Abstract
Key message
Antagonists and sonication treatment relieved the structural barriers of Agrobacterium entering into cells; hindered signal perception and transmission; alleviated defense responses and increased cell susceptibility to Agrobacterium infection.
Abstract
Soybean gene expression analysis was performed to elucidate the general response of soybean plant to Agrobacterium at an early stage of infection. Agrobacterium infection stimulated the PAMPs-triggered immunity (BRI1, BAK1, BZR1, FLS2 and EFR) and effector-triggered immunity (RPM1, RPS2, RPS5, RIN4, and PBS1); up-regulated the transcript factors (WRKY25, WRKY29, MEKK1P, MKK4/5P and MYC2) in MAPK pathway; strengthened the biosynthesis of flavonoid and isoflavonoid in the second metabolism; finally led to a fierce defense response of soybean to Agrobacterium infection and thereby lower transformation efficiency. To overcome it, antagonist α-aminooxyacetic acid (AOA) and sonication treatment along with Agrobacterium infection were applied. This novel method dramatically decreased the expression of genes coding for F3′H, HCT, β-glucosidase and IF7GT, etc., which are important for isoflavone biosynthesis or the interconversion of aglycones and glycon; genes coding for peroxidase, FLS2, PBS1 and transcription factor MYC2, etc., which are important components in plant–pathogen interaction; and genes coding for GPAT and α-l-fucosidase, which are important in polyesters formation in cell membrane and the degradation of fucose-containing glycoproteins and glycolipids on the external surface of cell membrane, respectively. This analysis implied that AOA and sonication treatment not only relieved the structural membrane barriers of Agrobacterium entering into cells, but also hindered the perception of ‘invasion’ signal on cell membrane and intercellular signal transmission, thus effectively alleviated the defense responses and increased the cell susceptibility to Agrobacterium infection. All these factors benefit the transformation process; other measures should also be further explored to improve soybean transformation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean [Glycine max (L.) Merrill.] transformation via Agrobacterium tumefaciens mediated methods is difficult, although a few reports claimed high efficiency ranging from 9.4 to 26.2 % (Olhoft et al. 2001, 2003; Dang and Wei 2007), these milepost workings in soybean genetic transformation were scarcely successfully replicated by other laboratory (Atif et al. 2013). Besides the strong dependence of transformation operation on personal practice, the lack of better understanding of transformation mechanism increased the difficulties of success. The mechanism underlying Agrobacterium-mediated transformation had been explored in both bacterium and plants (Gelvin 2010; Pitzschke and Hirt 2010; Liu et al. 2010; Lacroix and Citovsky 2013; Subramoni et al. 2014; Shi et al. 2014).
Agrobacterium infection is accompanied by pathogen defense of the host and the counter-defense launched by Agrobacterium. In the context of pathogen defense, the mitogen-activated protein kinase MPK3 has merited special attention in defense signaling pathway (Nakagami et al. 2005; Pitzschke et al. 2009). MPK3 phosphorylates the Arabidopsis VIP1 protein and thereby triggers the cyto-nuclear translocation of VIP1 protein, which increases T-DNA transfer and transformation efficiency (Djamei et al. 2007). The strategies utilized by Agrobacterium to transform the plant cell were well reviewed by Pitzschke (2013); while those for plant response to Agrobacterium were summarized by Gohlke and Deeken (2014). The host defense response is activated more or less strongly depending on the plant system and Agrobacterium genotype used for infection (Ditt et al. 2006; Lee et al. 2009; Gohlke and Deeken 2014), the timing and intensity of the microbe activated host defense reaction determine the success of transformation (Pitzschke 2013).
The development of full genome sequencing provides insights into Agrobacterium-induced host changes at the transcript level. Agrobacterium is capable of altering plant gene expression, specifically, the expression of plant defense related genes. Many Agrobacterium induced transcripts of Ageratum conyzoides plant cell cultures encoded putative defense factors (Ditt et al. 2001). Agrobacterium infection caused host defense response has also been reported by Veena Jiang et al. (2003), Zipfel et al. (2006) and Anand et al. (2008). The plant defense system has an important role in controlling infection and its gene expression level is negatively correlated with Agrobacterium-mediated transformation efficiency (Ditt et al. 2005). Deeken et al. (2006) found that the increased levels of anions, sugars and amino acids in metabolic solute of Arabidopsis thaliana tumors were correlated with changes in gene expression. The functional categories of up-regulated set of genes were very different from that of down-regulated (Ditt et al. 2006). Zhao et al. (2011) investigated the proteomic profile of grapevine embryogenic callus after co-cultivation with A. tumefaciens and found that the cellular reactive oxygen species (ROS) removal system, mitochondrial energy metabolism and the protein-degradation machinery for misfolded proteins were markedly inhibited by Agrobacterium transformation, while the apoptosis signaling pathway and hypersensitive response are strengthened. Zhou et al. (2013) analyzed the differentially expressed genes (DEGs) in wheat callus cells co-cultured with Agrobacterium and found that a big part of these DEGs was related to the process of stress or immunity response. We previously demonstrated that the high content of endogenous isoflavones is a serious obstacle in achieving high efficient Agrobacterium-mediated transformation in soybean (Zhang et al. 2015). Isoflavones are synthesized as part of the phenylpropanoid pathway. Phenylalanine ammonia-lyase (PAL) catalyzes the first step in the biosynthesis of phenylpropanoids. α-Aminooxyacetic acid (AOA), a specific inhibitor of PAL, had been successfully used in the inhibition of the isoflavonoid biosynthesis in cell suspension cultures of kudzu [Pueraria lobata (WILLD) Ohwi] (Li et al. 2009). Sonication treatment could disturb the synthesis of isoflavones by down regulating the expression of chalcone synthase (CHS) (Larkin 2001). Combined use of antagonist AOA and sonication treatments (novel method) greatly improved soybean’s T-DNA delivery efficiency (Zhang et al. 2015). In this study, soybean gene expression profile analysis has been performed to elucidate the general response of soybean plant to Agrobacterium at an early stage of infection and to uncover the mechanism underlie this novel method toward developing an efficient transformation protocols for recalcitrant soybean plant.
Materials and methods
Plant explant preparation and transformation
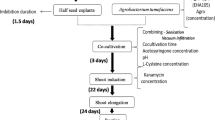
Soybean [Glycine max (L.) Merrill.] seeds of Jidou17 were surface sterilized with chlorine gas for 24 h and subsequently soaked in sterile distilled water overnight. The cotyledons were separated by a longitudinal cut along the hilum and the hypocotyl, seed coat and embryonic axis were removed. EHA105 (pCAMBIA2201) was used as A. tumefaciens strain and the infection solution preparation was as described (Zhang et al. 2015). Soybean transformation was prepared in groups: 15–20 cotyledonary-node explants were placed in a 50 ml glass tube containing 25 ml of infection solution and sonicated for 0 or 15 s in a bath sonicator (KH2200B, Kunshan Hechuang, China) and then incubated for another 20 min at ambient temperature. The infected explants were blotted dry on sterile filter paper and then transferred onto co-culture medium, which had the same ingredients as the infection medium plus 1 mM dithiothreitol (DTT), 1 mM sodium thiosulfate, 3.3 mM cysteine, and 5.0 g L−1 agar, with or without 20 μM AOA. Three treatments were designed: (a) without Agrobacterium infection and cultured on medium free of AOA; (b) infected by conventional Agrobacterium transformation method and cultured on medium free of AOA; (c) infected by Agrobacterium along with sonication treatment at 40 kHz for 15 s and cultured on medium with 20 μM AOA. Fifteen explants were cultured on one plate with a minimum of ten replicates per treatment. Co-cultivation was conducted for 3 days at 25 °C in dark.
Sample collection, cDNA library preparation and sequencing
Explants from three replicates were collected at 5 h after infection and pooled into two different parts, respectively. One part was used in the digital gene expression library preparation and Ion-Proton sequencing, and the other was used for quantitative PCR analysis. All samples were rapidly frozen in liquid nitrogen and stored at −80 °C until RNA extraction.
RNA was isolated from each sample using Trizol reagent (Invitrogen, Carlsbad, CA, USA). The total RNA samples were first treated with DNase I (RNase Free) to degrade any possible DNA contamination and then purified by magnetic beads (Invitrogen, Carlsbad, CA, USA). After that, the mRNA was enriched using the oligo (dT) conjugated magnetic beads (for eukaryotes), and then fragmented into short fragments (about 200 bp) using divalent cations at high temperature. Then cDNA was synthesized using random hexamer-primed reverse transcription (Invitrogen, Carlsbad, CA, USA). The double strand cDNA was purified with magnetic beads and then adaptors were ligated to the ends of these fragments. Ligation products were selected by size and purified on TAE-agarose gel. Finally, the fragments were enriched by PCR amplification, purified by magnetic beads and dissolved in the appropriate amount of Epstein–Barr solution. Agilent 2100 Bioanalyzer (Agilent Technologies, California, USA) was used to qualify and quantify the sample library. The qualified library was ready for sequencing via Ion Proton platform when necessary at BGI tech (Shenzhen, China).
Sequence analysis
Raw reads produced by the sequencer were transferred into clean data by removing some adaptor sequences and/or low quality reads present in the raw reads data. All high quality sequences were mapped to the reference genome (ftp://jgi-psf.org/pub/compgen/phytozome/v9.0/Gmax/assembly/Gmax_189.fa.gz) and reference gene set (ftp://jgi-psf.org/pub/compgen/phytozome/v9.0/Gmax/annotation/Gmax_189_transcript.fa.gz) using the short reads alignment software TMAP3.4.1 ( https://github.com/iontorrent/TMAPreads).
The quantification of gene expression was performed by mapping reads to the reference gene set; reads that were not mapped to the reference gene set might be mapped to the reference genome, which were important for novel transcript prediction. The expression level for each gene was determined by the numbers of reads uniquely mapped to the specific gene and the total number of uniquely mapped reads in the sample and was calculated by RPKM method (Mortazavi et al. 2008). A rigorous algorithm was developed to identify DEGs between different treatments. False discovery rate (FDR) was used to determine the threshold of P value in multiple tests and analysis. We used an FDR of 0.001 and the absolute value of log2Ratio ≥1 (twofold change) as the threshold to judge the significance of the gene expression difference.
With nr annotation, Blast2GO program was used to get GO annotation of DEGs. The DEGs in the GO database (http://www.geneontology.org/) were mapped into significantly enriched GO terms (Bonferroni corrected P value ≤0.05) by Go enrichment analysis. WEGO software was used to do GO functional classification of DEGs. KEGG pathway enrichment analysis was performed using KEGG database (http://www.kegg.jp/kegg/pathway.html) to identify significantly enriched metabolic pathways or signal transduction pathways in DEGs comparing with the whole genome background.
Validation of DEGs by real-time quantitative PCR, enzyme activity and metabolite content
Soybean cotyledonary-node explants collected at the same time as those used for gene expression profile analysis were used for real-time quantitative PCR validation. RNA extraction and cDNA synthesis were as described in the RNAseq part. The real-time quantitative PCR was performed on the 7500/7500 Fast Real-Time PCR System (Applied Biosystems, USA) as described (Zhang et al. 2015). The relative quantification of gene expression was calculated as 2−ΔΔCт. A total of 40 DEGs were selected; the primers were designed using online tools IDT-PrimerQuest Input (http://www.idtdna.com/primerquest/Home/Index) and were synthesized by Sangon Biotech (Shanghai, China) (Table S1). Triplicate per gene was included in each plate. Actin was used as an endogenous parallel control.
Explants 24 h after Agrobacterium infection were sampled for assaying the activity of phenylalanine ammonia-lyase [EC: 4.3.1.24], β-glucosidase [EC: 3.2.1.21] and peroxidase [EC: 1.11.1.7] as well as isoflavones content. PAL activity and isoflavones content determination were as previously described (Zhang et al. 2015). Peroxidase activity and β-glucosidase activity were determined following the methods described in “Guide for modern plant physiology experiments”(eds. Institute of Shanghai plant physiology, Chinese academy of sciences, Shanghai plant physiology society (China) 1999).
Histochemical assay of infected explants
The explants 3 dpi were used to perform GUS histochemical assay as described by Jefferson (1987). Percentage response was determined as the number of cotyledonary nodes of staining blue with X-gluc (Gold Biotechnology, Inc., USA) at the regenerable area divided by the number of cotyledonary nodes assayed. All other chemicals not specially stated were supplied by Sangon Biotech (Shanghai, China).
Results
Agrobacterium infection stimulated host defense related gene expression
10,586,715, 11,770,754 and 11,876,728 total reads were generated from A, B and C samples, respectively, with 98.85 % above mapped to reference gene set and 99.84 % above mapped to reference genome.
Compared with non-transformed control A, 2158 DEGs were identified from the Agrobacterium infected explants B, including 1190 up-regulated and 968 down-regulated DEGs. Among 2158 DEGs, 1304 DEGs were with pathway annotation (118 pathways), the 3 biggest pathway categories were plant–pathogen interaction (115 DEGs, 8.82 %), plant hormone signal transduction (112 DEGs, 8.59 %), and phenylpropanoid biosynthesis (94 DEGs, 7.21 %). Comparing with the whole genome background, the top three significantly enriched pathways, taking RichFactor coefficient as accounts, were phenylpropanoid biosynthesis (94 DEGs, 7.21 %), flavonoid biosynthesis (77 DEGs, 5.9 %) and isoflavonoid biosynthesis (19 DEGs, 1.46 %) (Fig. S1). The DEGs in significantly enriched pathway by Agrobacterium infection (A-vs-B) are listed in Table S2. Among 115 DEGs in plant–pathogen interaction categorization, 95 genes were up-regulated and 20 genes were down-regulated. The genes encoding the bacterial flagellin flg22 related LRR receptor-like serine/threonine-protein kinase FLS2, brassinosteroid insensitive 1-associated receptor kinase 1 (BAK1), and bacterial EF-Tu related EFR genes were stimulated greatly by Agrobacterium infection. The number of DEGs up-regulated was also significantly higher than that of down-regulated in phenylpropanoid biosynthesis (74 up-vs-20 down), flavonoid biosynthesis (64 up-vs-13 down) and isoflavonoid biosynthesis (16 up-vs-3 down), their detailed information was sketched into maps (Figs. S2, S3, S4). Based on pathway enrichment analysis, the peroxidase [EC: 1.11.1.7], WRKY transcription factor, LRR receptor-like serine/threonine-protein kinase FLS2 [EC: 2.7.11.1], brassinosteroid insensitive 1-associated receptor kinase 1 (BAK1) [EC: 2.7.10.1, 2.7.11.1], flavonoid 3′-monooxygenase (F3′H) [EC: 1.14.13.21], and chalcone synthase (CHS) [EC: 2.3.1.74], etc., were the most up-regulated metabolites in significantly enriched pathway by Agrobacterium infection (Table 1).
Antagonists and sonication changed the gene expression profile
Compared with explants B transformed with conventional method, the number (ratio) of down-regulated DEGs was greatly increased in explants C transformed with the novel method of antagonist AOA combined with sonication treatment (3524 DEGs down-regulated, being 69.6 % of the total 5062 DEGs). Among 5062 DEGs, 3018 DEGs were with pathway annotation (123 pathways), the four biggest pathway categories were plant hormone signal transduction (303 DEGs, 10.04 %), plant–pathogen interaction (285 DEGs, 9.44 %), starch and sucrose metabolism (143 DEGs, 4.74 %) and phenylpropanoid biosynthesis (89 DEGs, 2.95 %). From the view of whole genome background, the most significantly enriched pathway was fatty acid biosynthesis, flavone and flavonol biosynthesis, other glycan degradation, starch and sucrose metabolism (Fig. S5). The DEGs in significantly enriched pathway by antagonists and sonication treatment (B-vs-C) are listed in Table S3. Among 303 DEGs in plant hormone signal transduction, 196 DEGs were down-regulated, being 83 % higher than that of up-regulated, which was different from the situation in A-vs-B, where the number of up-regulated DEGs was similar to that of down-regulated (Table S2). Similar situation happened in many pathways such as plant–pathogen interaction (151 down-vs-134 up), phenylpropanoid biosynthesis (55 down-vs-34 up), isoflavonoid biosynthesis (14 down-vs-4 up), etc. Antagonist and sonication treatment alleviated the defense response elicited by Agrobacterium infection. Genes coding for peroxidase [EC: 1.11.1.7], β-glucosidase [EC: 3.2.1.21], flavonoid 3′-monooxygenase (F3′H) [EC: 1.14.13.21], shikimate O-hydroxycinnamoyltransferase (HCT) [EC: 2.3.1.133], isoflavone 7-O-glucosyltransferase (IF7GT) [EC: 2.4.1.170], LRR receptor-like serine/threonine-protein kinase FLS2 [EC: 2.7.11.1], serine/threonine-protein kinase PBS1 [EC: 2.7.11.1] and transcription factor MYC2, etc., which are important for isoflavone biosynthesis and plant–pathogen interaction, were all significantly down-regulated (Table 2).
Validation of enriched pathway by qPCR, enzyme activity and metabolite content
To evaluate the validity of gene expression profile results using high-throughput sequencing, we performed quantitative real-time PCR for 40 genes. Results of real-time PCR analysis verified the Ion Proton sequencing data (Table 3). Furthermore, the enzyme activity and metabolite content in soybean coty-node explants were detected 24 h after Agrobacterium inoculation. Compared with treatment A, the PAL activity, isoflavones content and peroxidase activity for treatment B were increased by 50, 7.5 and 37.2 %, respectively, which generally verified the sequencing data; but the β-glucosidase activity of treatment B was decreased by 14.8 % when compared with treatment A, showing a different trend from that of RNAseq analysis (Table 4). Antagonist AOA and sonication treatment greatly decreased the activity of above enzymes and isoflavones content, consisting with the sequencing results.
Antagonists and sonication made soybean easy to be transformed by Agrobacterium
As illustrated in Table 5, sonication at 40 kHz for 15 s along with Agrobacterium infection and cultured on medium of 20 μM AOA dramatically promoted the efficiency of T-DNA delivery in soybean, with the mean percentage of GUS transient expression as 54.2 %, significantly higher than that of not sonicated and co-cultured on AOA free medium (11.0 %). The ratio of coty-nodes with dark blue was greater in treatment C than that in treatment B (Fig. 1).
Discussion
Defense response in Agrobacterium infected soybean
Plant–pathogen interaction and plant hormone signal transduction are the two biggest categorized pathways by Agrobacterium infection when compared with non-transformed control. Some genes involved in both plant–pathogen interaction and plant hormone signal transduction, such as BAK1, PR1 (pathogenesis-related protein 1), JAZ (jasmonate ZIM domain-containing protein) and transcription factor MYC2. It was generally believed that the phytohormone salicylic acid (SA), jasmonic acid, and ethylene participate in plant defense regulation (Grant and Lamb 2006; Anand et al. 2008), but we failed to detect DEGs in SA metabolic pathway, this was in consistence with the results in barley where bacteria-triggered systemic immunity was associated with WRKY and ethylene responsive factors but not with salicylic acid (Dey et al. 2014). However, the expression of genes related to brassinosteroid metabolism was greatly changed in soybean (Table S2). Brassinosteroids are endogenous plant-growth regulators that modulate cell elongation and division (Clouse et al. 1996). BAK1 is a typical leucine-rich repeat sequence receptor kinase (LRR-RK) and was initially identified as a dual co-receptor of BRI1 and FLS2, which mediate BRs signaling and PAMP-triggered immunity (PTI) in plant, respectively (Tian et al. 2014). Subsequent works proved that BAK1 could associate with ligand-binding LRR-RLKs including BRI1, flagellin sensitive 2 (FLS2), EF-Tu receptor (EFR), receptors of BRs, bacterial flagellin, and bacterial elongation factor Tu (EF-Tu), respectively, to form receptor complexes, initiating corresponding phosphorylation cascades and eventually regulating downstream target gene expressions (reviewed in He et al. 2013).
Pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) was recognized as the first line of defense in plant and could be elicited by PRRs (Pattern Recognition Receptors) through specific cell surface located proteins, such as FLS2 and EFR, which recognize the oligopeptides flagellin and EF-Tu, respectively (Chinchilla et al. 2007). Each kind of PAMPs can be recognized by different PRRs with a very similar response pattern in plant cell. It was speculated that there are some common moleculs to contact these signals and BAK1 was the central regulator of innate immunity (Heese et al. 2007). In addition to be a co-receptor of many PRRs to regulate PTI, BAK1 also is the targeted protein of many effectors to regulate ETI reaction (Chen and Zhou 2013), so BAK1 plays a vital role in plant pathogen defense and immunity reaction. RNAseq results indicated that Agrobacterium infection greatly stimulated the PAMP-triggered immunity in soybean, the genes encoding for BRI1, BAK1, BZR1, the bacterial flagellin flg22 related FLS2, as well as bacterial EF-Tu related EFR genes were stimulated greatly by Agrobacterium infection. Effector-triggered immunity was also elicited, for example, disease resistance protein RPM1, RPS2, RPS5, RPM1-interacting protein 4 (RIN4), and serine/threonine-protein kinase PBS1, were all up-regulated by twofold, even though not so much as that in PTI (data not shown). The overexpression of BAK1 benefits the activation of defense associated MPK and WRKY transcription factors in the MAPK pathway and hence to regulate immunity through cascade amplification of MAPK signal (Asai et al. 2002; Yang et al. 2012). In this study, WRKY25, WRKY29, MEKK1P, MKK4/5P and MYC2 were significantly up-regulated by Agrobacterium infection, implying that soybean explants were sensitive to Agrobacterium infection and effective in signal perception and signal amplification to defense this kind of invasion. As a result of defense response, genes related to respiratory burst oxidase (RBOH) and peroxidase were up-regulated to eliminate ROS (Table S2).
Isoflavonoid metabolite is another reason for lower efficiency of Agrobacterium infection
The DEGs caused by Agrobacterium infection were most significantly enriched in phenylpropanoid, flavonoid and isoflavonoid biosynthesis in this study, this was different from the results in wheat (Zhou et al. 2013), where, although a big part of DEGs was categorized in the secondary metabolites and phenylpropanoid biosynthesis, but not in flavonoid and isoflavonoid biosynthesis. Phenylpropanoids may undergo different branches of metabolic pathway in wheat and soybean. Phenylalanine ammonia-lyase (PAL) catalyzes the first step in the biosynthesis of phenylpropanoids, 5 DEGs coding for PAL were found up-regulated with log2Ratio ≥1.5 and PAL activity increased by about 50 %, compared to that of non-infection control. Many genes coding for 4-coumarate-CoA ligase (4CL), ferulate-5-hydroxylase (F5H), HCT, CHS, F3′H, flavonol synthase (FLS), leucoanthocyanidin dioxygenase (LDOX), and 3,9-dihydroxypterocarpan 6a-monooxygenase (CYP93A1), important for flavonoid and isoflavonoid biosynthesis, were dramatically up-regulated in soybean by conventional Agrobacterium infection (Table S2). Soybean isoflavones play diverse roles in plant–microbe interactions; it significantly influenced soybean rhizosphere bacterial diversity (White et al. 2015). Many genes in isoflavone secondary metabolism are involved in plant defense responses. Soybean isoflavone includes two kinds of compounds, isoflavone aglycones and their glucosides, the latter accounts for about 97–98 % of the total isoflavones content and be of biological functions only in the situation that they turned into aglycones type through hydrolysis; β-glucosidase plays a role in this process (Sun et al. 2007). β-Glucosidase was up-regulated 5 h after Agrobacterium infection as revealed by RNAseq results, implying that soybean intended to increase its defense response to Agrobacterium invasion in virtue of active component ascension, but this phenomenon was not validated when p-nitrophenyl-β-d-glucopyranoside was used as substrate to determine the β-glucosidase activity (Table 5). It is also regretted that only total isoflavones content, not each isoflavone components, was determined in this study because of the constrain of experimental condition. Nevertheless, it does not interfere with the conclusions that isoflavones is an important factor in Agrobacterium infection elicited soybean defense response, which was in accordance with our previous conclusion (Zhang et al. 2015).
Improvement of soybean transformation by regulating plant defense response
Intense host defense response is often associated with reduced transformation efficiency, and thus, the attenuation of these responses by external measures can improve transformation efficiency (Pitzschke 2013). The improvement of transformation by manipulating the plant’s immune system to optimize plant–microbe interactions has recently been reported. AvrPto is an effector protein that suppresses plant immunity by interfering with plant immune receptors. Great improvement of transformation was achieved when AvrPto transgenic Arabidopsis plants were infected with Agrobacterium (Tsuda et al. 2012). Zhang et al. (2013) alleviated the defense responses by omission of myo-inositol in culture medium, combined with cold treatment before infection, which promoted Agrobacterium binding to the cell surface and inhibited the ROS after Agrobacterium infection, and ultimately improved Agrobacterium mediated transformation efficiency in perennial ryegrass (Lolium perenne L.). Salicylic acid is another important metabolin in plant defense regulation. To demonstrate the role of SA in Agrobacterium infectivity, Anand et al. (2008) used transgenic tomato plants overexpressing salicylate hydroxylase (NahG), which degrades SA to catechol, to perform GUS activity assays at 2 dpi, results indicated that NahG-expressing plants were more susceptible to Agrobacterium infection (Anand et al. 2008).
Soybean isoflavones had inhibitory effects on A. tumefaciens growth and respiration and was negatively correlated with T-DNA delivery efficiency (Zhang et al. 2015), which was consistent with the reports where silencing genes of isoflavone synthase (IFS) or chalcone reductase (CHR) led to a nearly complete (95 %) suppression of all isoflavone metabolites in soybean roots (Graham et al. 2007) and enhanced susceptibility to Phytophthora sojae (Subramanian et al. 2005; Graham et al. 2007). In this study, antagonist AOA and sonication treatment along with Agrobacterium infection dramatically decreased the expression of genes coding for F3′H, HCT, β-glucosidase and IF7GT, important for isoflavone biosynthesis (Figs. 2, 3, 4); isoflavones content was ultimately decreased by about 19.3 %. IF7GT converts the flavonoid aglycones into glycon in the last step of isoflavonoid biosynthesis. The down-regulation of IF7GT at transcription level maybe caused by the reduction of aglycones substrate, because the biosynthesis of isoflavone aglycon had been disturbed in the upper steps, such as F3′H, HCT and flavonol 3-O-methyltransferase [EC:2.1.1.76]. Genes coding for peroxidase, LRR receptor-like serine/threonine-protein kinase FLS2, serine/threonine-protein kinase PBS1 and transcription factor MYC2, etc., which are important components in plant–pathogen interaction, were also significantly down-regulated. All these factors benefit the transformation process.
Functional categorization of DEGs in phenylpropanoid biosynthesis pathway. Notes In a pairwise comparison (denote as A-vs-B for example), the former one (A) is considered as the control, and the latter one (B) is considered as the treatment, the same hereinafter. Each bar represents a functional group of transcripts with up-regulated (right bar) or down-regulated (left bar) expression in soybean tissue 5 h after Agrobacterium infection, the axis of abscissa represents the number of DEGs in that group
Functional categorization of DEGs in flavonoid biosynthesis pathway. Notes In a pairwise comparison (denote as A-vs-B for example), the former one (A) is considered as the control, and the latter one (B) is considered as the treatment, the same hereinafter. Each bar represents a functional group of transcripts with up-regulated (right bar) or down-regulated (left bar) expression in soybean tissue 5 h after Agrobacterium infection, the axis of abscissa represents the number of DEGs in that group
Functional categorization of DEGs in isoflavonoid biosynthesis pathway. Notes In a pairwise comparison (denote as A-vs-B for example), the former one (A) is considered as the control, and the latter one (B) is considered as the treatment, the same hereinafter. Each bar represents a functional group of transcripts with up-regulated (right bar) or down-regulated (left bar) expression in soybean tissue 5 h after Agrobacterium infection, the axis of abscissa represents the number of DEGs in that group
UDP-glycosyltransferase had been confirmed to participate in the response to pathogens (von Saint Paul et al. 2011). Transgenic A. thaliana expressing a barley UDP-glucosyltransferase exhibited resistance to the mycotoxin deoxynivalenol (Shin et al. 2012). Arabidopsis hat mutant over-expressing a UDP-glucosyltransferase gene was found to be resistant to Agrobacterium-mediated transformation (Gelvin 2010). Zhou et al. (2013) found UDP-glucosyltransferase were up-regulated at the level of transcription after infection by Agrobacterium and inferred that saccharide metabolism might affect the infection process. In this study, the DEGs were also significantly enriched in starch and sucrose metabolism and other glycan degradation in B-vs-C compare set (Table S3; Fig. 5). Glycerol-3-phosphate 1-O-acyltransferase (GPAT) [EC: 3.2.1.15], β-galactosidase [EC:3.2.1.23], alpha-l-fucosidase [EC:3.2.1.51], endoglucanase [EC:3.2.1.4], and β-glucosidase [EC:3.2.1.21], etc., were remarkably down-regulated as revealed by log2Ratio (B/C), but UDP-glucosyltransferase was not significantly enriched in these metabolic pathways. GPAT is a key enzyme in the triacylglycerol biosynthetic pathway, catalyzing triacylglycerol, chitin, suberin and other lipid biosynthesis (Shockey et al. 2015). Chitin and suberin are two important polyesters in plant, which constitute a protective guard to prevent pathogen invasion (Kolattukudy 1980), thus the down-regulation of GPAT may benefit Agrobacterium infection. 29 DEGs coding for α-l-fucosidase were down-regulated by AOA and sonication treatment (Table S3). α-l-Fucosidase is involved in the degradation of fucose-containing glycoproteins and glycolipids, which are located on the external surface of cell membrane and involved in the cell recognition, intercellular communication and immunity. The down-regulation of β-glucosidase impeded the conversion of isoflavones glycon into aglycones, thereby remitting the inhibitory effects of aglycones on Agrobacterium infection. Above analysis implied that AOA and sonication treatment not only relieved the structural membrane barriers of Agrobacterium entering into cells, but also hindered the perception of ‘invasion’ signal on cell membrane and intercellular signal transmission, making the plant unable to establish the corresponding defense system before transformation complement. This may best explain why the T-DNA delivery efficiency was greatly promoted by this novel method. The inhibitors of protein kinase K252a, actinomycin D and cycloheximide were also effective in inhibiting the increase of PAL activity and isoflavones biosynthesis in soybean (data not shown). The shortage of this paper was that the separate effects of antagonist AOA and sonication could not be distinguished owning to the problems in experimental design.
Functional categorization of DEGs in starch and sucrose metabolism and other glycan degradation. Notes In a pairwise comparison (denote as A-vs-B for example), the former one (A) is considered as the control, and the latter one (B) is considered as the treatment, the same hereinafter. Each bar represents a functional group of transcripts with up-regulated (right bar) or down-regulated (left bar) expression in soybean tissue 5 h after Agrobacterium infection, the axis of abscissa represents the number of DEGs in that group
In summary, the high content of endogenous isoflavones and fierce defense response to Agrobacterium infection result in the inefficiency of soybean transformation via Agrobacterium methods. Further improvement of the transformation efficiency lies in the manipulation of the plant itself to reduce the intensity of defense reaction or ignore this invasion signal, hence to increase the susceptibility of plant cells to Agrobacterium infection. This gene expression profile analysis has provided candidate external measures for improving soybean transformation efficiency. In addition to the presence of thiol compounds in co-culture medium, inhibition of isoflavones biosynthesis by sonication treatment and applying antagonists in co-culture medium relieved the structural barriers of Agrobacterium entering into cells and hindered the perception of ‘invasion’ signal on cell membrane and intercellular signal transmission, thus effectively alleviated the defense responses and increased the susceptibility of Agrobacterium infection. The antagonist used in this study was located in the forefront of isoflavones metabolic pathway; inhibitors for enzymes in the downstream of isoflavone biosynthesis pathway should be explored to reduce their impacts on other metabolic pathways, other measures should also be studied.
Author contribution statement
Z.Y.M. conceived, designed research and wrote paper. L.Z.H., Y.R.J., L.G.L. and G.X.L. conducted experiments. Z.H.N., Z.H.M. and D.R. analyzed data. Z.Q.S. contributed analytical tools. Z.M.C. and Z.H.M. provided capital. All authors read and approved the manuscript.
References
Anand A, Uppalapati SR, Ryu CM, Allen SN, Kang L, Tang Y, Mysore KS (2008) Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol 146:703–715
Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415(6875):977–983
Atif RM, Patat-Ochatt EM, Svabova L et al (2013) Gene transfer in legumes. Prog Bot 74:37–100
Chen XC, Zhou YL (2013) Molecular mechanism underlying BAK1 involved in plant immunity signal recognition and transduction. J Plant Genet Resour 14:1102–1107
Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T (2007) A flagellin induced complex of the receptor FLS2 and BAK1 initiates defense. Nature 448(7152):497–500
Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111:671–678
Dang W, Wei ZM (2007) An optimized Agrobacterium-mediated transformation for soybean for expression of binary insect resistance genes. Plant Sci 173:381–389
Deeken R, Engelmann JC, Efetova M et al (2006) An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell 18:3617–3634
Dey S, Wenig M, Langen G, Sharma S et al (2014) Bacteria-triggered systemic immunity in barley is associated with WRKY and ethylene responsive factors but not with salicylic acid. Plant Physiol 166:2133–2151
Ditt RF, Nester E, Comai L (2001) Plant gene expression response to Agrobacterium tumefaciens. Proc Natl Acad Sci USA 98:10954–10959
Ditt RF, Nester E, Comai L (2005) The plant cell defense and Agrobacterium tumefaciens. FEMS Microbiol Lett 247:207–213
Ditt RF, Kerr KF, de Figueiredo P, Delrow J, Comai L, Nester EW (2006) The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol Plant Microbe Interact 19:665–681
Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H (2007) Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science 318:453–456
Gelvin SB (2010) Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu Rev Phytopathol 48:45–68
Gohlke J, Deeken R (2014) Plant responses to Agrobacterium tumefaciens and crown gall development. Front Plant Sci. doi:10.3389/fpls.2014.00155
Graham TL, Graham MY, Subramanian S, Yu O (2007) RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues [OA]. Plant Physiol 144:728–740
Grant M, Lamb C (2006) Systemic immunity. Curr Opin Plant Biol 9:414–420
He K, Xu S, Li J (2013) BAK1 directly regulates brassinosteroid perception and BRI1 activation. J Integr Plant Biol 55:1264–1270
Heese A, Hann DR, Gimenez-Ibanez S et al (2007) The receptor like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104:12217–12222
Institute of Shanghai Plant Physiology, Chinese Academy of Sciences, Shanghai Plant Physiology Society (China) (1999) Guide for modern plant physiology experiments. Science Press, Beijing
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Kolattukudy PE (1980) Biopolyester membranes of plants: cutin and suberin. Science 208(4447):990–1000
Lacroix B, Citovsky V (2013) The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int J Dev Biol 57:467–481
Larkin KM (2001) Optimization of soybean transformation using SAAT and GFP. Wooster: OARDC/OSU 126p. (Thesis-Master)
Lee CW, Efetova M, Engelmann JC, Kramell R, Wasternack C, Ludwig-Müller J, Hedrich R, Deeken R (2009) Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell 21:2948–2962
Li HQ, Yang H, Zhang JJ, Wan XC, Fang CB (2009) The effect of specific inhibitors of phenylalanine ammonia-lyase and 4-coumarate-CoA ligase on isoflavone biosynthesis in Kudzu cell suspension culture. Chin J Trop Crops 30(1):47–52
Liu YK, Kong XP, Pan JW, Li DQ (2010) VIP1: linking Agrobacterium-mediated transformation to plant immunity? Plant Cell Rep 29:805–812
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628
Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signaling. Trends Plant Sci 10:339–346
Olhoft PM, Lin K, Galbraith J, Nielsen NC (2001) The role of thiol compounds in increasing Agrobacterium-mediated transformation of soybean cotyledonary-node cells. Plant Cell Rep 20:731–737
Olhoft PM, Flagel E, Donovan CM, Somers DA (2003) Efficient soybean transformation using hygromycin B selection in the cotyledonary-node method. Planta 216:723–735
Pitzschke A (2013) Agrobacterium infection and plant defense—transformation success hangs by a thread. Front Plant Sci. doi:10.3389/fpls.2013.00519
Pitzschke A, Hirt H (2010) New insights into an old story: Agrobacterium induced tumour formation in plants by plant transformation. EMBO J 29:1021–1032
Pitzschke A, Schikora A, Hirt H (2009) MAPK cascade signaling networks in plant defence. Curr Opin Plant Biol 12:421–426
Shi Y, Lee LY, Gelvin SB (2014) Is VIP1 important for Agrobacterium-mediated transformation? Plant J 79:848–860
Shin S, Torres-Acosta JA, Heinen SJ et al (2012) Transgenic Arabidopsis thaliana expressing a barley UDP-glucosyltransferase exhibit resistance to the mycotoxin deoxynivalenol. J Exp Bot 63:4731–4740
Shockey J, Regmi A, Cotton K, Adhikari N, Browse J, Bates PD (2015) Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol. doi:10.1104/pp.15.01563
Subramanian S, Graham MY, Yu O, Graham TL (2005) RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol 137:1345–1353
Subramoni S, Nathoo N, Klimov E, Yuan ZC (2014) Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front Plant Sci 5:322. doi:10.3389/fpls.2014.00322
Sun YM, Zhang YZ, Xu J, Li WB (2007) Study on hydrolysis of soybean isoflavone glucosides by β-glucosidases from Aspergillus niger. J Northeast Agric Univ 38(1):9–12
Tian R, Yang Y, Wang XF (2014) Research on BAK1 of a receptor kinase. Acta Bot Boreal-Occident Sin 34(3):636–644
Tsuda K, Qi Y, Nguyen LV, Bethke G, Tsuda Y, Glazebrook J, Glazebrook J, Katagiri F (2012) An efficient Agrobacterium-mediated transient transformation of Arabidopsis. Plant J 69:713–719
Veena Jiang H, Doerge RW, Gelvin SB (2003) Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J 35:219–236
von Saint Paul V, Zhang W, Kanawati B, Geist B, Faus-Keßler T, Schmitt-Kopplin P, Schäffner AR (2011) The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 23:4124–4145
White LJ, Jothibasu K, Reese RN, Brözel VS, Subramanian S (2015) Spatio temporal influence of isoflavonoids on bacterial diversity in the soybean rhizosphere. Mol Plant Microbe Interact 28:22–29
Yang H, Min DB, Huang J, Tang L, Huang Y (2012) Transformation and the defense role of BAK1 in Arabidopsis thaliana. Biotechnol Bull 8:71–75
Zhang WJ, Dewey RE, Boss W, Phillippy BQ, Qu R (2013) Enhanced Agrobacterium-mediated transformation efficiencies in monocot cells is associated with attenuated defense responses. Plant Mol Biol 81:273–286
Zhang YM, Zhang HM, Liu ZH, Guo XL, Li HC, Li GL, Jiang CZ, Zhang MC (2015) Inhibition of isoflavone biosynthesis enhanced T-DNA delivery in soybean by improving plant–Agrobacterium tumefaciens interaction. Plant Cell Tissue Organ Cult 121:183–193
Zhao FX, Chen LH, Perl A, Chen SW, Ma HQ (2011) Proteomic changes in grape embryogenic callus in response to Agrobacterium tumefaciens-mediated transformation. Plant Sci 181:485–495
Zhou XH, Wang K, Lv DW, Wu CJ, Li JR, Zhao P, Lin ZS, Du LP, Yan YM, Ye XG (2013) Global analysis of differentially expressed genes and proteins in the wheat callus infected by Agrobacterium tumefaciens. PLoS One 8(11):e79390. doi:10.1371/journal.pone.0079390
Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125:749–760
Acknowledgments
This work was supported by National Major Project for Transgenic Crops of Chinese Agriculture Ministry (Grant No. 2014ZX0800402B); Natural Science Foundation of Hebei Province, China (Grant No. C2013301033); Key project for fundamental research of Hebei Province, China (Grant No. 14962903D).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by H. Ebinuma.
Z.-H. Liu contributed equally as the first author.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, YM., Liu, ZH., Yang, RJ. et al. Improvement of soybean transformation via Agrobacterium tumefaciens methods involving α-aminooxyacetic acid and sonication treatments enlightened by gene expression profile analysis. Plant Cell Rep 35, 1259–1271 (2016). https://doi.org/10.1007/s00299-016-1958-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-1958-2