Abstract
Extraventricular neurocytoma (EVN) is a rare primary brain tumor occurring in brain parenchyma outside the ventricular system. Histopathological characteristics resemble those of central neurocytoma but exhibit a wider morphologic spectrum. Accurate diagnosis of these histologically heterogeneous tumors is often challenging because of the overlapping morphological features and the lack of defining molecular markers. Here, we explored the molecular landscape of 40 tumors diagnosed histologically as EVN by investigating copy number profiles and DNA methylation array data. DNA methylation profiles were compared with those of relevant differential diagnoses of EVN and with a broader spectrum of diverse brain tumor entities. Based on this, our tumor cohort segregated into different groups. While a large fraction (n = 22) formed a separate epigenetic group clearly distinct from established DNA methylation profiles of other entities, a subset (n = 14) of histologically diagnosed EVN grouped with clusters of other defined entities. Three cases formed a small group close to but separated from the epigenetically distinct EVN cases, and one sample clustered with non-neoplastic brain tissue. Four additional samples originally diagnosed otherwise were found to molecularly resemble EVN. Thus, our results highlight a distinct DNA methylation pattern for the majority of tumors diagnosed as EVN, but also indicate that approximately one third of morphological diagnoses of EVN epigenetically correspond to other brain tumor entities. Copy number analysis and confirmation through RNA sequencing revealed FGFR1–TACC1 fusion as a distinctive, recurrent feature within the EVN methylation group (60%), in addition to a small number of other FGFR rearrangements (13%). In conclusion, our data demonstrate a specific epigenetic signature of EVN suitable for characterization of these tumors as a molecularly distinct entity, and reveal a high frequency of potentially druggable FGFR pathway activation in this tumor group.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Extraventricular neurocytoma (EVN) is a rare neurocytic neoplasm arising outside the ventricular system with histopathological characteristics resembling central neurocytoma. Diagnosis of this usually well-circumscribed brain tumor is often challenging because of the wide range of histopathological appearances. Histologically, EVNs are composed of uniform neoplastic cells with small round nuclei and clear cytoplasm, embedded in a finely fibrillar neuropil-like matrix exhibiting neuronal/neurocytic differentiation [1, 14]. An oligodendroglioma-like honeycomb architecture, ganglion cell component and microcalcifications have been described as common features [14]. Immunohistochemistry demonstrates evidence of neuronal differentiation with consistent expression of synaptophysin. Rarely, focal expression of chromogranin-A and glial fibrillary acidic protein (GFAP) can be seen. The considerable histological overlap with other brain tumors, especially oligodendroglioma with neurocytic features and other neuronal and mixed glioneuronal entities, often pose diagnostic difficulties [1, 6, 15, 17, 19].
Over the past years, genome-wide DNA methylation profiling and identification of recurrent genomic alterations have become important tools in the diagnosis and classification of tumors of the central nervous system (CNS). Numerous studies have demonstrated the utility of DNA methylation profiling in defining distinct entities or subclasses of brain tumors that often improve diagnostic accuracy and has prognostic and therapeutic implications [2, 7, 8, 18, 26]. So far, no molecular signature has been described for EVN and the entity has not yet been implemented in the recently published DNA methylation-based brain tumor classification tool [2].
Here, we investigated the molecular landscape of this histologically heterogeneous entity using genome-wide DNA methylation data as a basis for molecular classification in order to facilitate more accurate diagnosis. Recurrent genetic alterations possibly underlying the formation of these tumors were also investigated. Moreover, we aimed to confirm the hypothesis that histologically diagnosed EVN may include a significant proportion of other unrelated entities.
Materials and methods
Tumor samples and clinical data
Tumor tissue and retrospectively determined clinical data from 40 patients with the institutional diagnosis of EVN (made between 2004 and 2018) were obtained from multiple international collaborating centers and collected at the Department of Neuropathology of the University Hospital Heidelberg (Heidelberg, Germany). For all cases a genotype check was performed to exclude the possibility that material from the same patient was received from more than one center. To compare molecular profiles in tumors diagnosed as EVN with those obtained for other glial or glioneuronal tumors, a molecular reference set was formed. The reference set included data from the following well-characterized entities: diffuse leptomeningeal glioneuronal tumor (n = 10); dysembryoplastic neuroepithelial tumor (n = 10); rosette-forming glioneuronal tumor (n = 10); central neurocytoma (n = 10); pilocytic astrocytoma (subclass posterior fossa, n = 10; subclass midline, n = 10; subclass hemispheric, n = 10); ganglioglioma (n = 10); cerebellar liponeurocytoma (n = 10); ependymoma (posterior fossa group A, n = 10; posterior fossa group B, n = 10; with RELA fusion, n = 10); oligodendroglioma, IDH-mutant and 1p/19q-codeleted (n = 10); astrocytoma, IDH-mutant (n = 10); diffuse midline glioma, H3 K27M-mutant (n = 10); glioblastoma, IDH-wildtype (subclass midline, n = 10; subclass mesenchymal, n = 10) and control tissue, white matter (n = 10). Detailed descriptions of the reference methylation classes are outlined under https://www.molecularneuropathology.org. To compare survival data of EVN patients with those of patients included in reference glioma groups, clinical data including information on progression-free survival (PFS) and overall survival (OS), from patients with pilocytic astrocytomas WHO grade I (PA I, n = 82), diffuse astrocytoma, IDH-mutant, WHO grade II (A IDH-mut II, n = 202) and anaplastic astrocytoma, IDH-mutant, WHO grade III (AA IDH-mut III, n = 157) were collected from our files. Research use of tissues and anonymization of data were in accordance with local ethical approvals.
DNA and RNA extraction
Representative tumor tissue with highest available tumor content was histologically identified and chosen for nucleic acid extraction. DNA as well as RNA was extracted from formalin-fixed and paraffin-embedded (FFPE) tissue samples using the automated Maxwell system with the Maxwell 16 FFPE Plus LEV DNA Purification Kit or Maxwell 16 LEV RNA FFPE Kit (Promega, Madison, WI, USA), according to the manufacturer’s instructions.
DNA methylation profiling
DNA methylation profiling of all samples was performed using the Infinium MethylationEPIC (850k) BeadChip (Illumina, San Diego, CA, USA) or Infinium HumanMethylation450 (450k) BeadChip (Illumina) array according to the manufacturer’s instructions at the Genomics and Proteomics Core Facility of the German Cancer Research Center (DKFZ). Filtering and genome-wide copy number analyses were performed as previously described [27], using the ‘conumee’ package in R (http://www.bioconductor.org).
Histopathology and immunohistochemistry
Cases were carefully reviewed by examining hematoxylin and eosin (H&E) sections according to the World Health Organization (WHO) 2016 classification of tumors of the central nervous system [14]. Further morphological workup was performed on tumors falling into a distinct DNA methylation class (n = 26) different from those previously established for human brain tumors [2]. Tumors were assessed histologically for the following features: cellularity, histomorphological growth pattern, vascular features (e.g., hyalinized vessels, microvascular proliferation), parenchymal infiltration, mitotic count [count per 10 high power fields (HPF)], presence of necrosis, ganglion cells and calcifications. For cases with available tissue (n = 16), immunohistochemistry with antibodies specific for synaptophysin and Ki67 was performed on a Ventana BenchMark ULTRA Immunostainer applying the OptiView DAB IHC Detection Kit for Ki67 or ultraView Universal DAB Detection Kit for synaptophysin (Ventana Medical Systems, Tucson, Arizona, USA). Immunohistochemistry was conducted on 1 μm-thick formalin-fixed, paraffin-embedded (FFPE) tissue sections mounted on Superfrost Plus slides (Thermo Scientific, Waltham, MA, USA) followed by drying at 80 °C for 10 min. Antibody dilution was 1:100 for Ki67 (clone MIB-1, Dako Agilent, Santa Clara, CA, USA) and 1:160 for synaptophysin (clone MRQ-40, Cell Marque Corp., Rocklin, CA, USA). For Ki67 analysis, tumor areas with the highest Ki67 labeling indices were evaluated for the fraction of positive cell nuclei by counting all cells excluding lymphocytes and vascular cells in one 200× microscopic field.
Sanger sequencing
Targeted sequencing of H3F3A and IDH1/2 was performed with 20 ng of DNA as previously described [21]. Primers for H3F3A were: forward 5′-CATGGCTCGTACAAAGCAGA-3′; reverse 5′-CAAGAGAGACTTTGTCCCATTTTT-3′, for IDH1: forward 5′-TGATGAGAAGAGGGTTGAGGA-3′; reverse 5′-GCAAAATCACATTATTGCCAAC-3′ and for IDH2: forward 5′-CTCCACCCTGGCCTACCT-3′; reverse 5′-GCTGCAGTGGGACCACTATT-3′. Sequences were determined using an ABI 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and the Sequence Pilot version 3.1 (JSI-Medisys, Kippenheim, Germany) software.
RNA sequencing
RNA sequencing of samples in methylation class EVN for which RNA of sufficient quality and quantity was available (n = 15) was performed on a NextSeq 500 (Illumina) as previously described [24]. Fusion discovery was done based on RNA sequencing data using three independent methods: TopHat [11], defuse [16] and Arriba (https://github.com/suhrig/arriba/).
Statistical analysis
DNA methylation array data were processed with the R/Bioconductor package minfi (version 1.20). For unsupervised hierarchical clustering of EVN and reference samples, the 20,000 most variable probes across the dataset were selected. The samples were hierarchically clustered using Euclidean distance and Ward’s linkage method. DNA methylation probes were reordered using Euclidian distance and complete linkage. The t-SNE plot was computed via the R package Rtsne using the 20,000 most variable CpG sites according to the standard deviation, 2000 iterations and a perplexity value of 5. Survival data were analyzed by Kaplan–Meier analysis and compared by log-rank test using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA). p values of less than 0.05 were considered significant.
Results
DNA methylation profiling segregates histologically diagnosed EVN into subgroups
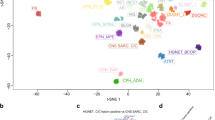
To explore the epigenetic landscape of EVN, we initially generated genome-wide DNA methylation profiles of 40 tumors histologically diagnosed as EVN. Additionally, four more cases initially diagnosed otherwise but with a high molecular similarity to EVN could be included through DNA methylation screening in an extended Heidelberg cohort comprising > 25,000 tumors. Three of these cases were initially diagnosed as (extraventricluar) ependymomas and one as dysembryoplastic neuroepithelial tumor. Reference samples were used to test whether EVN and other tumors with overlapping histological characteristics can be separated on the basis of their DNA methylation profiles. Unsupervised hierarchical clustering and t-SNE analysis, including 170 well-characterized reference tumors representing other CNS tumor entities, revealed that our histological defined tumor cohort molecularly segregated into different groups (Fig. 1 and supplementary Fig. 1). While the largest fraction (n = 26, 59%) of tumors formed a separate cluster (DNA methylation class EVN) clearly distinct from the reference entities, most of the remaining tumors (n = 14, 32%) grouped with clusters of other reference tumors and could be reclassified by the DNA methylation-based classification tool [2] and additional molecular characteristics: five cases were reclassified as pilocytic astrocytomas, one of which exhibited the typical KIAA1549–BRAF fusion and one an FGFR1–TACC1 fusion; three tumors turned out as glioblastomas exhibiting either gain of chromosome 7 combined with loss of chromosome 10, or MDM4 amplification; two tumors were diffuse leptomeningeal glioneuronal tumors with the typical combination of loss of 1p and KIAA1549–BRAF fusion [3]; and single cases each corresponded to IDH-mutant oligodendroglioma with co-deletion of 1p and 19q (IDH2 R172S confirmed by Sanger sequencing), IDH-mutant astrocytoma (IDH1 R132H confirmed by Sanger sequencing), H3 K27M-mutant diffuse midline glioma (H3F3A confirmed by Sanger sequencing), and rosette-forming glioneuronal tumor (Table 1). Three tumors sharing similar copy number changes could not be assigned (Table 1). One sample clustered with non-neoplastic brain tissue, possibly due to very low tumor cell content. Within the distinct EVN DNA methylation group, analysis of copy number profiles (CNPs) showed evidence in half of the cases (n = 13, 50%) for either an FGFR1–TACC1 (n = 11; Fig. 2) or an FGFR3–TACC3 fusion (n = 2). In one case an FGFR1 gain was detected which did not fit the typical pattern expected of a TACC1 fusion (see below). None of the tumors within the DNA methylation class EVN exhibited a co-deletion of chromosome arms 1p and 19q, which has been described in a subset of histologically diagnosed EVN [23].
Molecular classification of EVN by DNA methylation profiling. Unsupervised hierarchical clustering (a) and t-distributed stochastic neighbor embedding (t-SNE) analysis (b) of 40 histologically diagnosed EVN samples shows that the cohort segregates into different groups based on DNA methylation data. While a larger fraction forms a distinct group clearly separated from the reference entities (including four additional samples previously diagnosed otherwise), a subset of the original EVN tumors clusters with reference tumors or cannot be clearly classified. All EVN samples are colored in black, reference samples are colored according to their molecular reference entity
Histological and immunohistochemical characteristics of molecular EVN underline a wide range of histopathological appearances
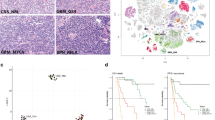
Sufficient FFPE tissue for a histological analysis was available from 22 of 26 tumors belonging to the DNA methylation class EVN. Histologically, all tumors showed a moderate to high cellularity of monomorphic cells with round nuclei and speckled chromatin (Fig. 3a, b). Tumor cells were embedded in a neuropil-like matrix (Fig. 3a, b). Neuropil islands (n = 17/22) as well as ganglion cell differentiation (n = 11/22) were seen in the majority of cases (Fig. 3b, e), matching observations in previous series [1]. Microcalcification (n = 7/22) and perinuclear clearing (n = 3/22) were less frequent (Fig. 3c, d). Tumor cells were arranged in sheets, clusters or rosettes. Necrosis was absent in all cases. Most tumors (n = 15/22) showed hyalinized vessels. Mitotic rates were generally low (between 0 and 2 mitosis/10 HPF). Three tumors exhibited a higher rate of up to 6 mitoses/10 HPFs. Diffuse synaptophysin immunoreactivity within the cytoplasm of the tumor cells was evident in all cases (Fig. 3f). Proliferation index (Ki67) ranged from 1 to 3%. Only three cases showed a higher Ki67 of 7, 10 and 20%. Although four cases would more likely have been favored as a typical differential diagnosis of EVN, all are histologically compatible with EVN as currently defined by the WHO. Details of histological and immunohistochemical characteristics of tumors belonging to the DNA methylation class EVN are given in supplementary Table 1.
Morphological and immunohistochemical features of EVN (a–d 200-fold, e, f 400-fold magnification). Sheets or clusters of isomorphic cells having round nuclei with speckled chromation that are embedded in a neuropil-like matrix (a, b). Neuropil islands (b), oligodendroglioma-like perinuclear clearing (c), microcalcifications (d) and ganglion cell differentiation (e). Diffuse synaptophysin immunoreactivity within the cytoplasm (f)
Transcriptome analysis identifies FGFR fusions as a frequent event in EVN
In light of the copy number data suggesting possible gene fusions on the short arm of chromosome 8 and 4 within the distinct EVN DNA methylation class, we performed transcriptome sequencing in a subset of cases with available material (n = 15). We identified gene fusions between the fibroblast growth factor receptor 1 (FGFR1) and transforming acidic coiled-coil containing protein 1 (TACC1) genes in nine samples (60%), as well as between the fibroblast growth factor receptor 3 (FGFR3) and transforming acidic coiled-coil containing protein 3 (TACC3) genes in one sample (7%) from RNA sequencing data. Moreover, a fusion between FGFR1 and ecotropic viral integration site 5 (EVI5) could be detected in one additional case. Interestingly, almost all cases with FGFR1-TACC1 fusion (78%) were located in the frontal lobe (Table 1). Details of structural variations identified in RNA sequencing are summarized in supplementary Table 2. Comparison of sequencing results and DNA copy number data revealed that 8 of 8 (100%) analyzed samples with suspected FGFR1–TACC1 fusion carried the respective rearrangement. The additional case with FGFR1 gain but not TACC1 fusion instead harbored an FGFR1–EVI5 fusion. In one sample, an FGFR1–TACC1 fusion was detected without clear evidence for a fusion by DNA copy number profiling. Additional material was available in only one of the two cases with suspected FGFR3–TACC3 fusion—here the fusion was validated using RNA sequencing. Altogether 11/15 (73%) of sequenced tumors within the DNA methylation class EVN showed rearrangements affecting members of the FGFR family.
Correlation with clinical data and patient outcome
Tumors in the DNA methylation class EVN were preferentially located supratentorially (95%). Median age at diagnosis was 21.5 years and sex distribution was balanced (male:female ratio 1.17 in DNA methylation class EVN). Outcome data were available for only ten patients with DNA methylation class EVN and ten patients whose tumors clustered molecularly with other entities, making an interpretation difficult. Analysis of progression-free survival (PFS) and overall survival (OS) according to the molecular classification or histology did not show significant differences (PFS p = 0.52; OS p = 0.64). Albeit based on small numbers, the molecularly defined EVN patient group showed a median PFS of 65.5 months. Median OS was not reached, but 5-year OS was 86%. Overall survival of DNA methylation class EVN patients in comparison to reference glioma group patients therefore appears broadly in line with the current WHO grade II designation (EVN vs PA I, p = 0.03; EVN vs A IDH-mut II p = 0.35; EVN vs AA IDH-mut III, p = 0.03; Fig. 4).
Kaplan–Meier curves for overall survival of EVN patients according to molecular classification or histology (a). Overall survival of methylation class EVN patients in comparison to reference glioma group patients (pilocytic astrocytoma WHO grade I; diffuse astrocytoma, IDH-mutant, WHO grade II and anaplastic astrocytoma, IDH-mutant, WHO grade III) (b)
Discussion
EVN represents a histologically heterogeneous CNS tumor that often poses diagnostic difficulties. A unifying morphological feature is the presence of rounded monomorphic cells with immunohistochemical evidence of neuronal differentiation. Although not systematically analyzed, the inter-observer variability in the histopathological diagnosis of EVN is high, presumably due to a wide overlap with other entities. To date, little is known about the molecular characteristics of this rare neoplasm. DNA methylation profiling of tumor tissue has recently been confirmed as a powerful technique for unbiased tumor classification [2]. Here, we subjected 40 tumors that were originally diagnosed by light microscopy as EVN to DNA methylation analysis. Analysis led to the identification of a core group (22/40, 55%) with a specific epigenetic signature, clearly different from established entities, considered as DNA methylation class EVN. Four additional tumors from our larger database also fell into this class. Our data clearly prove that this newly defined EVN entity differs from central neurocytoma. Moreover, a subset of tumors with the histological diagnosis of EVN (14/40, 35%) could be assigned to molecularly well-defined and established DNA methylation classes of other entities. Our findings, therefore, indicate that molecular profiling may refine the differential diagnosis of EVN and increase diagnostic accuracy when compared to previous approaches that were based on microscopy only.
A distinctive feature within the DNA methylation class EVN is evidence of an FGFR–TACC fusion, initially identified through copy number profiling (Fig. 2, Table 1). Subsequent transcriptome sequencing of tumors with sufficient material available confirmed the presence of FGFR1–TACC1 or FGFR3–TACC3 fusions in most tumors analyzed. Taken together, fusions affecting FGFR genes were detected by RNA sequencing in 11/15 (73%) of the tumors within the DNA methylation class EVN (Table 1), making it a strong additional biomarker. This contrasts with central neurocytoma, where characteristic molecular features are not known to date [10]. Transforming fusions of FGFR and TACC genes have been reported rarely in other tumors of the CNS, including glioblastoma (3%) [4, 25], IDH-wildtype glioma WHO grade II/III (3.5%) [4] and pediatric low-grade gliomas (7%) [28]. Moreover, FGFR–TACC rearrangements seem to be enriched in pediatric low-grade neuroepithelial tumors [9, 20, 22]. Their frequency in our EVN cohort, however, is much higher than that reported for any other entity to date. FGFR–TACC fusion proteins allow FGFR to dimerize, leading to autophosphorylation and constitutive FGFR tyrosine kinase activation [12]. Aberrant FGFR signaling results in increased cell proliferation and cancer progression. In one sample, a fusion between FGFR1 and EVI5 was detected. EVI5 acts as a regulator of cell cycle progression [5], but its role in cancer is not well understood [13], and it is not clear whether in this case it played a supplementary driving role or rather just a ‘carrier’ for FGFR1. Identification of fusions that activate FGFR signaling in a large proportion of tumors in the DNA methylation class EVN raises the possibility of targeted therapy. Highly potent FGFR inhibitors have recently been developed and are now under clinical investigation [12].
Due to the rarity of EVN and the lack of clinical follow-up data for most of the cases, it was not possible to collect meaningful survival data. At present, the overall survival of patients with DNA methylation class EVN tumors appears to be most consistent with that of patients with IDH-mutant astrocytoma, WHO grade II (Fig. 4); however, this finding will need confirmation in independent, molecularly defined cohorts.
In conclusion, our data demonstrate a specific epigenetic signature for a distinct molecular class of EVN, characterized by frequent FGFR fusions. We suggest restricting the designation EVN to tumors confirmed as having this profile, to avoid misinterpretation through current histological criteria. These findings represent a significant advance towards improved diagnostic accuracy of this rare group of CNS tumors, and points towards possible options for novel targeted therapies.
References
Brat DJ, Scheithauer BW, Eberhart CG, Burger PC (2001) Extraventricular neurocytomas: pathologic features and clinical outcome. Am J Surg Pathol 25:1252–1260
Capper D, Jones DTW, Sill M et al (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. https://doi.org/10.1038/nature26000
Deng MY, Sill M, Chiang J et al (2018) Molecularly defined diffuse leptomeningeal glioneuronal tumor (DLGNT) comprises two subgroups with distinct clinical and genetic features. Acta Neuropathol. https://doi.org/10.1007/s00401-018-1865-4
Di Stefano AL, Fucci A, Frattini V et al (2015) Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res 21:3307–3317. https://doi.org/10.1158/1078-0432.CCR-14-2199
Eldridge AG, Loktev AV, Hansen DV, Verschuren EW, Reimann JD, Jackson PK (2006) The evi5 oncogene regulates cyclin accumulation by stabilizing the anaphase-promoting complex inhibitor emi1. Cell 124:367–380. https://doi.org/10.1016/j.cell.2005.10.038
Giangaspero F, Cenacchi G, Losi L, Cerasoli S, Bisceglia M, Burger PC (1997) Extraventricular neoplasms with neurocytoma features. A clinicopathological study of 11 cases. Am J Surg Pathol 21:206–212
Hovestadt V, Jones DT, Picelli S et al (2014) Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510:537–541. https://doi.org/10.1038/nature13268
Hovestadt V, Remke M, Kool M et al (2013) Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol 125:913–916. https://doi.org/10.1007/s00401-013-1126-5
Huse JT, Snuderl M, Jones DT et al (2017) Polymorphous low-grade neuroepithelial tumor of the young (PLNTY): an epileptogenic neoplasm with oligodendroglioma-like components, aberrant CD34 expression, and genetic alterations involving the MAP kinase pathway. Acta Neuropathol 133:417–429. https://doi.org/10.1007/s00401-016-1639-9
Kane AJ, Sughrue ME, Rutkowski MJ, Tihan T, Parsa AT (2011) The molecular pathology of central neurocytomas. J Clin Neurosci 18:1–6. https://doi.org/10.1016/j.jocn.2010.06.004
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. https://doi.org/10.1186/gb-2013-14-4-r36
Lasorella A, Sanson M, Iavarone A (2017) FGFR-TACC gene fusions in human glioma. Neuro Oncol 19:475–483. https://doi.org/10.1093/neuonc/now240
Lim YS, Tang BL (2013) The Evi5 family in cellular physiology and pathology. FEBS Lett 587:1703–1710. https://doi.org/10.1016/j.febslet.2013.04.036
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (2016) WHO classification of tumours of the central nervous system, Revised 4 edn. IARC, Lyon
Makuria AT, Henderson FC, Rushing EJ, Hartmann DP, Azumi N, Ozdemirli M (2007) Oligodendroglioma with neurocytic differentiation versus atypical extraventricular neurocytoma: a case report of unusual pathologic findings of a spinal cord tumor. J Neurooncol 82:199–205. https://doi.org/10.1007/s11060-006-9268-0
McPherson A, Hormozdiari F, Zayed A et al (2011) deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 7:e1001138. https://doi.org/10.1371/journal.pcbi.1001138
Okada M, Yano H, Hirose Y et al (2011) Olig2 is useful in the differential diagnosis of oligodendrogliomas and extraventricular neurocytomas. Brain Tumor Pathol 28:157–161. https://doi.org/10.1007/s10014-011-0017-5
Pajtler KW, Witt H, Sill M et al (2015) molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27:728–743. https://doi.org/10.1016/j.ccell.2015.04.002
Perry A, Scheithauer BW, Macaulay RJ, Raffel C, Roth KA, Kros JM (2002) Oligodendrogliomas with neurocytic differentiation. A report of 4 cases with diagnostic and histogenetic implications. J Neuropathol Exp Neurol 61:947–955
Qaddoumi I, Orisme W, Wen J et al (2016) Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol 131:833–845. https://doi.org/10.1007/s00401-016-1539-z
Reuss DE, Kratz A, Sahm F et al (2015) Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol 130:407–417. https://doi.org/10.1007/s00401-015-1454-8
Rivera B, Gayden T, Carrot-Zhang J et al (2016) Germline and somatic FGFR1 abnormalities in dysembryoplastic neuroepithelial tumors. Acta Neuropathol 131:847–863. https://doi.org/10.1007/s00401-016-1549-x
Rodriguez FJ, Mota RA, Scheithauer BW et al (2009) Interphase cytogenetics for 1p19q and t(1;19)(q10;p10) may distinguish prognostically relevant subgroups in extraventricular neurocytoma. Brain Pathol 19:623–629. https://doi.org/10.1111/j.1750-3639.2008.00206.x
Sahm F, Schrimpf D, Stichel D et al (2017) DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol 18:682–694. https://doi.org/10.1016/S1470-2045(17)30155-9
Singh D, Chan JM, Zoppoli P et al (2012) Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337:1231–1235. https://doi.org/10.1126/science.1220834
Sturm D, Orr BA, Toprak UH et al (2016) New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164:1060–1072. https://doi.org/10.1016/j.cell.2016.01.015
Sturm D, Witt H, Hovestadt V et al (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22:425–437. https://doi.org/10.1016/j.ccr.2012.08.024
Zhang J, Wu G, Miller CP et al (2013) Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 45:602–612. https://doi.org/10.1038/ng.2611
Acknowledgements
We thank V. Zeller, U. Vogel, U. Lass, A. Habel, K. Lindenberg, S. Kocher and J. Meyer for excellent technical support and the microarray unit of the DKFZ Genomics and Proteomics Core Facility for providing Illumina DNA methylation array-related services. D. Jones is supported by the Everest Centre for Low-grade Paediatric Brain Tumours (The Brain Tumour Charity, UK).
Author information
Authors and Affiliations
Corresponding author
Additional information
David T. W. Jones and Andreas von Deimling share senior authorship.
Electronic supplementary material
Below is the link to the electronic supplementary material.
401_2018_1882_MOESM1_ESM.eps
Supplementary material 1 (EPS 15793 kb) Supplementary Fig. 1 Molecular Classification of EVN by DNA methylation profiling. Unsupervised hierarchical clustering analysis of 40 histologically diagnosed EVN samples shows that the cohort segregates into different groups based on DNA methylation data. While a larger fraction forms a distinct group clearly separated from the reference entities (including 4 additional samples previously diagnosed otherwise), a subset of the original EVN tumors clusters with reference tumors or cannot be clearly classified. All EVN samples are colored in black, reference samples are colored according to their molecular reference entity. In addition, fusion transcripts of samples within the methylation class EVN are indicated
Rights and permissions
About this article
Cite this article
Sievers, P., Stichel, D., Schrimpf, D. et al. FGFR1:TACC1 fusion is a frequent event in molecularly defined extraventricular neurocytoma. Acta Neuropathol 136, 293–302 (2018). https://doi.org/10.1007/s00401-018-1882-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-018-1882-3