Abstract
Medulloblastoma with extensive nodularity (MBEN) is a rare histological variant of medulloblastoma (MB). These tumors are usually occurring in the first 3 years of life and are associated with good prognosis. Molecular analyses of MBEN, mostly limited to single cases or small series, have shown that they always classify as sonic hedgehog (SHH)-driven MB. Here, we have analyzed 25 MBEN through genome-wide DNA methylation, copy-number profiling and targeted next-generation sequencing. Results of these analyses were compared with molecular profiles of other SHH MB histological variants. As expected, the vast majority of MBEN (23/25) disclosed SHH-associated epigenetic signatures and mutational landscapes but, surprisingly, two MBEN were classified as Group 3/4 MB. Most MBEN classified as SHH MB displayed SHH-related and mutually exclusive mutations in either SUFU, or PTCH1, or SMO at similar frequencies. However, only SUFU mutations were also identified in the germ-line. Most of SUFU-associated MBEN eventually recurred but patients were treated successfully with second-line high-dose chemotherapy. Altogether, our data show that risk stratification even for well-recognizable histologies such as MBEN cannot rely on histology alone but should include additional molecular analyses such as methylation profiling and DNA sequencing. For all patients with “MBEN” histology, we recommend sequencing SUFU and PTCH1 in the tumor as well as in the germ-line for further clinical stratification and choice of the optimal treatment strategy upfront.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The histological entity medulloblastoma (MB), one of the most common malignant brain tumors in childhood, comprises distinct histological variants and molecular subgroups. According to the current international consensus, four main molecular MB subgroups (WNT, SHH, Groups 3 and 4) with divergent histology, biology and clinical outcomes exist [10, 18]. Within the SHH MB subgroup, all histological variants of MB may occur, including classic, desmoplastic/nodular, large cell/anaplastic and medulloblastoma with extensive nodularity (MBEN), each associated with distinct outcomes [2, 9, 10, 14]. The other three subgroups mostly include classic or large cell anaplastic histologies [4, 9, 10, 14]. Among these different histological MB variants, MBEN is the rarest tumor variant previously also designated as “cerebellar neuroblastoma” [4, 5, 7, 9, 14, 17]. These tumors are usually occurring in the first 3 years of life and are associated with good to excellent prognosis [7, 10, 14, 17]. At a molecular level, MBEN show SHH pathway activation caused by germ-line and/or somatic inactivating mutations of PTCH1 or SUFU [3, 6, 8, 9, 15, 18]. Nevertheless, molecular analysis of MBEN has been limited to case reports or small series of samples. Previous clinical trials have suggested that MB patients can be risk stratified based on tumor histology and patients’ age as infant patients with histologically diagnosed desmoplastic/nodular MB or MBEN have a much better outcome than patients with classic or large cell/anaplastic MB variants [1, 2]. However, a recent infant MB study of the Children’s Oncology Group has been closed prematurely because of unexpected treatment failures [1], suggesting that risk stratification based on histology and other clinical parameters alone might not be sufficient. Indeed, several recent reports have shown that further molecular and clinical heterogeneity in MB exist, also within the infant MB population which is mainly restricted to the SHH and Group 3 subgroups, possibly explaining some of these unexpected treatment failures [1, 2, 14]. Some molecular subtypes of SHH MB were enriched with MBEN histology but also included other histological variants [4, 9, 13]. Consequently, a molecular classification of infant MBs is likely the preferred method for their risk stratification over histology-based classification, but the exact definition criteria are still lacking. In the current study, we have analyzed a representative MBEN cohort using genome-wide DNA methylation profiling, copy-number profiling and targeted next-generation sequencing to clarify the nosologic position of MBEN in the context of other SHH MB histological variants. Through improved understanding of the molecular basis of these rare tumors, we expect that stratification of MBEN patients according to tumor biology can be further improved leading to better risk-adapted treatments for these patients.
Materials and methods
Patient population
Tissue samples were obtained from 25 patients with histological diagnosis “Medulloblastoma with extensive nodularity (MBEN)” according to the 2016 WHO classification of tumors of the central nervous system (CNS) [14]. These neoplasms were diagnosed after pathological screening of 787 MB samples with various tumor histologies and a few of them were included in our previous studies [9, 10, 13]. The observed incidence of MBEN, accounting for 3.2% of all MB variants, is in line with other published data [1, 7, 14, 17]. All these patients were initially diagnosed between 1997 and 2016 at the Burdenko Neurosurgical Institute in Moscow and received combined treatment according to HIT_SKK protocols. Informed consent was obtained from all patients’ parents or other relatives.
DNA methylation analysis
DNA was extracted from tumors and analyzed using the Illumina Human Methylation 450 or 850 k/EPIC BeadChip array as previously described [11, 13, 14]. All DNA methylation analyses were performed in R version 3.3.0 (R Development Core Team). Raw signal intensities were obtained from IDAT-files using the minfi Bioconductor package version 1.18.2. Each sample was individually normalized by performing a background correction (shifting of the 5% percentile of negative control probe intensities to 0) and a dye-bias correction (scaling of the mean of normalization control probe intensities to 10,000) for both color channels. No further normalization or transformation steps were performed, and standard beta-values were used for downstream methylation analyses. The following criteria were applied to filter out probes prone to yield inaccurate methylation levels: Removal of probes targeting the X and Y chromosomes (n = 11,551), removal of probes that overlap common SNPs (dbSNP132 Common) within the CpG or the following base (n = 7998), and removal of probes not mapping uniquely to the human reference genome (hg19) (n = 3965). To enable comparability between the Illumina Infinium Human Methylation 450 and 850 k/EPIC arrays, we removed all probes not represented on the 450 k array (n = 32,260). In total, 428,799 probes were kept for analysis.
To consider a distinct molecular allocation of MBEN, we performed unsupervised clustering of epigenetic data generated for these tumors together with a cohort of 542 molecularly defined pediatric MB tumors encompassing all four molecular groups (WNT; SHH; Groups 3 and 4). For unsupervised hierarchical clustering, we selected the 10,000 most variably methylated probes across the dataset. Clustering was performed using the beta values of the n = 10,000 most variably methylated probes as measured by standard deviation. Samples were clustered using Pearson’s correlation coefficient as the distance measure and average linkage (x-axis). Methylation probes were reordered by hierarchical clustering using Euclidean distance and average linkage (y-axis). Additional analysis of tumor subgroups was performed using a t-distributed stochastic neighbor embedding (t-SNE)-based approach. This was computed using the R-package Rtsne, version 0.13, with a perplexity of 15 and 20 iterations [13]. Copy number profiles were generated using the ‘conumee’ package for R (https://www.bioconductor.org/packages/release/bioc/html/conumee.html) [11]. To evaluate the methylation status of the MGMT promoter region, we evaluated beta values of probes cg12434587 and cg12981137 using the MGMT_STP27 logistic regression model [13].
Targeted next-generation sequencing (NGS)
Molecular barcode-indexed ligation-based sequencing libraries were constructed using 200 ng of sheared DNA. Twenty-three paired tumor and blood samples yielded sufficient DNA amounts for analysis. Libraries were enriched by hybrid capture with custom biotinylated RNA oligo pools covering exons of 130 cancer-associated genes [16]. Paired-end sequencing was performed using the NextSeq 500 (Illumina). Sequence data were mapped to the reference human genome using the Burrows–Wheeler Aligner and were processed using the publicly available SAM tools. Recurrent gene mutations were also assessed with residual DNA from the same pool used for sequencing by polymerase chain reaction followed by direct Sanger sequencing of the corresponding exons.
Statistics
The distribution of overall survival (OS) was calculated according to the Kaplan–Meier method using the log rank test for significance. OS was calculated from the date of diagnosis until death of patient from disease or last contact for patients who were still alive. For multivariate analysis, Cox proportional hazards regression models were used. Estimated hazard ratios are provided with 95% confidence intervals and a p value from the Wald test. Tests with a p value below 0.05 were considered significant.
Results
Clinical parameters of the MBEN patients
Basic clinical characteristics of all 25 MBEN patients included in this study are summarized in Table 1. Tumors occurred predominately in very young children with a median age of 11 months (range 3–28 months) all but one younger than 2 years. There was a small preponderance of male patients in this cohort (59 vs. 41% for female, male: female ratio = 1.4:1). All MBEN were located in the cerebellar vermis also involving neighboring hemispheres. At neuroradiological examination, the tumors appeared as large multinodular lesions with “prototypic” grape-like structures. Most (22/25; 88%) of the patients disclosed M0 stage and only three (12%) patients were diagnosed with M3 stage. At the time of diagnosis, no patients met the diagnostic criteria of a naevoid basal cell carcinoma syndrome (NBCCS) [6]. All patients were operated upon and gross total tumor resection was achieved in 15/25 (60%) of cases. After operation, all patients with MBEN received chemotherapy in two regimens: 22 M0 patients were treated with HIT_SKK protocols (97; 2000 or 2014) with intraventricular methotrexate (MTX) injection, while the remaining three M3 patients received high-dose chemotherapy (HD/CHT) with stem cell rescue (SCR) [12, 15]. The median follow-up for all patients was 54 months (range 14–168 months). Tumor recurred as a local relapse in five patients (median progression-free survival 46 months; range 8–168 months; 5-year PFS—80%) and all these patients received salvage HD/CHT (followed with a complete response for relapsed tumor) without re-operation and/or radiotherapy. No post-treatment metastatic dissemination was developed. All 25 patients with MBEN are still alive at the follow-up closure, regardless of postoperative clinical course including five patients in remission after relapse (median overall survival 54 months; range 14–168 months; 5-year OS—100%). During follow-up period, no patients developed additional tumors.
Histopathology of the MBEN cohort
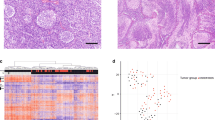
Histopathologically, all 25 tumors analyzed disclosed similar microscopic appearance typical for MBEN, as described before [1, 5, 7, 14, 17]. A clear lobular architecture composed of two components was seen in all tumors (Fig. 1a, b). One component consisted of large reticulin-free zones containing populations of small cells disclosing features of neurocytic differentiation. These cells exhibited stream-like patterns, were embedded in neuropil-like matrix and also disclosed strong nuclear expression of NeuN (Fig. 1a, b). The other component consisted of internodular reticulin-rich zones containing densely packed undifferentiated and mitotically active cells. An extension of this component was varied from one area to another and looked as markedly reduced in some places. MIB1 nuclear expression was almost absent in nodular zones but was counted as relatively high in internodular areas.
Pathological similarity of MBEN clustered with SHH MB (a) and Group 3 MB (b) (H&E; NeuN immunostaining as insertion). Both samples show a clear lobular architecture composed of two components: large reticulin-free zones with intense NeuN nuclear expression and internodular reticulin-rich zones. c Unsupervised clustering of DNA methylation patterns for 542 tumors diagnosed as medulloblastoma (MB). Shown is a two-dimensional representation of pairwise sample correlations using the 10,000 most variably methylated probes by t-distributed stochastic neighbor embedding (tSNE) dimensionality reduction. Most of MBEN samples (black spots) were clustered together with SHH MB (red spots) but two MBEN were allocated to Group 3/4 MB (yellow and green spots). d, e CNV profile for SHH MBEN discloses only loss of 10 (D), whereas CNV profile for MBEN clustered with Group 3 MB shows trisomy 17
DNA methylation patterns, cytogenetic alterations and data of mutational analysis
DNA of all 25 MBEN samples was analyzed on Illumina 450 k (17) and 850 k/EPIC (8) Bead Chip methylation arrays to investigate their epigenetic signatures and cytogenetic profiles [11, 13]. Clustering analyses of the DNA methylation profiles of these 25 MBENs together with 542 other MBs representing all 4 molecular MB subgroups and using t-Distributed Stochastic Neighbor Embedding (t-SNE) algorithms, showed that, as expected, the vast majority of the MBEN (23/25; 92%) clustered together with the SHH MB tumors (Fig. 1c). They did not form any separate cluster. Surprisingly, methylation profiles of two MBEN cases did not cluster with the SHH MB tumors, but with either Groups 3 or 4 (Fig. 1c). Histopathological re-investigation of these two non-SHH MBEN cases revealed no microscopic differences between these 2 tumors and all remaining 23 neoplasms that were classified as SHH MB (see Fig. 1b). Immunohistochemistry with GAB1 protein (marker SHH pathway activation [14, 18]) revealed no staining in these tumor samples in contrast to the remaining 23 SHH MBEN which were strongly GAB1 positive. However, these two MBEN samples with unusual epigenetic signatures disclosed no OTX2 immunoexpression.
All these molecular allocations remained stable even when we used more or less differentially methylated CpG sites (7000; 5000; 2000 and 800; SD > 0.30, respectively (data not shown), confirming that the obtained DNA methylation data are robust and reliable to discriminate between different molecular variants of MBEN. MGMT was unmethylated in all MBEN samples.
Using SNP information contained on the DNA methylation arrays, we also analyzed copy number aberrations (CNA) in all MBENs (Table 2). A vast majority of MBENs (16/25; 64%) revealed no chromosomal aberrations at all, showing absolutely balanced genomes. In the remaining nine MBENs, the number of CNAs varied from one to nine per tumor (mean 1.2 + 0.3). For SHH MBEN, only 10q loss (Fig. 1d) and gain of chromosome 2 were detected as recurrent aberrations (22 and 12%, respectively). In contrast, the two MBEN classified as non-SHH MB both had a relatively high number of CNAs (7 and 10, respectively), and in both cases trisomy of chromosome 17 was detected (Fig. 1e). No high-level amplifications or homozygous deletions were detected across the entire MBEN cohort.
Using targeted NGS, we analyzed accessible amounts of tumor DNA for 23 MBEN, including both non-SHH MBENs. The panel sequencing analysis also included matched normal DNA controls for all these samples. After filtering for common polymorphisms (see “Methods”), nonsynonymous and non-polymorphic variants were called across the series within the coding regions of 130 sequenced genes. Single somatic nucleotide variants (SNVs) and small insertions/deletions were detected across all 23 MBEN samples, with individual tumor SNV counts ranging from one to three per tumor (mean 1.5; Table 2). Recurrent gene mutations (present in more than 10% of samples sequenced) included only SUFU (5/23; 24%); PTCH1 (5/23; 24%); SMO (4/23; 20%), and KMT2D (4/23; 20%). All other non-recurrent mutations were scattered throughout this MBEN cohort. All SHH-associated mutations (SUFU, PTCH1 and SMO) were mutually exclusive and they were detected only in MBEN with SHH signature, covering up to 70% of these tumors. There were no differences in cytogenetic profiles for SHH MBEN with and without SHH pathway mutations. All five SUFU mutations were also present in the germ-line DNA of these patients. However, there were no clinical patterns of NBCCS in affected patients at the time of diagnosis. On the other hand, all PTCH1 or SMO mutations were identified as exclusively somatic alterations. Both MBEN samples molecularly classified as non-SHH MB had no SHH pathway mutations, supporting their classification as non-SHH.
Comparing detected molecular findings with MBEN patients’ outcome, we revealed that all five recurrent tumors were molecular SHH MBEN and four of them were associated with germ-line SUFU alterations. Both patients carrying Group 3/4 MBEN were treated with HD/CHT with SCR, and are currently in complete remission at 26 and 63 months follow-up, respectively. Univariate PFS analysis for clinical and molecular parameters across the whole MBEN cohort and SHH MBEN subset only revealed that presence of SUFU alterations was significantly associated with tumor recurrence (log-rank test; p < 0.01). None of the other tested clinical or molecular variables (age older or younger 1 year, M stage, volume of tumor resection, recurrent CNVs and other SHH-associated mutations) were related with patients’ outcome. Multivariate analysis did not identify any significant prognostic parameter. This may perhaps be due to the small number of the cases available.
Clinico-molecular characterization of SHH MBs with various histology
We also performed comparative clinico-molecular analyses of SHH MBEN with three other histological variants of pediatric SHH MB—desmoplastic/nodular (DNMB; n = 34), classic (n = 27) and large cell/anaplastic (n = 21). Clustering and three-dimensional t-SNE analyses of DNA methylation data showed two distinct SHH MB subgroups (Fig. 2a). One of them included all MBEN and vast majority of DNMB thus suggesting an epigenetic similarity of these SHH MB variants, whereas another cluster was composed of classic and large cell SHH MB predominantly. Patients from MBEN/DNMB cluster were significantly younger (median age 2 years, range 0–8) than their classic/large cell counterparts (median age 9 years, range 3–17), confirming previous reports showing distinct methylation profiles between infant SHH MB and non-infant SHH MB [9].
a Unsupervised clustering of DNA methylation patterns for tumors diagnosed as SHH MB with various histological variants. Shown is a three-dimensional representation of pairwise sample correlations using the 10,000 most variably methylated probes by t-distributed stochastic neighbor embedding (tSNE) dimensionality reduction. MBEN samples were clustered together with desmoplastic nodular tumors (DNBM) whereas most of classic SHH MB were grouped with large cell TP53 mutant tumors. b Summarizing cytogenetic profiles for various SHH MB histological subtypes show almost balanced profiles for MBEN, obvious cytogenetic similarity for DNMB and classic SHH MB, and, also, genomic instability for large cell and TP53 mutant SHH MB (LCA)
Summarizing cytogenetic profiles for these four SHH MB histological subtypes (n = 132) are presented in Fig. 2b and Table 3. Surprisingly, despite the epigenetic differences between DNMB and classic SHH MB, the copy number profiles of these two histological subtypes are quite similar with a high frequency of “prototypic” aberrations like chromosome 2 gain, 9p gain, 9q loss and 10q loss. In contrast, MBEN and large-cell tumors occupied opposite ends of this “cytogenetic spectrum” disclosing either predominantly balanced (MBEN) or extremely instable CNVs profiles with chromothripsis patterns (large cell SHH MB), respectively. The mutational landscape for DNMB and classic SHH MB was also highly similar with a significant prevalence of PTCH1 alterations (about 70% of the cases) over other SHH-associated mutational events (Table 3). On the other hand, all three SHH-associated mutations were evenly distributed among MBEN (PTCH1—24%; SUFU—24%; SMO—20%), whereas all large cell SHH MB showed TP53 mutations only, which were not detected in any of the other three histologies. Survival analysis revealed PFS differences between large cell/TP53 mutant SHH MB and all three remaining TP53 wt histological variants (Fig. 3). However, in terms of OS, MBEN disclosed an excellent prognosis and treatment response (even in the second line treatment), whereas results of salvage therapy for patients with DNMB or classic SHH MB were not looking as being extremely successful.
Discussion
MBEN is a clearly defined histological variant of MB, which has been introduced into the current version of the WHO CNS tumor classification [7, 14, 17, 18]. Given their rarity, real incidence data, the distinct nosologic position and optimal treatment strategy for these tumors remain under discussion. Nevertheless, the biological behavior of MBEN is considered as favorable and, in general, quite similar to DNMB of infants [7, 17]. Moreover, long-term survivors and even occasional cases with neuronal maturation have been reported [1, 2, 5, 7, 17]. MBEN frequently carry mutations in genes encoding different members of the SHH pathways, and up to 30% of patients with MBEN could be associated with SUFU-related or PTCH1-related naevoid basal cell carcinoma syndrome [1, 3, 4, 6, 8, 9, 15].
In the current study, we have investigated a cohort of histologically diagnosed MBEN using genome-wide DNA methylation profiling as well as NGS mutational analysis [11, 16]. We also compared methylation profiles, copy number patterns and mutational landscapes of MBEN with those in other histological variants of pediatric SHH MB. Clinical characteristics of MBENs were similar to published before with very young patients’ age, low frequency of M2–3 tumor stages, and excellent overall survival [2, 7, 14, 17]. However, our findings suggest that histologically similar MBEN disclosed some molecular variability and they are not related with any single mutation or other unique molecular event.
Epigenetically, MBEN did not form a separate molecular cohort and were nearly all classified as MB with SHH signatures, in line with previous findings [4, 9, 13, 14]. Unexpectedly, two MBEN were classified as either Groups 3 or 4 MB, and it has not been reported previously. Both cases indeed also disclosed a distinct biological behavior (M3 stage), cytogenetic aberrations (trisomy 17), and absence of SHH-associated mutations, when compared to all other MBEN classified as SHH MB. Both patients received upfront intense chemotherapy (due to advanced tumor stage at diagnosis) and showed quite satisfactory treatment response. Of course, the incidence of such MBEN harboring unusual molecular signatures is extremely low, but such possibility should be kept in mind. An application of GAB1 immunohistochemistry might be helpful for diagnostic of these rare MBEN molecular variants and obviate the need for epigenetic analysis, which remains beyond the capability of most routine laboratories. Moreover, deciphering the biological mechanisms underlying such unusual/enigmatic association between “favorable” MBEN histology and “aggressive” tumor molecular profile is clearly warranted in future studies.
MBEN classified as SHH MB disclosed predominately balanced DNA profiles and similar frequencies of SHH-related gene alterations (either missense mutations or frameshift insertions/deletions) involving PTCH1, SUFU and SMO. The high incidence of germ-line SUFU alterations is associated strongly with postoperative local regrowth, which, however, responded well after application of intense CHT protocols. It is well known, that most cases of familial SHH MB (including MBEN) could be associated with SUFU mutations [3, 6, 8, 15]. Therefore, SUFU mutational screening (both tumor and blood) was recommended in all children presenting SHH MB before 5 years of age [3, 8]. Nevertheless, the best therapeutic strategy for infants with MB is still under discussion and the question of whether SUFU-altered patients require a specific therapeutic approach remains to be defined [1, 2, 12, 15]. Radiotherapy is still debated due to a risk of second malignancies. On the other hand, SUFU is a SHH pathway gene which is downstream of SMO, and it could rise an intrinsic resistance to SMO targeting inhibitors, and, moreover, these patients could suffer from severe bone-marrow toxicity [6, 8, 9]. Our data suggest a benefit of HD/CHT for SUFU mutant MBEN, which could be considered as an initial treatment option in the future trials. Two other SHH-associated mutations (PTCH1 and SMO) were exclusively somatic and not related with clinical outcomes. It is also interest to note a high frequency of SMO mutations in MBEN, which previously were predominantly identified among the adult SHH MB [9]. Thus, the above-mentioned findings led us to propose a provisional model for stratification of patients with MBEN into two risk groups. Patients with M2–3 stages (and perhaps non-SHH tumor affiliation) and/or somatic/germ-line SUFU alterations could be considered as a “high-risk MBEN” requiring more intense CHT upfront. Remaining MBEN patients could initially benefit from the “standard” HIT SKK regimens accompanying with de-escalating toxicity [2, 12, 15]. Further clarification of this provisional stratification scheme through detailed analysis of additional MBEN cohorts included in future clinical trials would, therefore, be of significant value.
Pediatric SHH MB could represent all histological MB subtypes with variable molecular appearance, including almost cytogenetically balanced MBEN at its one end and genetically instable, TP53 mutant large cell MB at another one [1, 4, 9, 14]. These variants of SHH MB are also extremely distinct in terms of their biological behavior and clinical outcomes. However, molecular relationships of DNMB and classic SHH MB are looking as being more complex. On the one hand, these SHH MB variants disclosed obvious methylome differences, because DNMB showed a clear epigenetic kinship with MBEN. On the other hand, DNMB and classic SHH MB disclosed apparent similarities for their cytogenetic profiles (“prototypic” 9q loss), mutational landscape (over-representing with PTCH1 alterations) and, finally, coincidence in the clinical outcomes, including relatively poor response for salvage treatments applied. Perhaps, DNMB and classic SHH MB variants could have different either cell sources or time of origin, such suggesting that epigenetic state of MB is dictated at the time of tumor initiation and remains stable during tumor development. However, further progression of these SHH MB variants could be associated with acquisition of similar events, such as PTCH1 mutation and/or aberrations on chromosome 9. Similarity of DNMB and classic SHH MB molecular biology could, in turn, equate their clinical behavior. SHH MB histological variants should be additionally molecularly stratified in future studies.
In conclusion, the vast majority of MBEN disclosed SHH-associated molecular profiles and mutational landscapes but a minority of tumors with MBEN histology could be classified as non-SHH MB. SHH MBEN cohort showed a wide spectrum of SHH-related mutations at similar frequency but only SUFU mutations were identified as germ-line and clinically relevant alterations. SUFU-associated MBEN eventually recurred but patients were treated successfully with the second-line HD/CHT. Consequently, we could also recommend an integrated molecular analysis (including screening for germ-line SUFU alterations) for all patients with “MBEN” histological diagnosis for their further clinical stratification and choice of the optimal treatment strategy upfront.
References
Abdel-Baki MS, Boué DR, Finlay JL, Kieran MW (2017) Desmoplastic nodular medulloblastoma in young children: a management dilemma. Neuro Oncol. https://doi.org/10.1093/neuonc/nox222 (Epub ahead of print)
Aristizabal P, Burns L, Rivera-Gomez R et al (2017) Medulloblastoma with extensive nodularity: tailored therapy in a low-resource setting. J Pediatr Hematol Oncol 39:299–301. https://doi.org/10.1097/MPH.0000000000000798
Brugières L, Remenieras A, Pierron G et al (2012) High frequency of germline SUFU mutations in children with desmoplastic/nodular medulloblastoma younger than 3 years of age. J Clin Oncol 30:2087–2093. https://doi.org/10.1200/JCO.2011.38.7258
Cavalli FMG, Remke M, Rampasek L et al (2017) Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31:737–754. https://doi.org/10.1016/j.ccell.2017.05.005
Chelliah D, Sarfo-Poku CM, Stea BD, Gardetto J, Zumwalt J (2010) Medulloblastoma with extensive nodularity undergoing post-therapeutic maturation to a gangliocytoma: a case report and literature review. Pediatr Neurosurg 46:381–384
Garrè ML, Cama A, Bagnasco F et al (2009) Medulloblastoma variants: age-dependent occurrence and relation to Gorlin syndrome—a new clinical perspective. Clin Cancer Res 15:2463–2471. https://doi.org/10.1158/1078-0432.CCR-08-2023
Giangaspero F, Perilongo G, Fondelli MP et al (1999) Medulloblastoma with extensive nodularity: a variant with favorable prognosis. J Neurosurg 91:971–977
Guerrini-Rousseau L, Dufour C et al (2017) Germline SUFU mutation carriers and medulloblastoma: clinical characteristics, cancer risk and prognosis. Neuro Oncol. https://doi.org/10.1093/neuonc/nox228 (Epub ahead of print)
Kool M, Jones DT, Jäger N et al (2014) Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25:393–405. https://doi.org/10.1016/j.ccr.2014.02.004
Kool M, Korshunov A, Remke M et al (2012) Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 123:473–484. https://doi.org/10.1007/s00401-012-0958-8
Korshunov A, Chavez L, Northcott PA et al (2017) DNA-methylation profiling discloses significant advantages over NanoString method for molecular classification of medulloblastoma. Acta Neuropathol 134(6):965–967. https://doi.org/10.1007/s00401-017-1776-9
Lafay-Cousin L, Smith A, Chi SN et al (2016) Clinical, pathological, and molecular characterization of infant medulloblastomas treated with sequential high-dose chemotherapy. Pediatr Blood Cancer 63:1527–1534. https://doi.org/10.1002/pbc.26042
Northcott PA, Buchhalter I, Morrissy AS et al (2017) The whole-genome landscape of medulloblastoma subtypes. Nature 547:311–317. https://doi.org/10.1038/nature22973
Pietsch T, Schmidt R, Remke M et al (2014) Prognostic significance of clinical, histopathological, and molecular characteristics of medulloblastomas in the prospective HIT2000 multicenter clinical trial cohort. Acta Neuropathol 128:137–149. https://doi.org/10.1007/s00401-014-1276-0
Rutkowski S, von Hoff K, Emser A et al (2010) Survival and prognostic factors of early childhood medulloblastoma: an international meta-analysis. J Clin Oncol 28:4961–4968. https://doi.org/10.1200/JCO.2010.30.2299
Sahm F, Schrimpf D, Jones DT et al (2016) Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol 131:903–910
Suresh TN, Santosh V, Yasha TC et al (2004) Medulloblastoma with extensive nodularity: a variant occurring in the very young—clinicopathological and immunohistochemical study of four cases. Childs Nerv Syst 20:55–60
Taylor MD, Northcott PA, Korshunov A et al (2012) Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 123(4):465–472. https://doi.org/10.1007/s00401-011-0922-z
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Korshunov, A., Sahm, F., Stichel, D. et al. Molecular characterization of medulloblastomas with extensive nodularity (MBEN). Acta Neuropathol 136, 303–313 (2018). https://doi.org/10.1007/s00401-018-1840-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-018-1840-0