rate. In E. monticola, the B chromosome is mitotically unstable and B number varies between cells within a same individual. A comparison of B frequency between round and elongating spermatids of a same individual revealed a significant 12.3% decrease. We conclude that B chromosome elimination during spermiogenesis is a defense weapon of the host genome to get rid of parasitic chromosomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Regular chromosome elimination from somatic cells has been reported in nematodes, insects, mites, finches, bandicoots, and hagfish, and has been interpreted as a mechanism for gene silencing, dosage compensation, sex determination, or germ line and soma differentiation (for review, see Wang and Davis 2014). Of course, this variety of adaptations is evolutionarily viable provided that the somatically eliminated chromosomes have granted their presence in the germ line. For the same reason, chromosome elimination from germ cells is most likely the result of a genetic conflict where the standard genome tries to get rid of a disturbing harmful element, e.g., a parasitic chromosome.
B chromosomes are considered genomic parasites which prosper in natural populations because they show an advantage in transmission (drive) counteracting their detrimental effects on host genome fitness (for review, see Camacho et al. 2000; Camacho 2005; Burt and Trivers 2006). The presence of B chromosomes evokes an evolutionary response in the host genome leading to suppress drive, and the two contending parts develop a true coevolutionary arms race (Camacho et al. 1997; Frank 2000) which may elicit the emergence of new adaptations in the host genome. A suggestive example of these adaptations are germ-line restricted B chromosomes, such as those in the marsupial Echymipera kalabu (Hayman et al. 1969) and the ant Leptothorax spinosior (Imai 1974), as this minimizes their harm to the somatic cells while still assuring their transmission to future generations.
The formation of aberrant meiotic products during spermatogenesis has been a recurrent subject in the literature on B chromosomes. The post-meiotic part of spermatogenesis is called spermiogenesis, during which the round-shaped spermatids resulting from meiosis generate a tail and undergo drastic nuclear changes to become spermatozoa. In grasshoppers, spermatozoa possess extremely elongated heads showing almost the same width as the tail. During spermatid nuclear elongation, DNA packaging changes to a highly condensed state facilitated by histone replacement with protamines. Electron microscopy studies have shown that grasshopper spermiogenesis can be divided into ten developmental stages (Szöllösi 1975). Under optical microscopy, however, it is only possible to differentiate between the immature round spermatids, the mature spermatozoa (with fibrillar heads), and the intermediate stages with elongating spermatids at several degrees of elongation.
In addition to the temporally differentiated round (immature) and elongating (maturing) spermatids, optical microscopy allows identifying other types of spermatids, on the basis of size. In addition to standard haploid spermatids, polyploid spermatids have frequently been reported in grasshoppers (named macrospermatids). For instance, Cabrero et al. (2013) showed the presence of 2C, 4C, 8C, and 16C macrospermatids in males that had been RNAi knocked-down for the Ku70 gene. In addition, tiny microspermatids can sometimes appear within cysts of standard spermatids. However, the frequency of macro- and microspermatids has shown to be significantly higher in B-carrying males of several species (see below).
Nur (1969) was the first in claiming that the production of macro- and microspermatids could be related with the presence of B chromosomes in the grasshopper Camnula pellucida. This author suggested that lagging B chromosomes could block cytokinesis in both meiotic divisions leading to the formation of restitution nuclei and thus 2C or 4C macrospermatids. Alternatively, lagging B chromosomes could be excluded from the standard meiotic products giving rise to microspermatids. Other authors have later found aberrant spermatid formation in other species carrying B chromosomes (for review, see Teruel et al. 2009). Partial support to Nur’s claiming and slightly different explanations were later given by other authors. For instance, Bidau (1986) reported that unequal cytokinesis in Metalaptea brevicornis gave rise to macrospermatids and a small nuclear bud which sometimes could include the B chromosome. Likewise, Suja et al. (1989) “observed the presence of condensed Bs outside the nuclei in both recently formed secondary spermatocytes and early spermatids” thus supporting the hypothesis that lagging Bs can be eliminated from “standard nuclei.” However, based on the fact that spermatocytes within a same cyst are connected by cytoplasmic bridges as a result of incomplete cytokinesis (Phillips 1970), Suja et al. (1989) suggested that macrospermatids could also derive from B-provoked impairment of spermatid differentiation during early spermiogenesis, which would explain the lack of correspondence they observed between the number of centriolar adjuncts and ploidy level in spermatids of the grasshopper Eyprepocnemis plorans. In addition, Loray et al. (1991) found that the presence of B chromosomes in Dichroplus elongatus was associated with an increase in the frequency of macrospermatids even in testis tubules lacking this mitotically unstable B chromosome, and claimed for physiological effects of B’s affecting meiosis even in cells lacking them. This kind of systemic response could also be explained through some kind of gene expression change due to B presence (included gene expression in the B itself) whose effects would be exported to B-lacking testis tubules. This kind of effect would be compatible with the spermiogenesis impairment suggested by Suja et al. (1989), but not with the odd-even effect frequently reported for the frequency of aberrant spermatids in the case of mitotically unstable B chromosomes, as they are most abundant in testis tubules carrying odd numbers of B chromosomes (see Camacho et al. 2004; Teruel et al. 2009) whereas the physiological effect should erase this difference.
It is, however, unknown whether microspermatids actually contain B chromosomes, as no direct evidence has hitherto been provided in animals. In contrast, using DNA probes specific to A or B chromosomes, Chiavarino et al. (2000) showed that “micronuclei formed during male meiosis in maize can include both A and B chromosomes.” On this basis, and given that B-carrying males showed higher frequency of microspermatids than 0B ones in E. plorans (0.73 and 0.22%, respectively), Teruel et al. (2009) suggested that most microspermatids in this species presumably include B chromosomes, with a consequent decrease in B transmission rate.
The finding of repetitive DNAs which B chromosomes are very enriched for (e.g., ribosomal DNA) or else are specific to them (e.g., some satellite DNAs) allows getting an easy estimation of their transmission rate by simply visualizing them in the meiotic products by fluorescence in situ hybridization (FISH) for DNA probes being highly specific to B chromosomes. For instance, Milani et al. (2016) found U2 repeats in a B chromosome in the grasshopper Abracris flavolineata being useful for B-chromosome identification in interphase cells, and they can be also useful for B transmission studies.
Here, we analyze the presence of B chromosomes in standard and aberrant spermatids in two grasshopper species harboring B chromosome systems differing in mitotic stability. In E. plorans, B chromosomes are mitotically stable, meaning that they show the same number of Bs in all cells from a same individual. In Eumigus monticola, however, B chromosomes are mitotically unstable, so that the number of B chromosomes differs between the cells from different testis tubules but not between cells within a same tubule (Ruiz-Ruano et al. 2016). In each species, we have used DNA probes for FISH analysis which allowed scoring the number of B chromosomes in standard spermatids and demonstrated the presence of B chromosomes in most microspermatids observed in both species. Remarkably, the standard spermatids showed a significant decrease in the frequency of B chromosomes between their round and elongating stages, suggesting that B chromosomes are eliminated during spermiogenesis.
Materials and methods
Adult males of the grasshoppers E. plorans and E. monticola were collected in natural populations from Spain, the former species in Alhama de Murcia (Murcia province), Salobreña (Granada), Otívar (Granada), and Torrox (Málaga), and the latter in Hoya de la Mora (Sierra Nevada, Granada). For the present analysis, we chose E. plorans males carrying a single B chromosome belonging to several different variants: B1 (four males from Alhama de Murcia and one from Torrox), B2 (four males from Salobreña and six from Otívar), and B24 (four males from Torrox). A description of these B chromosomes can be found in Cabrero et al. (2014). In the case of E. monticola, we used here one male carrying a mitotically unstable B chromosome, thus showing different B number in different cells.
Males were anesthetized with ethyl acetate vapors before dissection. Testes were fixed in 3:1 ethanol/acetic acid and stored at 4 °C. The number of B chromosomes was analyzed in squash preparations of testis tubules stained with acetic orcein. FISH, including DNA probe preparation and FISH reaction, was performed following the protocols described in Camacho et al. (2015a, b) and Ruiz-Ruano et al. (2016). The DNA probes employed in E. plorans were 18S ribosomal DNA (rDNA), which shows the largest cluster on B1 and B2 variants, and a B-specific satellite DNA recently found by us (Martín-Peciña et al., in preparation) which shows FISH signals only on B chromosomes. In E. monticola, we used a B-specific satellite DNA (EmoSat26-41) previously reported by Ruiz-Ruano et al. (2016). The electron microscope (Fig. 2f) was obtained by the methods reported in Teruel et al. (2009).
Statistical analysis of spermatid counts in E. plorans was performed by a goodness-of-fit χ 2 test with null hypothesis predicting that 1B males produce 0B and 1B standard spermatids at Mendelian 1:1 proportion. This test was separately applied to round and elongating spermatids and a heterogeneity χ 2 test was also employed to analyze within-population heterogeneity before testing the 1:1 proportion at the population level. In E. monticola, however, the mitotic instability of the B chromosome did not allow applying the same null hypothesis and we compared the number of spermatids with 0–3 B chromosomes, between round and elongating ones, by the RxC software (provided by G. Carmody, Ottawa, Ontario, Canada), which performs χ 2 tests in contingency tables, with permutation, and calculates P values by Monte Carlo methods. Twenty batches of 2500 replicates were performed.
Results
Mitotically stable B chromosomes in E. plorans
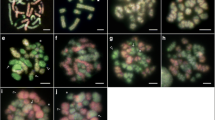
The B1 and B2 variants carry the largest block of rDNA in B-carrying genomes (Fig. 1a), so that FISH with an rDNA probe allows easy identification of B-carrying and B-lacking round and elongating spermatids (Fig. 1b–d). B24, however, carries a smaller rDNA block (see Cabrero et al. 2014), and this marker does not discriminate properly between B24+ and B24− meiotic products. However, the B-specific satellite DNA (EplSat115-11) shows conspicuous clusters on both ends of the B24 chromosome (Fig. 1e) and is clearly apparent in spermatids as one or two small dots (Fig. 1f–h).
Detection of B chromosomes in primary spermatocytes at diplotene (a, e), round spermatids (b, f), and elongating spermatids (c, d, g, h) of the grasshopper Eyprepocnemis plorans by means of FISH for 18S rDNA (a–d) and the B-specific EplSat115-11 satellite DNA (e–h) as DNA probes. Hybridization signals in (a)–(e) are merged with DAPI staining. Inset in (e) depicts the same B in the diplotene cell, at higher magnification, showing satellite location. In (a), note that the B1 chromosome in the Alhama de Murcia population carries the largest cluster for 18S rDNA and that this allows identifying B-carrying spermatids in (b)–(d). In (f)–(h), note that B24-carrying spermatids are identified by the presence of the B-specific satellite (small dots) in Torrox males. Bar in (a) indicates 5 μm for (a) and (e), and 10 μm for the remaining photographs
We analyzed the presence of B chromosomes in round (strictly circular) spermatids (Fig. 1b, f) and also in elongating ones (i.e., showing elliptic to spearhead shape) (Fig. 1c–h). The four males from the Alhama de Murcia population, carrying one B1 chromosome, showed about similar proportions of B-carrying and B-lacking spermatids at both round and elongating stages (Table 1), thus showing a Mendelian rate of B chromosome transmission (k B ). In the Salobreña and Torrox populations, which harbor the B2 and B24 variants, respectively, no significant difference was observed between B-carrying and B-lacking round spermatids (Table 1). However, two males in each population showed a significant deficit of B-carrying elongating spermatids, and χ 2 tests applied to the totals in each population (supported by the heterogeneity χ 2 test) yielded significant decreases in the transmission rate of these 1B males (k B being 0.455 for B2 in Salobreña and 0.463 for B24 in Torrox) (Table 1).
In the Otivar population, which also harbors the B2 variant, we scored only elongating spermatids in six males and all of them showed k B lower than 0.5, but the difference with the Mendelian one was not significant. However, as a whole, they showed a significant tendency to B elimination (Table 1).
In the Torrox population, we found a male carrying one B1 chromosome, a very unusual event in this population where B24 is the most frequent variant. Remarkably, this male showed about similar proportions of B-carrying and B-lacking round spermatids, but a significantly lower proportion of B-carrying elongating spermatids rendering a low B transmission rate (k B = 0.375) (Table 1).
The observed k B in elongating spermatids implied only residual B loss in Alhama for the B1 variant (2.23%), but it was higher in Otívar (4.9%) and Salobreña (9.04%) for B2, as well as in Torrox for B24 (7.41%) and B1 (25.1%).
FISH analysis showed that these decreases in B transmission rate (k B ) were paralleled by the presence of B-carrying macro- and microspermatids (Fig. 2a, b), and we scored them in cysts containing round spermatids in 13 males (excepting those from Otivar) and in cysts of elongating ones in all 19 males analyzed (Table 2). Multiple regression analysis, with k B as dependent variable and the proportion of B-carrying macro- and microspermatids as independent variables, showed that k B was independent of the frequency of these two types of aberrant gametes in the cysts containing round spermatids (round microspermatids—r = 0.08, N = 13, t = 0.24, df = 10, P = 0.82; round macrospermatids—r = −0.12, N = 13, t = 0.35, df = 10, P = 0.73). However, in the cysts of elongating spermatids, k B was significantly negatively correlated with the frequency of B-carrying microspermatids (r = −0.54, N = 19, t = 2.47, df = 16, P = 0.025) but not with the frequency of B-carrying macrospermatids (r = −0.25, N = 19, t = 1.12, df = 16, P = 0.28). This suggests that microspermatid formation is related with a decrease in k B whereas macrospermatids are not, confirming predictions by Teruel et al. (2009).
Presence of macro- and microspermatids in the grasshopper E. plorans. a Two B-carrying (B+) and two B-lacking (B−) standard spermatids, and one macrospermatid (M). b Six standard elongating spermatids, four of which lack B chromosomes (B−) and two carry the B chromosome (B+). Note the presence of a B-carrying microspermatid (m). c Three round spermatids (on the left) showing a large chromosome being apparently extruded from the nucleus. Note that this chromosome carries a small rDNA cluster, which allows identifying it as the X chromosome (arrow). Note in the nucleus at the center that the centromere region, indicated by rDNA location, is still contacting the nucleus, whereas in the nucleus on the right, the whole X chromosome has lost contact with the main nucleus. The elongating B-carrying standard spermatid, on the right, was exceptional by lying beside a B-lacking microspermatid (m). d Examples of microspermatids (m) lying at different distances from a B-lacking standard spermatid. Note the presence of a small FISH signal in the main nucleus on the left (arrow). e Additional examples of microspermatids (m) laying by a B-lacking standard spermatid, showing DAPI staining (upper row) and FISH + DAPI (lower row). f Electron microscope photograph of cross-sectioned standard spermatid nuclei (dense bodies), showing the presence of small dense bodies appearing to be microspermatids, some of which share the cytoplasm with the main nucleus (arrows) and one is outside (arrowhead). Bar in (a) indicates 10 μm for (a)–(e), and that in (f) equals 1 μm
As Table 2 shows, not all microspermatids carried a B chromosome, the main exception being 29 round spermatids, all found in the m16 male from Salobreña, showing a nuclear bud containing a long chromosome carrying a small cluster of rDNA, which allowed identifying it as the X chromosome (Fig. 2c). All these 29 spermatids carried the B chromosome, and most of them showed the X chromosome still stuck to the nucleus, excepting one which was partially separated but still contacting by its end carrying rDNA, i.e., its centromeric region, and another nucleus showing the X chromosome completely separated from it (Fig. 2c). In elongating spermatids, we only observed four B-lacking microspermatids (Fig. 2c), one in m16 from Salobreña and three in m27 from Torrox, the 31 remaining microspermatids carrying the B chromosome.
It was highly remarkable that all B-carrying microspermatids observed by us were placed very close to a standard B-lacking spermatid (Fig. 2d, e), suggesting that the former derived from the same nucleus as the latter and that both share the same cytoplasm. This is also inferred from the fact that our preparations were made by squashing, so that the likelihood that the 47 B-carrying microspermatids were adjacent to a B-lacking standard spermatid would be negligible unless they share the same cytoplasm. In fact, some of the observed B-carrying microspermatids were physically in contact with an adjacent B-lacking nucleus whereas others did not contact with the nucleus and were found at different distance from the B-lacking nucleus (Fig. 2d, e), suggesting that microspermatids are finally expelled from the standard spermatids. Remarkably, a review of the photographs made by us in a previous analysis of spermatogenesis under electronic microscope (Teruel et al. 2009) revealed the presence of microspermatids sharing the same cytoplasm as standard spermatids and also the presence of very similar dense bodies outside spermatids which could correspond to remains of microspermatids extruded from the cytoplasm (Fig. 2f).
Assuming that every B-carrying microspermatid implied the conversion of a B-carrying standard spermatid into a B-lacking one due to B chromosome loss, we can calculate the expected frequency of B+ and B− standard spermatids and test whether this would explain the observed k B in round and elongating ones. In the case of round ones, the analysis of 3272 standard spermatids indicated k B = 0.497, and we found 16 B-carrying microspermatids (see Table 2). The expected frequencies of B+ and B− round spermatids is thus 3272 × 0.5 − 16 = 1620 B+ and 3272 × 0.5 + 16 = 1652 B− (k B = 0.495), and a goodness-of-fit χ 2 test comparing these expected frequencies with the observed ones (1627 and 1645, respectively) indicated the absence of significant difference (χ 2 = 0.06, df = 1, P = 0.8066). On the contrary, we observed 2833 B+ and 3234 B− elongating spermatids (k B = 0.467) plus 31 B-carrying microspermatids, and the expected frequencies, namely 6067 × 0.5 − 31 = 3002.5 B+ and 6067 × 0.5 + 31 = 3064.5 B− (k B = 0.495), differed significantly from the observed ones (χ 2 = 18.94, df = 1, P = 0.00001). This indicates that the observed amount of microspermatids does not explain the decrease in k B observed in elongating spermatids. A possible explanation is that a fraction of the microspermatids produced are finally degraded and lost, so that we are able to visualize only part of those actually formed. We calculated that the loss of B chromosomes in 200 microspermatids (instead of the 31 B-carrying ones observed) would have yielded the observed k B = 0.467, implying that we detected only 16% of the B chromosomes lost as microspermatids.
Mitotically unstable B chromosomes in E. monticola
The exclusive presence of the EmoSat26-41 satellite DNA in the B chromosome of the grasshopper E. monticola (Fig. 3a, b) (see also Ruiz-Ruano et al. 2016) allows scoring the number of B chromosomes in spermatids submitted to FISH (Fig. 3c–h). B chromosomes in this species are mitotically unstable, implying that B number varies among cells within a same individual, but not within a same testis tubule. For this reason, we analyzed round and elongating spermatids in the same six testis tubules and compared B frequency between these two kinds of standard spermatids. In total, we analyzed 911 round spermatids (355 with 0B, 465 with 1B, 89 with 2B, and 2 with 3B) and 442 elongating spermatids (210 with 0B, 193 with 1B, 34 with 2B, and 5 with 3B) and found a significant decrease in the mean number of B chromosomes between round (0.71) and elongating (0.62) spermatids (RxC contingency test—P = 0.0004, SE = 0.0002). This suggests that B chromosomes in E. monticola undergo about 12.3% elimination during spermiogenesis [(0.71 − 0.62)/0.71 = 0.123], as was also evidenced by the presence of 3% of B-carrying round microspermatids (Fig. 3c, d) and 5% of B-carrying elongating microspermatids (Fig. 3e, f). Likewise in E. plorans, the observed frequency of microspermatids was lower than the 12.3% decrease in B frequency, implying that we observed only about 42% (0.05/0.123) of B losses in the form of microspermatids, presumably because many of them are finally degraded.
Detection of B chromosomes in primary spermatocytes at metaphase I (a, b) and spermatids (c–h) of the grasshopper Eumigus monticola, submitted to FISH for the EmoSat26-41 B-specific satellite DNA. Examples of B-carrying (B+) and B-lacking (B−) standard round and elongating spermatids are shown in (c)–(h). In (c)–(e), note the presence of microspermatids (m) beside a B-lacking standard spermatid. Mitotic instability of B chromosomes in this species explains the presence of B-carrying standard spermatids beside a B-carrying microspermatid (f) since standard spermatids in this species can carry two (g) or three (h) B chromosomes. The number of B chromosomes within a sperm nucleus is indicated by the number of plus signs. Bar in (a) indicates 5 μm for (a) and (b), that in (c) indicates 10 μm for (c), (f), and (g), and that in (e) indicates 10 μm for (d), (e), and (h)
Discussion
Population invasion by a parasitic B chromosome needs some kind of drive (Camacho et al. 1997). In E. plorans, we observed that B chromosomes show drive in some populations (Zurita et al. 1998) but not in others (López-León et al. 1992), as a consequence of drive suppression (Herrera et al. 1996; Camacho et al. 1997). In E. monticola, however, nothing is known at this respect. However, our present results suggest that the loss during spermiogenesis would have impeded its birth as a B chromosome. It is thus likely that this B chromosome show drive at other stages of the reproductive cycle. Its mitotic instability suggests possible B accumulation based on mitotic non-disjunction during early cleavage divisions, with preferential destiny of mitotic products carrying more B chromosomes toward the germ line. This kind of accumulation has been reported for mitotically unstable B chromosomes of grasshopper species such as Calliptamus palaestinensis (Nur 1963), Camnula pellucida (Nur 1969), and Locusta migratoria (Nur 1969; Kayano 1971; Viseras et al. 1990). In the latter species, pre-meiotic accumulation of B chromosomes represents about a 30% increase in male B transmission, but it is counteracted by a 20% decrease during subsequent stages of the reproductive cycle, including the formation of microspermatids, the net B transmission thus implying about 10% accumulation in males (Pardo et al. 1994). In addition, this B chromosome shows 62% accumulation during female transmission (Pardo et al. 1994), which explains the worldwide distribution of B chromosomes in L. migratoria.
Our results have shown a significant decrease in B transmission rate (k B ) during spermiogenesis in two species of grasshopper carrying B chromosomes. In the case of E. plorans, males carrying one mitotically stable B chromosome yielded meiotic products at the Mendelian rate, given that about half of round spermatids carried the B chromosome. Therefore, spermiogenesis in these males begins with 1:1 proportion of B-carrying and B-lacking round spermatids. In contrast, most males showed a tendency to a decreased proportion of B-carrying elongating spermatids, which was significant in five males and, as a whole, in Salobreña, Otivar, and Torrox populations (see Table 1). Therefore, the k B decrease takes place necessarily during spermiogenesis. We also demonstrate here that k B was negatively correlated with the frequency of B-carrying microspermatids, suggesting that B chromosomes are lost during spermiogenesis in the form of microspermatids. Our FISH visualization of B chromosomes within microspermatids constitutes the first direct demonstration of Nur’s claiming that microspermatids are a way of B chromosome loss (Nur 1969). Remarkably, we only observed B-carrying macrospermatids even though some B-lacking ones might be expected if cytokinesis failures would take place in B-lacking secondary spermatocytes. This suggests a direct role of lagging B chromosomes in the formation of macrospermatids, as was also suggested by Nur (1969).
In addition, the fact that B-carrying microspermatids were always found beside a B-lacking standard spermatid, even in squash preparations, along with the presence of microspermatids sharing a common cytoplasma with standard spermatids at electronic microscope images, suggests a causal relationship between microspermatid formation, B chromosome loss, and the decrease in k B in standard spermatids. The conventional explanation for microspermatids is that they contain B chromosomes lagged during the precedent meiotic divisions, as was first suggested by Nur (1969) and later supported by other authors (Pearse and Ehrlich 1979; Viseras and Camacho 1985; Bidau 1987; Teruel et al. 2009; Abdel-Haleem et al. 2009). Our present results, however, challenge this hypothesis. Of course, we cannot rule out that some of the 16 B-carrying microspermatids found within cysts containing round standard spermatids could have derived from B chromosomes lagged during previous meiosis which failed to properly integrate into the main nucleus, and even some of the 31 B-carrying microspermatids observed in the cysts of elongating spermatids could actually have derived from them. However, if meiosis were the only source of microspermatids, we should observe similar values of k B in round and elongating spermatids, and this was not the case in the two species analyzed here, thus clearly implying microspermatid formation during spermiogenesis.
The finding that micronuclei can be formed by nuclear budding in interphase cells could provide a mechanistic support to B chromosome elimination during spermiogenesis. Nuclear budding and micronucleus formation are common characteristics to many cell cultures frequently leading to chromosome elimination (Elston 1963; Longwell and Yerganian 1965). The classical mechanism of micronucleus formation claims that they incorporate lagging chromosomes during mitosis (Heddle and Carrano 1977; Schubert and Oud 1997; Fenech et al. 2011). However, recent findings have shown that nuclear budding and micronucleus formation can also occur in interphase cells (Gernand et al. 2005, 2006; Utani et al. 2011; Ishii et al. 2016). Similarly, round and elongating spermatids can form micronuclei (called here microspermatids) during spermiogenesis without the involvement of any additional cell division. In fact, our results show remarkable similarities with some characteristics of interphase micronucleus formation described by the former authors.
For instance, Gernand et al. (2005, 2006) reported that the chromosomes destined to elimination occupied a peripheral location in interphase cells of interspecific hybrids. This appears to be a general tendency since other chromosomes being regularly eliminated also occupy peripheral locations, such as E chromosomes in Cecidomyiidae (Kloc and Zagrodzinska 2001), the germ-line restricted chromosomes (GRC) in the zebra and the Bengalese finches (Schoenmakers et al. 2010; Del Priore and Pigozzi 2014), and even acentric, autonomously replicating extrachromosomal structures called double-minute chromosomes (Shimizu et al. 1998).
Interestingly, a tendency of B chromosomes to occupy peripheral locations in the nucleus during cell division was early noted by Avdulow (1933) in maize (see also Randolph 1941; Darlington and Upcott 1941; Carlton and Cande 2002). Subsequent research has reached the same conclusion for B chromosomes in Poa alpina (Hakansson 1948), Dactylis (Williams and Barclay 1972), and rye (Jones 1995; Morais-Cecílio et al. 1996; Langdon et al. 2000). In animals, the paternal sex ratio (PSR) is an extremely parasitic B chromosome which localizes to the outer periphery of the paternal nucleus and at the tip of the sperm nucleus, but in this case the B chromosome escapes from elimination which is focused on the paternal standard set (Swim et al. 2012). These authors visualized PSR by FISH in spermatids and mature sperm and about 98% of them, in both cases, carried the B chromosome, so that we can infer that PSR is not eliminated at all during spermiogenesis.
Sex chromosomes in animals also occupy peripheral locations (see Turner 2007; Finch et al. 2008; Calvente et al. 2013) and are inactivated during meiosis by means of epigenetic marks (Vaskova et al. 2010). Likewise, in E. plorans, X and B chromosomes are heterochromatic; they show frequent non-homologous association during the first meiotic prophase (Camacho et al. 1980) and are hypoacetylated for H3K9 during entire meiosis (Cabrero et al. 2007). They also tend to occupy peripheral location in meiotic nuclei, which probably facilitates their elimination in the form of microspermatids. It is tempting to speculate that the high similarity between X and B chromosomes during meiosis may lead to eventual X chromosome elimination, presumably because some of the epigenetic marks used for microspermatid formation are common to these two chromosomes.
Another resemblance of our present results with those in interspecific hybrids is that centromeric regions of pearl millet chromosomes are the last in being eliminated in wheat-pearl millet hybrids (Gernand et al. 2005). We observed this same fact in the case of the X chromosome elimination in m16 from Salobreña (see Fig. 2c). Interestingly, Gernand et al. (2005) suggested that micronucleus formation can eventually leave the centromeric region of the expulsed chromosome in the main nucleus, thus opening the possibility to de novo formation of B chromosomes in interspecific crosses. Our Fig. 2d shows a round microspermatid, beside a round standard spermatid, which harbors most of the B-specific satellite except a small FISH signal remaining in the main nucleus, indicating that B chromosome extrusion can be incomplete, thus giving indirect support to Gernand et al.’s claims.
In addition, Gernand et al. (2005) suggested that post-translational histone modification might play a role in chromosome elimination, as differential acetylation of histones H3 and H4 and methylation of histone H3 had been reported in chromosome elimination in sciarid flies (Goday and Ruiz 2002) and in programmed DNA elimination in Tetrahymena (Taverna et al. 2002). In addition, the GRC chromosome in the zebra finch is silenced from early leptotene onwards and is eliminated through micronucleus formation following metaphase I (Schoenmakers et al. 2010). It is thus presumable that the observed H3K9 hypoacetylation of X and B chromosomes in E. plorans (Cabrero et al. 2007) may serve as a signal for elimination through the evolutionary conserved mechanism suggested by Gernand et al. (2005, 2006).
Our present results suggest that even in organisms where chromosome elimination occurs only sporadically, e.g., B chromosome loss during spermiogenesis, interphase cells appear to show the ability to eliminate chromosomes through micronucleus formation. The parasitic nature of B chromosomes makes them an elimination target with high fitness reward for the host genome. Ideally, the best situation for a B chromosome would be to remain limited to the germ line by being eliminated from somatic cells, thus minimizing harmful effects on the host. Examples of germ-line restricted B chromosomes have been found, for example, in the marsupial E. kalabu (Hayman et al. 1969) and the ant L. spinosior (Imai 1974). Even in this case, B chromosome presence in the germ line is still a load for the host genome, as it has to replicate extra DNA without a reward, except in the case that the B chromosome carries a gene whose activity result is profitable for the host (e.g., see Miao et al. 1991). In most cases, however, it is expected that the host genome continues trying to get rid of the parasitic element. The existence of post-meiotic elimination mechanisms like that shown here might help in this task, but it does not always work. Suggestive examples are germ-line restricted chromosomes like those reported in diptera (Bauer and Beermann 1952; Staiber 1988; Herrick and Seger 1999; Goday and Esteban 2001) or zebra finches (Pigozzi and Solari 2005; Schoenmakers et al. 2010; Del Priore and Pigozzi 2014), as they could actually be the last face of obstinately resistant parasitic B chromosomes.
References
Abdel-Haleem AA, Sharaf HM, El-Kabbany AI (2009) New record of B-chromosome through meiosis in the Egyptian locust Anacridium aegyptium (Acrididae) with indication to its origin. J King Saud Univ Sci 21:163–166. doi:10.1016/j.jksus.2009.10.002
Avdulow NP (1933) On the additional chromosomes in maize. Bull Appl Bot Ser 2:101–130
Bauer H, Beermann W (1952) Der chromosomencyclus der Orthocladiinen (Nematocera, Diptera). Z Naturforsch 7:557–563
Bidau CJ (1986) Effects on cytokinesis and sperm formation of a B-isochromosome in Metaleptea brevicornis adspersa (Acridinae, Acrididae). Caryologia 39:165–177. doi:10.1080/00087114.1986.10797778
Bidau CJ (1987) Influence of a rare unstable B-chromosome on chiasma frequency and nonhaploid sperm production in Dichroplus pratensis (Melanoplinae, Acrididae). Genetica 73:201–210. doi:10.1007/BF00055275
Burt A, Trivers R (2006) Genes in conflict: the biology of selfish genetic elements. Belknap Press of Harvard University Press, Cambridge
Cabrero J, Teruel M, Carmona FD, Jiménez R, Camacho JPM (2007) Histone H3 lysine 9 acetylation pattern suggests that X and B chromosomes are silenced during entire male meiosis in a grasshopper. Cytogenet Genome Res 119:135–142. doi:10.1159/000109630
Cabrero J, Bakkali M, Navarro-Domínguez B, Ruíz-Ruano FJ, Martín-Blázquez R, López-León MD, Camacho JPM (2013) The Ku70 DNA-repair protein is involved in centromere function in a grasshopper species. Chromosom Res 21:393–406. doi:10.1007/s10577-013-9367-7
Cabrero J, López-León MD, Ruíz-Estévez M, Gómez R, Petitpierre E, Rufas JS, Massa B, Kamel Ben Halima M, Camacho JPM (2014) B1 was the ancestor B chromosome variant in the western Mediterranean area in the grasshopper Eyprepocnemis plorans. Cytogenet Genome Res 142:54–58. doi:10.1159/000356052
Calvente A, Viera A, Parra MT, De La Fuente R, Suja JA, Page J, Santos JL, García de la Vega C, Barbero JL, Rufas JS (2013) Dynamics of cohesin subunits in grasshopper meiotic divisions. Chromosoma 122:77–91. doi:10.1007/s00412-012-0393-6
Camacho JPM (2005) B chromosomes. In: Gregory TR (ed) The evolution of the genome. Academic Press, New York, pp 223–286
Camacho JPM, Carballo AR, Cabrero J (1980) The B-chromosome system of the grasshopper Eyprepocnemis plorans subsp. plorans (Charpentier). Chromosoma 80:163–176. doi:10.1007/BF00286298
Camacho JPM, Shaw MW, López-León MD, Pardo MC, Cabrero J (1997) Population dynamics of a selfish B chromosome neutralized by the standard genome in the grasshopper Eyprepocnemis plorans. Am Nat 149:1030–1050. doi:10.1086/286037
Camacho JPM, Sharbel TF, Beukeboom LW (2000) B-chromosome evolution. Phil Trans Roy Soc Lond B 355:163–178. doi:10.1098/rstb.2000.0556
Camacho JPM, Perfectti F, Teruel M, López-León MD, Cabrero J (2004) The odd-even effect in mitotically unstable B chromosomes in grasshoppers. Cytogenet Genome Res 106:325–331. doi:10.1159/000079307
Camacho JPM, Cabrero J, López-León MD, Cabral-de-Mello DC, Ruiz-Ruano FJ (2015a) Grasshoppers (Orthoptera). In: Sharakhov IV (ed) Protocols for cytogenetic mapping of arthropod genomes. CRC Press, pp 381–438
Camacho JPM, Ruiz-Ruano FJ, Martín-Blázquez R, López-León MD, Cabrero J, Lorite P, Cabral-de-Mello DC, Bakkali M (2015b) A step to the gigantic genome of the desert locust: chromosome sizes and repeated DNAs. Chromosoma 124:263–275. doi:10.1007/s00412-014-0499-0
Carlton PM, Cande WZ (2002) Telomeres act autonomously in maize to organize the meiotic bouquet from a semipolarized chromosome orientation. J Cell Biol 157:231–242. doi:10.1083/jcb.200110126
Chiavarino AM, Rosato M, Manzanero S, Jiménez G, González-Sánchez M, Puertas MJ (2000) Chromosome nondisjunction and instabilities in tapetal cells are affected by B chromosomes in maize. Genetics 155:889–897
Darlington CD, Upcott MB (1941) The activity of inert chromosomes in Zea mays. J Genet 41:275–296
Del Priore L, Pigozzi MI (2014) Histone modifications related to chromosome silencing and elimination during male meiosis in Bengalese finch. Chromosoma 123:293–302. doi:10.1007/s00412-014-0451-3
Elston RN (1963) Nuclear budding and micronuclei formation in human bone marrow, skin and fascia lata cells in vitro and in oral mucosa cells in vivo. Acta Pathol Microbiol Immunol Scand B 59:195–199
Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD, Thomas P (2011) Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 26:125–132. doi:10.1093/mutage/geq052
Finch KA, Fonseka KGL, Abogrein A, Ioannou D, Handyside AH, Thornhill AR, Hickson N, Griffin DK (2008) Nuclear organization in human sperm: preliminary evidence for altered sex chromosome centromere position in infertile males. Hum Reprod 23:1263–1270. doi:10.1093/humrep/den112
Frank SA (2000) Polymorphism of attack and defense. Trends Ecol Evol 15:167–171. doi:10.1016/S0169-5347(99)01814-5
Gernand D, Rutten T, Varshney A, Rubtsova M, Prodanovic S, Brüβ KCJ, Matzk F, Houben A (2005) Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell 17:2431–2438. doi:10.1105/tpc.105.034249
Gernand D, Rutten T, Pickering R, Houben A (2006) Elimination of chromosomes in Hordeum vulgare x H. bulbosum crosses at mitosis and interphase involves micronucleus formation and progressive heterochromatinization. Cytogenet Genome Res 114:169–174. doi:10.1159/000093334
Goday C, Esteban MR (2001) Chromosome elimination in sciarid flies. BioEssays 23:242–250. doi:10.1002/1521-1878(200103)23:3<242::AID-BIES1034>3.0.CO;2-P
Goday C, Ruiz MF (2002) Differential acetylation of histones H3 and H4 in paternal and maternal germline chromosomes during development of sciarid flies. J Cell Sci 115:4765–4775. doi:10.1242/jcs.00172
Hakansson A (1948) Embryology of Poa alpina plants with accessory chromosomes. Hereditas 34:233–247. doi:10.1111/j.1601-5223.1948.tb02838.x
Hayman DL, Martin PG, Waller PF (1969) Parallel mosaicism of supernumerary chromosomes and sex chromosomes in Echymipera kalabu (Marsupialia). Chromosoma 27:371–380. doi:10.1007/BF00325676
Heddle JA, Carrano AV (1977) The DNA content of micronuclei induced in mouse bone marrow by gamma irradation: evidence that micronuclei arise from acentric chromosomal fragments. Mutat Res 44:63–69. doi:10.1016/0027-5107(77)90115-4
Herrera J, López-León M, Cabrero J, Shaw M, Camacho JPM (1996) Evidence for B chromosome drive suppression in the grasshopper Eyprepocnemis plorans. Heredity 76:633–639. doi:10.1038/hdy.1996.90
Herrick G, Seger J (1999) Imprinting and paternal genome elimination in insects. Results Probl Cell Differ 25:41–71. doi:10.1007/978-3-540-69111-2_3
Imai HT (1974) B-chromosomes in the Myrmicine ant, Leptothorax spinosior. Chromosoma 45:431–444. doi:10.1007/BF00283387
Ishii T, Karimi-Ashtiyani R, Houben A (2016) Haploidization via chromosome elimination: means and mechanisms. Ann Rev Plant Biol 67:1–18. doi:10.1146/annurev-arplant-043014-114714
Jones RN (1995) B chromosomes in plants. Tansley Review No. 85. New Phytol 131:411–434. doi:10.1111/j.1469-8137.1995.tb03079.x
Kayano H (1971) Accumulation of B chromosomes in the germ line of Locusta migratoria. Heredity 27:119–123. doi:10.1038/hdy.1971.76
Kloc M, Zagrodzinska B (2001) Chromatin elimination—an oddity or a common mechanism in differentiation and development? Differentiation 68:84–91. doi:10.1046/j.1432-0436.2001.680202.x
Langdon T, Seago C, Jones RN, Ougham H, Thomas H, Forster JW, Jenkins G (2000) De novo evolution of satellite DNA on the rye B chromosome. Genetics 154:869–884
Longwell AC, Yerganian G (1965) Some observations on nuclear budding and nuclear extrusions in a Chinese hamster cell culture. J Natl Cancer I 34:53–69. doi:10.1093/jnci/34.1.53
López-León M, Cabrero J, Camacho JPM, Cano M, Santos JL (1992) A widespread B chromosome polymorphism maintained without apparent drive. Evolution 46:529–539. doi:10.2307/2409869
Loray MA, Remis MI, Vilardi JC (1991) Parallel polymorphisms for supernumerary heterochromatin in Dichroplus elongatus (Orthoptera): effects on recombination and fertility. Genetica 84:155–163. doi:10.1007/BF00127242
Miao VP, Covert SF, VanEtten HD (1991) A fungal gene for antibiotic resistance on a dispensable (“B”) chromosome. Science 254:1773–1776. doi:10.1126/science.1763326
Milani D, Palacios-Gimenez OM, Cabral-de-Mello DC (2016) The U2 snDNA is a useful marker for B chromosome detection and frequency estimation in the grasshopper Abracris flavolineata. Cytogenet Genome Res (in press)
Morais-Cecílio L, Delgado M, Jones RN, Viegas W (1996) Painting rye B chromosomes in wheat: interphase chromatin organization, nuclear disposition and association in plants with two, three or four Bs. Chromosom Res 4:195–200. doi:10.1007/BF02254959
Nur U (1963) A mitotically unstable supernumerary chromosome with an accumulation mechanism in a grasshopper. Chromosoma 14:407–422. doi:10.1007/BF00326786
Nur U (1969) Mitotic instability leading to an accumulation of B-chromosomes in grasshoppers. Chromosoma 27:1–19. doi:10.1007/BF00326108
Pardo MC, López-León MD, Cabrero J, Camacho JPM (1994) Transmission analysis of mitotically unstable B chromosomes in Locusta migratoria. Genome 37:1027–1034. doi:10.1139/g94-146
Pearse FK, Ehrlich PR (1979) B chromosome variation in Euphydryas colon (Lepidoptera, Nymphalidae). Chromosoma 73:263–274. doi:10.1007/BF00288691
Phillips DM (1970) Insect sperm: their structure and morphogenesis. J Cell Biol 44:243–277
Pigozzi MI, Solari AJ (2005) The germ-line-restricted chromosome in the zebra finch: recombination in females and elimination in males. Chromosoma 114:403–409. doi:10.1007/s00412-005-0025-5
Randolph LF (1941) Genetic characteristics of the B chromosomes in maize. Genetics 26:608–631
Ruiz-Ruano FJ, Cabrero J, López-León MD, Camacho JPM (2016) Satellite DNA content illuminates the ancestry of a supernumerary (B) chromosome. Chromosoma (in press) doi: 10.1007/s00412-016-0611-8
Schoenmakers S, Wassenaar E, Laven JSE, Grootegoed JA, Baarends WM (2010) Meiotic silencing and fragmentation of the male germline restricted chromosome in zebra finch. Chromosoma 119:311–324. doi:10.1007/s00412-010-0258-9
Schubert I, Oud JL (1997) There is an upper limit of chromosome size for normal development of an organism. Cell 88:515–520. doi:10.1016/S0092-8674(00)81891-7
Shimizu N, Itoh N, Utiyama H, Wahl GM (1998) Selective entrapment of extrachromosomally amplified DNA by nuclear budding and micronucleation during S phase. J Cell Biol 140:1307–1320. doi:10.1083/jcb.140.6.1307
Staiber W (1988) G-banding of germ line limited chromosomes in Acricotopus lucidus (Diptera, Chironomidae). Chromosoma 97:231–234. doi:10.1007/BF00292966
Suja JA, de la Vega CG, Rufas JS (1989) Mechanisms promoting the appearance of abnormal spermatids in B-carrier individuals of Eyprepocnemis plorans (Orthoptera). Genome 32:64–71. doi:10.1139/g89-412
Swim MM, Kaeding KE, Ferree PM (2012) Impact of a selfish B chromosome on chromatin dynamics and nuclear organization in Nasonia. J Cell Sci 125:5241–5249. doi:10.1242/jcs.113423
Szöllösi A (1975) Electron microscope study of spermiogenesis in Locusta migratoria (Insect Orthoptera). J Ultrasruct Res 50:322–346. doi:10.1016/S0022-5320(75)80064-5
Taverna SD, Coyne RS, Allis CD (2002) Methylation of histone H3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110:701–711. doi:10.1016/S0092-8674(02)00941-8
Teruel M, Cabrero J, Perfectti F, Alché JD, Camacho JPM (2009) Abnormal spermatid formation in the presence of the parasitic B24 chromosome in the grasshopper Eyprepocnemis plorans. Sex Dev 3:284–289. doi:10.1159/000253307
Turner JMA (2007) Meiotic sex chromosome inactivation. Development 134:1823–1831. doi:10.1242/dev.000018
Utani K-i, Okamoto A, Shimizu N (2011) Generation of micronuclei during interphase by coupling between cytoplasmic membrane blebbing and nuclear budding. PLoS One 6(11):e27233. doi:10.1371/journal.pone.0027233
Vaskova EA, Pavlova SV, Shevchenko AI, Zakian SM (2010) Meiotic inactivation of sex chromosomes in mammals. Russ J Genet 46:385–393. doi:10.1134/s1022795410040010
Viseras E, Camacho JPM (1985) The B-chromosome system of Omocestus bolivari: changes in B-behaviour in M4-polysomic B-males. Heredity 54:385–390. doi:10.1038/hdy.1985.55
Viseras E, Camacho J, Cano M, Santos J (1990) Relationship between mitotic instability and accumulation of B chromosomes in males and females of Locusta migratoria. Genome 33:23–29. doi:10.1139/g90-005
Wang J, Davis RE (2014) Programmed DNA elimination in multicellular organisms. Curr Opin Genet Dev 27:26–34. doi:10.1016/j.gde.2014.03.012
Williams E, Barclay PC (1972) Transmission of B-chromosomes in Dactylis. New Zeal J Bot 10:573–584. doi:10.1080/0028825X.1972.10430247
Zurita S, Cabrero J, López-León M, Camacho J (1998) Polymorphism regeneration for a neutralized selfish B chromosome. Evolution 52:274–277. doi:10.2307/2410945
Acknowledgments
We thank M. Teruel and J.D. Alché for their help to obtain the electron microscope photograph in Fig. 2f. This study was supported by a grant from the Spanish Secretaría de Estado de Investigación, Desarrollo e Innovación (CGL2015-70750-P) and was partially performed by FEDER funds.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Integrity of research
All experiments comply with the current Spanish laws.
Conflict of interest
The authors declare that they have no competing interests.
Human and animal rights and informed consent
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Rights and permissions
About this article
Cite this article
Cabrero, J., Martín-Peciña, M., Ruiz-Ruano, F.J. et al. Post-meiotic B chromosome expulsion, during spermiogenesis, in two grasshopper species. Chromosoma 126, 633–644 (2017). https://doi.org/10.1007/s00412-017-0627-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-017-0627-8