Abstract
Desmoplastic/nodular medulloblastomas (DNMB) and medulloblastomas with extensive nodularity (MBEN) were outlined in the current WHO classification of tumors of the nervous system as two distinct histological MB variants. However, they are often considered as cognate SHH MB entities, and it is a reason why some clinical MB trials do not separate the patients with DNMB or MBEN histology. In the current study, we performed an integrated DNA/RNA-based molecular analysis of 83 DNMB and 36 MBEN to assess the etiopathogenetic relationship between these SHH MB variants. Methylation profiling revealed “infant” and “children” SHH MB clusters but neither DNMB nor MBEN composed separate epigenetic cohorts, and their profiles were intermixed within the “infant” cluster. In contrast, RNA-based transcriptional profiling disclosed that expression signatures of all MBEN were clustered separately from most of DNMB and a set of differentially expressed genes was identified. MBEN transcriptomes were enriched with genes associated with synaptic transmission, neuronal differentiation and metabolism, whereas DNMB profiling signatures included sets of genes involved in phototransduction and NOTCH signaling pathways. Thus, DNMB and MBEN are distinct tumor entities within the SHH MB family whose biology is determined by different transcriptional programs. Therefore, we recommend a transcriptome analysis as an optimal molecular tool to discriminate between DNMB and MBEN, which may be of benefit for patients’ risk stratification in clinical trials. Molecular events identified in DNMB by RNA sequencing could be considered in the future as potent molecular targets for novel therapeutic interventions in treatment-resistant cases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The histological entity medulloblastoma (MB), one of the most common malignant brain tumors in childhood, comprises distinct histological variants and molecular subgroups. According to the current international consensus, four main molecular MB subgroups (WNT, SHH, Group 3 and Group 4) with divergent histology, biology and clinical outcomes exist [4, 12, 18, 21, 23, 30]. Within the SHH-activated MB subgroup, a wide spectrum of histological variants may occur, including desmoplastic/nodular (DNMB), medulloblastoma with extensive nodularity (MBEN), classic, and large cell/anaplastic, each associated with specific molecular signatures and clinical course [1, 6, 11, 13, 18, 19, 24, 30]. Although DNMB and MBEN were outlined in the current WHO classification of tumors of the nervous system as two distinct histological MB variants [18], they are often considered as cognate SHH MB entities; therefore, most of the clinical MB trials do not separate the patients with DNMB or MBEN histology [1, 14, 23, 24]. Indeed, both these SHH MB variants disclose similar bi-compartmental histological composition, frequent SHH-activating mutations (PTCH1, SUFU, SMO) and epigenetic commonality [1, 11, 13, 18, 21, 24]. However, clinico-pathological associations for these SHH MB histological variants are considered as controversial. Some studies revealed no survival differences between DNMB and MBEN suggesting that patients’ risk stratification based on SHH MB histology is not sufficient for an optimal management [1, 23, 24]. In other trials, however, MBEN histology was associated with good to excellent survival, whereas the prognostic significance of pediatric DNMB was considered as controversial [7, 25, 26]. Thus, the question of an intrinsic clinico-biological relationship between DNMB and MBEN still remains open. Recently, we have found that DNMB and MBEN showed a paradoxical combination of epigenetic similarity with simultaneous cytogenetic dissimilarity [13]. In the current study, we further investigated the etiopathogenetic relationship between DNMB and MBEN by applying an integrated DNA/RNA-based analysis, thereby aiming to answer the question of whether these SHH MB variants are “close relatives or distant cousins”.
Materials and methods
Patient population
Tissue and blood samples were obtained from 119 pediatric patients (age 0–16 years) with histological diagnoses either “desmoplastic nodular medulloblastoma” (DNMB; 83 cases) or “medulloblastoma with extensive nodularity” (MBEN; 36 cases) according to the 2016 WHO classification of tumors of the central nervous system (CNS) [18]. All these patients were initially diagnosed between 01.01.1997 and 31.12.2017 at the Burdenko Neurosurgical Institute in Moscow and all received combined treatment according to various protocols (see “Results”). Two previously reported tumor samples which were diagnosed histologically as MBEN but disclosed epigenetic profiles of group 3/4 MB were not included in this analysis [13]. Informed consent was obtained from all patients´ parents or other relatives/caregivers. This retrospective study was conducted under the auspices of the Ethics Committee of the Burdenko Neurosurgical Institute (Ethical vote number 563/6-16) and those of the University of Heidelberg, in compliance with the Russian Federation and German rules and regulations of the Health Insurance Portability, and in adherence to the tenets of the Declaration of Helsinki. The follow-up analysis was stalled on 01.01.2019 (the end point of follow-up).
DNA and RNA extraction
Representative tissue samples with highest available tumor content were histologically identified, microdissected, and chosen for nucleic acid extraction. DNA and RNA were extracted from formalin-fixed and paraffin-embedded (FFPE) tissue samples using the automated Maxwell system with the Maxwell 16 FFPE Plus LEV DNA Purification Kit or Maxwell 16 LEV RNA FFPE Kit (Promega, Madison, WI, USA), according to the manufacturer’s instructions. To evaluate FFPE RNA quality, we used the percentage of RNA fragments > 200 nt fragment determination value (DV200). Only RNA samples with DV200 > 70% were included in the further analysis.
DNA methylation analysis
DNA was analyzed using the Illumina Human Methylation 450k or 850k/EPIC BeadChip array as previously described. All DNA methylation analyses were performed in R version 3.3.0 (R Development Core Team) as described [10, 13, 21, 23]. Raw signal intensities were obtained from IDAT-files using the minfi Bioconductor package version 1.18.2. Each sample was individually normalized by performing a background correction (shifting of the 5% percentile of negative control probe intensities to 0) and a dye-bias correction (scaling of the mean of normalization control probe intensities to 10,000) for both color channels. No further normalization or transformation steps were performed, and standard beta-values were used for downstream methylation analyses. The following criteria were applied to filter out probes prone to yield inaccurate methylation levels: removal of probes targeting the X and Y chromosomes (n = 11,551), removal of probes that overlap common SNPs (dbSNP132 Common) within the CpG or the following base (n = 7998), and removal of probes not mapping uniquely to the human reference genome (hg19) (n = 3965). To enable comparability between the Illumina Infinium HumanMethylation 450k and 850k/EPIC arrays, we removed all probes not represented on the 450k array. In total, 428,799 probes were kept for analysis.
For unsupervised hierarchical clustering, we selected the 10,000 most variably methylated probes across the dataset as measured by standard deviation. Samples were clustered using Pearson correlation coefficient as the distance measure and average linkage (x-axis). Methylation probes were reordered by hierarchical clustering using Euclidean distance and average linkage (y-axis). Additional analysis of tumor subgroups was performed using a t-distributed stochastic neighbor embedding (t-SNE)-based approach [13, 21]. This was computed using the R-package R-tsne, version 0.13, with a perplexity of 15 and 20 iterations. Copy number profiles were generated using the ‘conumee’ package for R [10, 23].
Targeted next-generation sequencing (NGS)
Molecular barcode-indexed ligation-based sequencing libraries were constructed using 200 ng of sheared DNA. All 119 paired tumor and blood samples yielded sufficient DNA amounts for analysis. Libraries were enriched by hybrid capture with custom biotinylated RNA oligo pools covering exons of 130 cancer-associated genes [27,28,29]. Paired-end sequencing was performed using the NextSeq 500 (Illumina). Sequence data were mapped to the reference human genome using the Burrows–Wheeler Aligner [27,28,29] and were processed using the publicly available SAM tools. Only variants annotated as “exonic” or “splicing” were included, and “intergenic” and other untranslated regions were excluded. Recurrent gene mutations of PTCH1, SUFU, and SMO were also assessed with residual DNA from the same pool used for sequencing by polymerase chain reaction followed by direct Sanger sequencing of the corresponding exons.
RNA sequencing
RNA sequencing of DNMB samples for which RNA of sufficient quality and quantity was available (n = 75/63%) was performed on a NextSeq 500 (Illumina) as previously described [28, 29]. The reads were aligned to hg19 reference using STAR version 2.5.2b and for each sample; gene expression was quantified by the feature counts module of the Subread package version 1.4.6 using Gencode version 19 annotations considering uniquely mapped reads only [5, 17]. To identify potential low-quality or outlier samples, quality control and multi-sample comparisons were done using Qualimap version 2.2.1 [5]. In general, reads alignment rate is high per sample and the mean proportion of mapped reads within exons across all samples was around 90%. Unsupervised tumor samples comparison was performed with principal component analysis and hierarchical clustering based on the selection of the top 1000 and 500 most variable genes with log2 RPKM gene expression normalization. Subgroup-specific differential gene expression analysis was performed by comparing one molecular class against the other using DESeq2 R package (adjusted p < 0.05) and the identified differentially upregulated genes were grouped according to their previously assigned molecular classes. Gene ontology analysis was done using ClueGO (Cytoscape) by combining the top 250 specifically expressed genes in the individual molecular classes, with visualization using Cytoscape version 3.4 [2]. Fusion discovery was done based on RNA sequencing data using five independent algorithms: FusionCatcher, InFusion, TopHat, deFuse, and Arriba [20, 22, 29]. Only gene fusions detected with all these algorithms together were estimated as the “real fusions”. Direct RNA sequencing was performed for confirmation of the detected fusions. Levels of mRNA for 2 selected candidate genes (RIMS1 and MYO7A) were also comparatively measured using a quantitative reverse transcription real-time polymerase chain reaction (RQ-PCR) as described [29].
Statistics
The distributions of overall survival (OS) and progression free survival (PFS) were calculated according to the Kaplan–Meier method using the log rank test for significance. PFS was calculated from the date of diagnosis until tumor recurrence or last contact for patients who were free of disease. OS was calculated from the date of diagnosis until death of patient from disease or last contact for patients who were still alive. For multivariate analysis, Cox proportional hazards regression models were used. Estimated hazard ratios are provided with 95% confidence intervals and a p value from the Wald test. Tests with a p value below 0.05 were considered significant.
Results
Histopathological appearance of DNMB and MBEN
Among 119 samples studied, 83 specimens were diagnosed as DNMB and 36 as MBEN using consensus histological criteria [6, 13, 15, 16]; both these MB variants disclosed prototypic bi-compartmental microscopic appearance. DNMB included nodular reticulin-free zones containing cells with various patterns of neuronal maturation (so-called “pale islands”), and an interstitial, reticulin-rich component composed of densely packed cells with mitotic activity (Fig. 1a). In MBEN, the reticulin-free lobulated component contained populations of small cells disclosing clear features of neurocytic differentiation (Fig. 1b) and the extent of inter-nodular reticulin-rich zones varied from one sample/area to another.
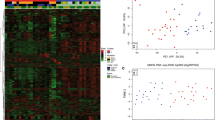
Microscopic appearance of DNMB (a) shows two “canonical” components: reticulin-free “pale islands” and interstitial zones. Histology of MBEN (b) also comprises two structural components revealing a clear lobular architecture (H&E; a; b × 200; Scale bar—1µk). c Heatmap of unsupervised hierarchical cluster analysis across a combined set of 83 DNMB and 36 MBEN based on the 10,000 most variable methylation probes; SD > 0.30; beta values: 0–0.5 blue; 0.5–1 red; patient age and tumor histology are indicated above the heatmap DNA methylation profiles of DNMB (black bars) and MBEN (red bars) are not distinct and distributed across the tumor cohort. Two epigenetic clusters that differ in terms of the patients’ age (infants; black bars vs. children; white bars) were disclosed. d Two-dimensional t-distributed stochastic neighbor embedding (tSNE) analysis of infant SHH MB revealed two additional tumor subgroups but MBEN were clustered as interspersed with DNBM samples
GAB1 protein immunoreactivity (a marker of SHH pathway activation) was found in all DNMB and MBEN samples studied. Nuclear expression of NeuN was also identified within nodular components of both these histological variants (Suppl. Figure 1a, b). However, NeuN expression was homogeneously strong in MBEN but variable in terms of intensity and extent in DNMB. MIB1 nuclear expression was absent in nodular zones of MBEN and observed in inter-nodular areas only. In contrast, both components of DNMB disclosed variable MIB1 immunoreactivity and the median labeling index was significantly higher for these tumors (38% vs. 17% for MBEN; p < 0.01; Suppl. Figure 1c, d).
Clinical characteristics of DNMB and MBEN
Basic clinical characteristics of all 119 DNMB/MBEN patients included in this study are summarized in Table 1. Patients with DNMB were older: 6.7 years vs. 1.6 years for MBEN patients, although the male:female ratio was equal for both tumor cohorts. A similar proportion of patients with DNMB (21/83; 25%) and MBEN (7/36; 19%) disclosed metastatic M2-3 stages at the time of diagnosis (p = 0.63). All patients were operated upon and gross total tumor resection was achieved in 71 cases (60%). After the operation, all patients received combined treatment in the three following regimens: (1) chemotherapy alone: HIT-SKK with intraventricular methotrexate (MTX) injection for 79 infant patients with any M stage (43 DNMB and all 36 MBEN). (2) HIT-based radio-chemotherapy protocol in standard doses for 29 patients with M0 at diagnosis and DNMB histology. (3) Eleven patients with M2–3 stage DNMB received initially HIT-SKK with MTX injection followed by hyperfractioned radiotherapy and HIT maintenance chemotherapy.
Tumor recurrence was observed in 27 patients (33%) with DNMB: eight as a local re-growth but in the other 19 cases—as combined local relapse/spinal metastases. Eighteen patients died from their disease (22%). Disease relapses also developed in eight patients (22%) with MBEN, but all were diagnosed as an isolated local regrowth and no MBEN patients died during the follow-up period. Progression-free survival times (PFS) were similar for both histological variants (log-rank test; p = 0.23), but overall survival (OS) was significantly worse for patients harboring DNMB (log-rank test, p < 0.01; see Tables 2 and 3, and Fig. 3). All eight patients with recurrent MBEN received salvage high-dose chemotherapy (HD/CHT) with stem cell rescue (SCR), resulting in a complete response of the relapsed tumor without re-operation or radiotherapy. Patients with recurrent DNMB were treated with various modalities. Fourteen patients were re-operated and received second-line CSI. All these 27 patients received chemotherapy as a second-line treatment (12—HD/CHT with SCR and 14—HIT-REZ 2005 chemotherapy protocol). However, the applied salvage treatment regimens were not different in terms of their effect on the DNMB patients’ final outcomes.
DNA methylation patterns and copy number alterations
DNA of all 119 DNMB and MBEN samples was analyzed on either Illumina 450k (n = 57) or 850k/EPIC (n = 62) Bead Chip methylation arrays [13]. Unsupervised hierarchical clustering of 119 DNMB/MBEN DNA profiles performed together with other MB samples representing all four molecular subgroups showed that these tumors were grouped together within the SHH MB cluster as it has been reported before [4, 11, 13, 21]. These results confirm an epigenetic commonality of all histopathological variants within the SHH MB molecular cohort.
Unsupervised clustering of 119 DNMB/MBEN samples revealed two main clusters which were clearly associated with the patients’ age; “infant” cluster (n = 86; 50 DNMB and 36 MBEN) and “children” cluster (n = 33; DNMB only) (Fig. 1c). In line with previous reports, neither DNMB nor MBEN composed separate epigenetic cohorts, and their profiles were intermixed within the “infant” cluster [11, 13, 23, 24]. These data were also confirmed with t-SNE analysis (not shown). Also, attempts to find differentially methylated sites and annotated pathways between DNMB and MBEN histological variants were unsuccessful, indicating that they are epigenetically highly similar. Finally, two additional epigenetic subsets were recognized within the “infant” SHH MB cohort, which did not differ in terms of tumor histology (Fig. 1d).
Using raw intensity values from the DNA methylation arrays, we also analyzed copy number aberrations (Table 1). For the DNMB cohort, the mean number of CNVs per tumor was 3.88 ± 2.1 and only 12/83 disclosed balanced genomes (14%). DNMB showed a high frequency of 9q loss (60%) with various oncogene amplifications detected in 26 samples (31%). Among the recurrently amplified oncogenes were MYCN (9 samples), PPM1D (5), TERT (4), CCND2 (3), MYCL1 (3), and MLH1 (3). Additionally, eight DNMB disclosed PTCH1 homozygous deletions. In contrast, a vast majority of MBEN (23/36; 64%) revealed no chromosomal aberrations at all. In the remaining 12 MBEN, the number of CNAs varied from one to five per tumor (mean 1.1 ± 0.8); 10q loss and gain of chromosome 2 were detected as recurrent aberrations. No high-level amplifications or homozygous deletions were detected across the entire MBEN cohort. Cytogenetic profiles of “infant” DNMB only were similar to the whole and “children” DNMB cohorts, but distinct from MBEN (Table 1).
Mutational landscape detected with targeted NGS
Using targeted NGS, we analyzed tumor and matched normal DNA for all 119 tumors as described [22, 23]. Single somatic nucleotide variants (SNVs) and small insertions/deletions were detected across all 119 samples studied. The mean number of alterations did not differ for DNMB and MBEN: 3.6 ± 1.2 and 3.7 ± 1.4 per tumor respectively (p = 0.74). Recurrent mutations affected three “prototypic” SHH-associated genes: PTCH1 (65/119; 55%), SUFU (17/119; 14%), and SMO (12; 10%), which all were mutually exclusive. Frequent mutations of KMT2D (23/119; 19%) were also found; all other non-recurrent mutations were scattered throughout the DNMB/MBEN cohort. There were no TP53 mutations in this tumor set. All 17 SUFU mutations but only 8/65 PTCH1 alterations (12%) were also present in patients’ germ-line DNA. However, there were no clinical patterns of nevoid basal cell carcinoma syndrome (Gorlin syndrome) at the time of diagnosis. PTCH1 mutations prevailed in DNMB (66% vs. 33% for MBEN; p < 0.01) with similar proportions for “infant” samples analyzed separately (62% vs 33%). Eight non-mutant “children” DNMB harbored homozygous PTCH1 deletion and, thereby, PTCH1 alterations were detected in 73% of these tumors. In contrast, SUFU mutations were frequent in MBEN (33% vs. 4% for DNMB; p < 0.01) as well as SMO and KMT2D alterations (17% vs. 7% for SMO and 33% vs. 15% for KMT2D;p < 0.01). Germ-line SUFU/PTCH1 mutations were identified in “infant” tumors only (25/86; 29%; 9 DNMB and 16 MBEN).
Gene fusions detected by RNA sequencing
We performed transcriptome sequencing in a subset of 75 cases with available material for DNMB (52) and MBEN (23), respectively. Reliable gene fusion transcripts were detected in 18/75 (24%) samples by various algorithms (see “Materials and methods” and Table 2) [21, 23, 30] and confirmed with direct RNA sequencing (Fig. 2). A vast majority of these gene fusions (17/18) were detected in DNMB (17/52 cases; 33%); they were frequently associated with CNVs/DNA rearrangements at the affected chromosomal regions and accompanied by changes in expression of the involved genes. Thus, three DNMB samples with MYCL1 amplifications disclosed recurrent RLF-FOXJ3 fusions at 1p34 (Fig. 2a, b), whereas two samples with MYCN amplification revealed FAM49A-NBAS (neuroblastoma amplified sequence) fusion at 2p24. Seven non-recurrent gene fusions were also identified in the other altered DNA regions (see Table 2). Notably, one DNMB with a prototypic 9q loss disclosed PTCH1-c9orf3 fusion (deletion) at 9q22 (Fig. 2c, d). This SHH MB revealed neither mutation nor homozygous deletion of PTCH1 suggesting a possible additional mechanism for its inactivation via deleterious gene fusion involving the coding sequence. In addition, some gene fusions were not tied to altered DNA regions (see Table 2) and had unclear functional roles. For example, a fusion between glioblastoma-associated genes GBAS and EGFR at balanced 7p11 (Fig. 2e, f) was accompanied with an elevated EGFR over-expression and aggressive tumor behavior (patient died after 8 months), suggesting a possible driving role. In contrast, only one MBEN with trisomy 2 (1/23; 4%) showed a fusion between LTBP1 and BIRC6 at 2p22.
Gene fusions detected in DNMB and confirmed by RNA direct sequencing. a DNMB with MYCL1 amplification at 1p34 (arrow) disclosed fusion (b) between RLF and FOXJ3 genes. c DNMB with “prototypic” 9q loss (arrow) revealed deleterious fusion between (d) PTCH1 and c9orf3.e DNMB with MYCN amplification (arrow) showed fusion (f) between glioblastoma-associated genes GBAS and EGFR at the unaffected 7p11 region
Transcriptome analysis by RNA sequencing
In 75 samples with sufficient data available, we also did RNA sequencing-based gene expression profiling to assess transcriptional differences between DNMB and MBEN and consider the possible role in oncogenesis of these SHH MB variants. Notably, almost all transcriptome profiles of DNMB and MBEN were clustered separately from RNA signatures generated for Group 3 MB at the same way, thus confirming a reliability of RNA sequencing data and excluding batch effects (Suppl. Figure 2). Initially, we performed unsupervised hierarchical clustering (Fig. 3a), and principal component (Fig. 3b) and tSNE (Suppl. Figure 3) analyses based on top 500 high-variance genes which disclosed that expression signatures of all 23 MBEN were clustering separately from most of the DNMB. Only 4 RNA profiles generated for tumors diagnosed as “DNMB” in very young patients (mean age—1.2; all still alive) were clustered together with MBEN, thus suggesting some shortcomings in distinct identification of SHH MB histology. All clusters remained stable when we used various amounts of most highly variable expressed genes (1000 and 250; not shown), thus confirming that transcriptome profiling data are reliable to discriminate between DNMB and MBEN. This trend remained stable when we analyzed RNA profiles of 53 tumors from patients younger than four years (Suppl. Figure 4). Nevertheless, we were unable to identify clear cut transcriptomic discrimination between the “infant” and “children” DNMB cohorts which, in turn, revealed a significant variability in distribution of their RNA profiles. We also compared expression levels of the gene sets differentially activated between DNMB and MBEN and performed a supervised clustering with these selected candidates (Fig. 3c; Suppl. Table 1). Thus, a set of the most-confident 20 genes overexpressed either in MBEN or DNMB allowed one to discriminate between these SHH MB variants. Consistent with the RNA sequencing experiments, the nine MBEN analyzed by Real-Time Quantitative Reverse-Transcription PCR (RQ-PCR) displayed statistically significant higher expression levels of RIMS as compared to ten DNMB (Mann–Whitney U test, p < 0.01). Within the DNMB cohort, higher MYO7A expression measured by RQ-PCR as compared to MBEN was identified (Mann–Whitney U test, p < 0.01) (Suppl. Figure 5). Comparing methylome and RNA sequencing data, we did not find any changes in methylation status for genes differentially expressed between DNMB and MBEN, thus suggesting that transcriptional activity of these discriminatory genes is not regulated by epigenetic events. Further, to delineate characteristic signaling patterns for each of these SHH MB variants, we performed pathway annotation using Gene Ontology analysis (Fig. 3d and Suppl. Tables 2 and 3) [2, 29]. MBEN were enriched with gene sets associated with neurotrophic regulation, neuronal and synaptic transmission, and neuroendocrine secretion. In contrast, transcriptome signatures of DNMB were characterized by genes involved in NOTCH signaling which is associated with self-renewal of MB cells and metastases initiation and, also, in phototransduction pathway which has been detected in Group 3 MB [12, 21, 30]. We were unable to identify pathways specifically annotated for children and infant DNMB, respectively.
Gene expression profiling data obtained after RNA sequencing. a Heatmap of unsupervised hierarchical cluster analysis (Row Z Scores: green: from 0 to − 6; red: from 0 to 6). b Scale of principal component analysis (rhombuses—samples with SUFU/PTCH1 germ-line alterations; circles—non-mutant samples). Both methods revealed expression signatures of MBEN (red) clustering separately from DNMB profiles (black). Notably, transcriptional profiles generated for DNMB revealed a significant variability in their distribution. c A set of 20 top most-confident genes differentially overexpressed in MBEN (red) and DNMB (black), respectively, allows one to discriminate clearly between these tumor cohorts (Row Z Scores; green: from 0 to − 6; red: from 0 to 6). d Gene ontology analysis disclosed that MBEN transcriptional profiles (blue) were associated with neuronal metabolism, synaptic transmission, and neuroendocrine secretion. In contrast, expression profiling signatures of DNMB (red) were characterized by genes involved in NOTCH signaling, phototransduction and sucrose metabolism pathways
Survival analysis
Correlation of clinical and molecular parameters with patient outcomes disclosed some significant associations (Tables 3 and 4). Univariate survival analysis of all 119 patients revealed that MBEN histology was associated with favorable OS but not PFS (Fig. 4a, b). However, advanced metastatic stages (M2–3), the presence of oncogene amplification, germ-line PTCH1/SUFU mutations, and 17p loss were associated with poor outcomes in terms of both PFS and OS; all reached an independent significance level. Survival analysis performed for all DNMB and “infant” DNMB, respectively, disclosed that most of these variables were also associated with patients’ PFS and OS. However, loss of 9q was associated with favorable survival within the “infant” DNMB cohort only. Survival analysis across the MBEN cohort revealed that a presence of germ-line PTCH1/SUFU mutations was significantly associated with PFS, whereas other variables showed no prognostic relevance. In addition, we revealed no PFS and OS differences for the patients from two different epigenetic clusters outlined within the “infant” SHH MB cohort (See Figs. 1d, 4c, d). However, molecular subgroups of SHH MB identified during expression profiling analysis as a “cluster MBEN plus DNMB” revealed favorable patients’ outcomes in terms of the PFS (p < 0.01) and OS (p < 0.01) in comparison to transcriptome clusters composed of “pure DNMB” samples (Fig. 4e, f).
a, b Survival analysis for all cohort of DNMB and MBEN shows that overall survival (OS) (a) for DNMB patients was significantly worse (log-rank; p < 0.01). However, progression-free survival (PFS) (b) did not differ significantly between these SHH MB variants (log-rank; p = 0.23) revealing only a trend to better event-free outcomes for MBEN patients. (c, d ) Survival analysis for two epigenetic “infant” SHH MB subgroups revealed no differences in terms of (c) OS (log-rank; p = 0.42) and (d) PFS (log-rank; p = 0.34). (e, f) Survival analysis for SHH MB subgroups detected by transcriptome profiling revealed significantly better (e) OS (log-rank; p < 0.01) and (f) PFS (log-rank; p < 0.01) for cluster “MBEN and DNMB” in comparison to samples composed of “pure DNMB” clusters
Discussion
SHH-activated MB histologically present with a wide spectrum of tumor morphologies; however, the DNMB and MBEN patterns are more frequent, accounting for 70–80% of these tumors [1, 6, 11, 13, 18, 23]. Moreover, both DNMB and MBEN are quite often considered as kindred SHH MB variants which overlap both histologically and molecularly [1, 15, 19, 23, 24]. Indeed, the “canonical” histological appearance of both DNMB and MBEN includes an obligatory presence of two structural components and sometimes it can be difficult to discriminate between these tumors microscopically, especially in small biopsy samples [19]. Besides, both these histological variants harbor similar SHH-associated gene mutations and epigenetic signatures that also point to their biological affinity [11, 13, 21, 24].
Most of the MB clinical trials did not subdivide between DNMB and MBEN including them as a common histological variant and emphasizing similar recurrence rates [7, 23, 24, 30]. However, some other studies reported on better overall survival for MBEN, wherein metastatic disease at presentation did not affect the favorable prognosis, in contrast to DNMB [16, 25, 26]. Our data are in line with these findings, because MBEN patients in the current series disclosed only local disease relapses and an excellent long-term survival. In contrast, DNMB showed a more aggressive clinical behavior recurring frequently as disseminated disease with unfavorable final outcomes. Such variability in the treatment results of patients with DNMB and MBEN raises some doubts on their histo-molecular similarity announced before and, in turn, necessitates further unveiling of biological mechanisms which could explain these clinical differences.
Epigenetically, neither DNMB nor MBEN formed a distinct molecular subtype and all MBEN were clustered in close proximity to the “infant” DNMB samples, thus suggesting a clear epigenetic kinship within this age-related cohort of SHH MB. These findings are in concordance with the previously promoted suggestion that the epigenetic state of any MB is dictated principally at the time of tumor initiation and remains stable during tumor development and clinical progression [11, 13, 21, 31]. Also, in line with the previously reported data, two histologically intermixed epigenetic subsets were identified within the “infant” SHH MB cohort [24]. However, event-free survival for these outlined molecular subtypes did not differ significantly and this is in contradiction to the data obtained recently for the SJYC07 clinical trial [24]. This discrepancy could be partly explained by therapeutic differences, because all our patients received standard HIT-SKK protocol with MTX injection [24, 25] but not the risk-adapted approach applied in the SJYC07 multicenter trial.
Integrated molecular analysis revealed that the biological relationship of DNMB and MBEN is quite complicated, with their DNA and RNA profiles showing differences in terms of tumor cytogenetics (various CNVs, DNA breakpoints and gene fusions in DNMB vs. predominately flat and stable genomes in MBEN), dominant SHH-activating mutations (PTCH1 in DNMB vs. SUFU in MBEN) and, finally, in transcriptional landscapes (NOTCH signaling and phototransduction gene signatures in DNMB vs. neuron-associated pathways in MBEN). Moreover, DNA methylation profiles cannot recapitulate DNMB/MBEN gene expression signatures suggesting that disclosed transcriptional variability is not associated with epigenetic dysregulation. These findings are convincing evidence that, despite an apparent histo-molecular resemblance, DNMB and MBEN are biologically different tumor entities within the SHH MB family explaining the striking variability of their posttreatment outcomes. However, the spatio-temporal sequence of these molecular events and their distinct role in DNMB/MBEN pathogenesis are still unclear. Perhaps, both infant DNMB and MBEN could have a similar source/cell of origin, but their initiation and further progression are associated with different molecular events which, in turn, may activate variable transcriptional programs resulting in biologically divergent tumor phenotypes. On the other hand, two age-associated DNMB subgroups based on epigenetic differences revealed similarities in terms of other molecular and clinical features. It suggests that although DNMB could have different sources and/or time of origin, their further development is associated with acquisition of similar mutational events, DNA aberrations and activation of cancer-associated signaling pathways which, in total, results in similar clinical courses of all DNMB patients.
Disclosed clinico-biological differences between DNMB and MBEN may indicate the necessity of different approaches for patients’ risk stratification and choice of optimal treatment in future. Thus, germ-line PTCH1/SUFU mutation is a single prognosticator of MBEN local regrowth. Consequently, these patients could be stratified on the presence of germline alterations to receive more intense CHT upfront (instead of “standard” HIT-SKK) with the aim of optimal local disease control [3, 5, 8, 9, 11, 13, 31]. Clinically relevant molecular stratification of the pediatric DNMB patients is currently less clear and the question of whether outlined patients’ subgroups require specific therapeutic approaches remains to be defined. A few clinic-molecular patterns (M stage, oncogene amplifications, germ-line SUFU/PTCH1 mutations) could be considered as prognostic markers for DNMB, but their optimal exploitation must to be determined in upcoming clinical trials. Furthermore, other molecular aberrations identified in DNMB, such as gene fusions or expression profiling signatures, may also have potential as either outcome prognosticators or even targets for molecular therapy, and their further evaluation is clearly warranted.
Taking into an account the certain clinical importance of accurate MBEN identification and sometimes insufficient yield of tiny biopsy samples, we could recommend transcriptome analysis as a molecular tool for reliable DNMB/MBEN discrimination. For example, a NanoString gene panel (or RQ-PCR-based tools) applying top genes differentially expressed for DNMB and MBEN might be developed for diagnostically challenging samples. Alternatively, proteins encoded by differentially expressed genes may be also considered as surrogate immunohistochemical markers for DNMB/MBEN discrimination, but their application for small tumor samples is difficult.
In conclusion, well-established histological SHH MB variants traditionally designated as DNMB and MBEN exhibit a quite similar methylation profiles but biologically are driven by different transcriptional programs. Pathways associated with synaptic transmission, neuronal differentiation and metabolism are activated in MBEN suggesting some possible rationale behind the now obsolete term “cerebellar neurocytoma”, and potentially explaining the ganglionic maturation that can be observed in MBEN. In contrast, NOTCH signaling and phototransduction pathways are activated in DNMB, possibly contributing to their aggressive behavior and a tendency towards post-treatment dissemination. Consequently, we recommend a transcriptome analysis as an optimal molecular tool to discriminate accurately between DNMB and MBEN for patients’ risk stratification in clinical trials and choice of treatment strategy. Moreover, molecular events recently identified in DNMB such as gene fusions or expression profiling signatures could be considered in the future as potential molecular targets for novel therapeutic interventions in treatment-resistant cases.
References
Abdel-Baki MS, Boué DR, Finlay JL, Kieran MW (2018) Desmoplastic nodular medulloblastoma in young children: a management dilemma. Neuro Oncol 20:1026–1033. https://doi.org/10.1093/neuonc/nox222
Bindea G, Mlecnik B, Hackl K, Charoentong P, Tosolini M, Kirilovsky A et al (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093. https://doi.org/10.1093/bioinformatics/btp101
Brugières L, Remenieras A, Pierron G, Varlet P, Forget S, Byrde V et al (2012) High frequency of germline SUFU mutations in children with desmoplastic/nodular medulloblastoma younger than 3 years of age. J Clin Oncol 30:2087–2093. https://doi.org/10.1200/JCO.2011.38.7258
Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B et al (2017) Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31:737–754. https://doi.org/10.1016/j.ccell.2017.05.005
Dobin A, Gingeras TR (2016) Optimizing RNA-seq mapping with STAR. Methods Mol Biol 1415:245–262. https://doi.org/10.1007/978-1-4939-3572-7_13
Eberhart CG, Kepner JL, Goldthwaite PT, Kun LE, Duffner PK, Friedman HS et al (2002) Histopathologic grading of medulloblastomas: a Pediatric Oncology Group study. Cancer 94:552–560
Ellison DW, Kocak M, Dalton J, Felice G, Catherine G, Pietsch T et al (2011) Definition of disease-risk stratification groups in childhood medulloblastoma using combined clinical, pathologic, and molecular variables. J Clin Oncol 29:1400–1407
Garrè ML, Cama A, Bagnasco F, Morana G, Giangaspero F, Brisigotti M et al (2009) Medulloblastoma variants: age-dependent occurrence and relation to Gorlin syndrome—a new clinical perspective. Clin Cancer Res 15:2463–2471. https://doi.org/10.1158/1078-0432.CCR-08-2023
Guerrini-Rousseau L, Dufour C, Varlet P, Masliah-Planchon J, Bourdeaut F, Guillaud-Bataille M et al (2018) Germline SUFU mutation carriers and medulloblastoma: clinical characteristics, cancer risk and prognosis. Neuro Oncol 20:1122–1132. https://doi.org/10.1093/neuonc/nox228
Hovestadt V, Remke M, Kool M, Pietsch T, Northcott PA, Fischer R et al (2013) Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol 125:913–916. https://doi.org/10.1007/s00401-013-1126-5
Kool M, Jones DT, Jäger N, Northcott PA, Pugh TJ, Hovestadt V et al (2014) Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25:393–405. https://doi.org/10.1016/j.ccr.2014.02.004
Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA et al (2012) Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 123:473–484. https://doi.org/10.1007/s00401-012-0958-8
Korshunov A, Sahm F, Stichel D, Schrimpf D, Ryzhova M, Zheludkova O et al (2018) Molecular characterization of medulloblastomas with extensive nodularity (MBEN). Acta Neuropathol 136:303–313. https://doi.org/10.1007/s00401-018-1840-0
Lafay-Cousin L, Smith A, Chi SN, Wells E, Madden J, Margol A et al (2016) Clinical, pathological, and molecular characterization of infant medulloblastomas treated with sequential high-dose chemotherapy. Pediatr Blood Cancer 63:1527–1534. https://doi.org/10.1002/pbc.26042
Lamont JM, McManamy CS, Pearson AD, Clifford SC, Ellison DW (2004) Combined histopathological and molecular cytogenetic stratification of medulloblastoma patients. Clin Cancer Res 10:5482–5493
Lannering B, Rutkowski S, Doz F, Pizer B, Gustafsson G, Navajas A et al (2012) Hyperfractionated versus conventional radiotherapy followed by chemotherapy in standard-risk medulloblastoma: results from the randomized multicenter HIT-SIOP PNET 4 trial. J Clin Oncol 30:3187–3193. https://doi.org/10.1200/JCO.2011.39.8719
Liao Y, Smyth GK, Shi W (2014) FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK et al (2016) The 2016 WHO classification of tumours of the nervous system: a summary. Acta Neuropathol 131:803–820. https://doi.org/10.1007/s00401-016-1545-1
McManamy CS, Pears J, Weston CL, Hanzely Z, Ironside JW, Taylor RE et al (2007) Nodule formation and desmoplasia in medulloblastomas-defining the nodular/desmoplastic variant and its biological behavior. Brain Pathol 17:151–164
McPherson A, Hormozdiari F, Zayed A (2011) deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 7:e1001138. https://doi.org/10.1371/journal.pcbi.1001138
Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T et al (2017) The whole-genome landscape of medulloblastoma subtypes. Nature 547:311–317. https://doi.org/10.1038/nature22973
Okonechnikov K, Imai-Matsushima A, Paul L (2016) InFusion: advancing discovery of fusion genes and chimeric transcripts from deep RNA-sequencing data. PLoS ONE 11:e0167417. https://doi.org/10.1371/journal.pone.0167417
Pietsch T, Schmidt R, Remke M, Korshunov A, Hovestadt V, Jones DT et al (2014) (2014) Prognostic significance of clinical, histopathological, and molecular characteristics of medulloblastomas in the prospective HIT2000 multicenter clinical trial cohort. Acta Neuropathol 128:137–149. https://doi.org/10.1007/s00401-014-1276-0
Robinson GW, Rudneva VA, Buchhalter I, Billups CA, Waszak SM, Smith KS et al (2018) Risk-adapted therapy for young children with medulloblastoma (SJYC07): therapeutic and molecular outcomes from a multicentre, phase 2 trial. Lancet Oncol 19:768–784. https://doi.org/10.1016/S1470-2045(18)30204-3
Rutkowski S, Bode U, Deinlein F, Ottensmeier H, Warmuth-Metz M, Soerensen N et al (2005) Treatment of early childhood medulloblastoma by postoperative chemotherapy alone. N Engl J Med 352:978–986
Rutkowski S, von Hoff K, Emser A, Zwiener I, Pietsch T, Figarella-Branger D et al (2010) Survival and prognostic factors of early childhood medulloblastoma: an international meta-analysis. J Clin Oncol 28:4961–4968. https://doi.org/10.1200/JCO.2010.30.2299
Sahm F, Schrimpf D, Jones DT, Meyer J, Kratz A, Reuss D et al (2016) Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol 131:903–910. https://doi.org/10.1007/s00401-015-1519-8
Sahm F, Schrimpf D, Stichel D, Jones DTW, Hielscher T, Schefzyk S et al (2017) DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol 18:682–694
Sievers P, Stichel D, Schrimpf D, Sahm F, Koelsche C, Reuss DE et al (2018) FGFR1:TACC1 fusion is a frequent event in molecularly defined extraventricular neurocytoma. Acta Neuropathol 136:293–302. https://doi.org/10.1007/s00401-018-1882-3
Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC et al (2012) Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 123(4):465–472. https://doi.org/10.1007/s00401-011-0922-z
Waszak SM, Northcott PA, Buchhalter I, Robinson GW, Sutter C, Groebner S et al (2018) Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol 19(6):785–798. https://doi.org/10.1016/S1470-2045(18)30242-0
Acknowledgements
The authors thank I. Leis, U. Lass, A. Habel, K. Lindenberg, and S. Kocher for excellent technical support and the microarray unit of the DKFZ Genomics and Proteomics Core Facility for providing Illumina DNA methylation array-related services. A. Korshunov is supported by the Helmholtz Association Research Grant (Germany).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Korshunov, A., Sahm, F., Okonechnikov, K. et al. Desmoplastic/nodular medulloblastomas (DNMB) and medulloblastomas with extensive nodularity (MBEN) disclose similar epigenetic signatures but different transcriptional profiles. Acta Neuropathol 137, 1003–1015 (2019). https://doi.org/10.1007/s00401-019-01981-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-019-01981-6