Abstract
Agrobacterium tumefaciens is the most efficient vehicle used today for the production of transgenic plants and plays an essential role in basic scientific research and in agricultural biotechnology. Previously, plant VirE2-interacting protein 1 (VIP1) was shown to play a role in Agrobacterium-mediated transformation. Recent reports demonstrate that VIP1, as one of the bZIP transcription factors, is also involved in plant immunity responses. Agrobacterium is able to activate and abuse VIP1 for transformation. These findings highlight Agrobacterium-host interaction and unveil how Agrobacterium hijacks host cellular mechanism for its own benefit. This review focuses on the roles played by VIP1 in Agrobacterium-mediated transformation and plant immunity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Agrobacterium tumefaciens is a soil borne, Gram-negative bacterium and is the causative agent of crown gall tumors in plants by transferring and integrating its DNA segment (T-DNA) into the host genome. Recombinant Agrobacterium strains, in which the native T-DNA has been replaced with genes of interests, are the most efficient vehicles used today for the production of transgenic plants (Gelvin 2003; Tzfira and Citovsky 2006). Furthermore, the findings that Agrobacterium was capable, at least under laboratory conditions, of genetically transforming yeast, filamentous fungi, cultivated mushrooms and human cultured cells, places Agrobacterium at the forefront of future biotechnological applications (Lacroix et al. 2006). The basic biological processes underlying Agrobacterium-mediated transformation have been intensively studied. However, the biological processes are not fully understood.
Similar to many other plant pathogens, A. tumefaciens is a disease-causing bacterium (Escobar and Dandekar 2003). Accumulating evidence indicate that A. tumefaciens activates the induction of various defense genes at early stages of the infection process. At later stages, however, this pathogen may actually repress some of the defense genes for T-DNA transformation (Veena et al. 2003; Ditt et al. 2006). Furthermore, A. tumefaciens modulates plant hormone balance which is known to be involved in plant defense against pathogens (Lee et al. 2009). However, the interactions between Agrobacterium-mediated transformation and plant immunity are largely unknown. VirE2-interacting protein 1 (VIP1) was originally shown to be a VirE2-interacting protein and required for VirE2 nuclear import and Agrobacterium tumorigenicity (Tzfira et al. 2001). Further studies demonstrated that VIP1 interacts with more Agrobacterium and plant proteins (Table 1). Recently, the VIP1, as one of the bZIP transcription factors, was shown to play a role in plant immunity signaling (Djamei et al. 2007; Pitzschke et al. 2009a). Here, we discuss the roles played by VIP1 in Agrobacterium-mediated transformation and plant immunity.
Role of VIP1 in nuclear import of T-complex
Agrobacterium-mediated plant transformation is a complex process involving numerous steps. These include (1) bacterial attachment; (2) induction of virulence genes; (3) T-strand generation; (4) T-strand and virulence factor export to the host cytoplasm; (5) cytoplasmic trafficking and nuclear import of T-complex; (6) uncoating of the T-complex; (7) T-DNA integration and (8) T-DNA gene expression (for comprehensive understanding of Agrobacterium-mediated plant transformation, see recent reviews, Gelvin 2010b; Pitzschke and Hirt 2010).
Processed from the bacterial tumor-inducing plasmid, T-DNA fragment is a single-stranded DNA molecular (T-strand). Then, T-strand, with virulence (vir) protein VirD2 remains covalently attached to the 5′ end (Durrenberger et al. 1989), transports into the host-cell cytoplasm via T4SS (type IV secretion system) (Chandran et al. 2009; Fronzes et al. 2009a, b). Inside the host cytoplasm, T-strand is thought to be covered with numerous virulence protein VirE2 molecules, which translocate into the host cells independently of the T-strand (Gelvin 1998). T-strand covered with VirE2 is a mature T-strand-protein complex (T-complex) and that is then imported into the host-cell nucleus by an active mechanism mediated by the nuclear import machinery of the host cell. It is established that both VirD2 and VirE2 are required for the efficient import of the T-complex (Citovsky et al. 1994; Ziemienowicz et al. 2001; Pelczar et al. 2004).
Involvement of VIP1 in the transformation process was indicated by the observation that plants expressing a VIP1 anti-sense gene showed reduced transformation rate, and that over-expression of VIP1 increased the transformation efficiency of plant cells by Agrobacterium (Tzfira et al. 2001, 2002). VIP1 was shown to interact with VirE2 but not with VirD2 (Tzfira et al. 2001). The nuclear import of VirE2 was induced by co-expression of VIP1. VIP1 contains a conventional NLS and interacts directly with karyopherin α (Tzfira et al. 2002; Citovsky et al. 2004). Thus, VIP1 probably functions by forming a molecular adaptor between the T-complex and the karyopherin α-mediated nuclear import machinery of the host cell. Indeed, the ternary complexes, VIP1–VirE2–ssDNA and VIP1–VirE2–karyopherin α were observed in vitro (Tzfira et al. 2001; Citovsky et al. 2004) (Table 1).
Although homologs of Arabidopsis VIP1 are found in several diverse plant species, there is no yeast or animal homolog of VIP1, suggesting a role for VIP1 in plant-specific nuclear import of VirE2 (Tzfira et al. 2001). VIP1, as well as other plant proteins, may thus represent a set of limiting host factors for Agrobacterium-mediated transformation. Their absence in a functional form may explain the recalcitrance of animal cells and some plant species. However, VIP1 is not an abundant cellular protein (Tzfira et al. 2001; Avivi et al. 2004) and its function in the T-complex nuclear import is augmented by the bacterial effector, VirE3 (Lacroix et al. 2005). In addition, VirD2 and VirE2 are likely to have different and complementary functions in nuclear import of T-complex (Ziemienowicz et al. 2001; Pelczar et al. 2004). Further, VirE2 interacts with numerous importin α isoforms (Bhattacharjee et al. 2008), suggesting in addition to via VIP1, other mechanism may exist for nuclear import of the T-complex (Gelvin 2010a). The function of VIP1 in this process needs further to be elucidated.
Role of VIP1 in intranuclear transport of T-complex
Although little is known about movement of the T-complex inside the nucleus, it is speculated that some mechanism must exist, as macromolecule trafficking in the nucleus is suggested to be under tight regulation (Phair and Misteli 2000). Arabidopsis vip1-1 mutant encodes N-terminal portion of VIP1. Studies on VIP1-N164 (a recombinant protein corresponding to the N-terminal 164 amino acid of VIP1 sequence in the vip1-1 mutant) indicated that VIP1-N164 and wild-type VIP1 showed no difference in interacting with VirE2, subcellular localization and transient expression of T-DNA (Li et al. 2005), suggesting N-terminal portion of VIP1 generated by the vip1-1 mutation most likely retains its ability to facilitate nuclear import of VirE2 in plant cells. However, C-terminal portion of VIP1 is involved in stable transformation (Li et al. 2005), supporting the involvement of VIP1 in intranuclear function.
As a bZIP protein, VIP1 is also involved in transcription (Pitzschke et al. 2009a) and this transcriptional process associates with the chromosomal DNA either directly or through other components of transcription complex. Thus, VIP1 may perform second function: facilitating nuclear targeting of T-complex and playing a role in the intranuclear transport of VirE2 and its cognate T-complex to the site of integration. If this holds true, VIP1 may resemble the yeast karyopherin Kap114p, of which functions are to import the TBP (TATA-binding protein) into the cell nucleus, and to target it to the promoters of genes to be transcribed (Pemberton et al. 1999).
In addition to be involved in stable transformation, C-terminal parts of VIP1 are required for VIP1 dimerization and interaction with H2A (Li et al. 2005). It seems that this potential intranuclear targeting of VIP1 requires protein multimerization that is inhibited by the C-terminal truncation. This inhibition may block the ability of VIP1 to transport T-complexes to the plant cell chromatin, thereby reducing the stable transformation. Indeed, VIP1 has been shown to interact with the plant histone H2A in planta, as well as with purified Xenopus core histones H2A, H2B, H3 and H4 in vitro (Li et al. 2005; Loyter et al. 2005) (Table 1). Consistent with interaction of VIP1 with histones, formation of bZIP protein dimers is thought to play a role in their interactions with the host chromatin (Baxevanis and Vinson 1993).
Role of VIP1 in T-DNA integration
T-DNA integration is an important step in Agrobacterium-mediated transformation process. Compared with T-strand generation and nuclear import of T-complex, T-DNA integration process is much less known, although several different T-DNA integration mechanisms have been suggested (Tzfira et al. 2004a; Ziemienowicz et al. 2008). It is possible that T-DNA molecules integrate into plant-cell genomes by not one, but several mechanisms.
VIP1 is also thought to be involved in uncoating of the T-complex and perturbing the host chromatin before T-DNA integration. This function links to the A. tumefaciens F-box protein, VirF (Tzfira et al. 2004b). VirF contains an F-box motif and interacts with several members of the ASK (Arabidopsis SKP1-LIKE) protein family of Arabidopsis homologs of yeast Skp1 (S-phase kinase-associated protein 1) protein (Schrammeijer et al. 2001). F-box and Skp1 represent the conserved components of E3 ubiquitin ligases called SCF (Skp1-Cullin-F-box protein) complexes that specify protein destabilization by targeted proteasomal degradation (Ho et al. 2008). VIP1 represents one of the cellular substrates for VirF. VirF interacts and destabilizes VIP1 when coexpressed in yeast cells or in planta. In yeast, this destabilization requires the presence of Skp1 (Tzfira et al. 2004b).
VirF, VIP1 and VirE2 colocalize to the plant cell nucleus, the cellular compartment in which the T-DNA uncoating is expected to occur. VirF was shown to destabilize both VIP1 and VirE2. Interaction and destabilization of VIP1 by VirF play crucial roles in the stripping of proteins from the T-DNA. Destabilization of VirE2 by VirF was VIP1-dependent, suggesting that although VirF does not recognize VirE2, proteolysis of VirE2 is affected indirectly through the VIP1–VirE2 interaction (Tzfira et al. 2004b). The function of VirF in T-DNA integration is based on the assumption that binding of VIP1-associated T-complex can occur at the site of integration, i.e. at the host chromatin. This notion gained support from the observations that VirF can exist in complex with VIP1, VirE2 and nucleosomes (Table 1), in which VIP1 represents the link for all of the other components (Lacroix et al. 2008). Furthermore, because VirF can promote degradation of VIP1-bound VirE2 and because VIP1 interacts with nucleosome, it is tempting to speculate that the degradation of VIP1 by VirF may effectively perturb the host chromatin and facilitate integration of the bacterial T-DNA. It should be noted, however, that the T-complex–chromatin interactions is based on an in vitro system. How well the in vitro assay can reflect their in vivo function needs further assessment.
Interestingly, although phosphorylation is known to enhance VIP1 nuclear import (Djamei et al. 2007), this phosphorylation seems not involved in its chromatin targeting, since VIP1 and its phosphorylation mutant showed no difference in their capacity to bind nucleosomes or promote the association of VirE2 with nucleosomes (Lacroix et al. 2008). Interesting question about VirF is that it is required for transformation of some but not all plant species (Hirooka et al. 1987; Regensburg-Tuink and Hooykaas 1993). Recent research demonstrated plant F-box protein VBF (VIP1-binding F-box protein) can functionally replace the Agrobacterium VirF in regulating levels of the VirE2 and VIP1 proteins (Zaltsman et al. 2010), suggesting Agrobacterium may harness the host SCF pathways for its own benefit. Mutation in F-box-encoding Arabidopsis gene resulted in decreased susceptibility of plant roots to Agrobacterium-mediated transformation (Zhu et al. 2003), confirming the importance of protein degradation mediated by host factors.
VIP1 mediates induction of plant immunity
Although VIP1’s functions in Agrobacterium-mediated transformation process have been studied, its function in plant biological process is largely unknown. Does the only purpose of this plant protein merely lie in assisting pathogen invasion? Recently, VIP1 was demonstrated to participate in plant immunity signaling and was phosphorylated by MPK3 (mitogen-activated protein kinase 3). Phosphorylation of VIP1 by MPK3 is required for VIP1 translocation into the host-cell nucleus and for activation of defense gene expression (Djamei et al. 2007).
VIP1 belongs to subgroup I of the bZIP family of A. thaliana (Jakoby et al. 2002). Recently, VRE (VIP1 response element), a DNA hexamer motif (ACNGCT), was demonstrated to be direct target of VIP1 (Pitzschke et al. 2009a). VRE represents a novel bZIP-binding DNA motif, as it does not match any characterized regulatory DNA motif, nor does it contain the TGAC core sequence commonly targeted by various bZIP proteins (Foster et al. 1994). Further study showed that Trxh8 and MYB44 (harboring 6 and 3 VREs, respectively) are direct VIP1 target genes and that direct binding through the bZIP domain to these cognate VRE sequences is necessary for promoter activation (Pitzschke et al. 2009a). Thus, VIP1, upon phosphorylation by MPK3, translocates into the nucleus where it induced stress gene expression via binding VREs of gene promoter. Consistently, stress-induced Trxh8 and MYB44 expression were lower in mpk3 mutant than in wild-type Arabidopsis. Furthermore, VIP1-overexpressing Arabidopsis showed pronounced accumulation of MYB44 and Trxh8 transcripts. VREs are also more abundant in promoters of genes differentially expressed in mkp1 (MAPK phosphatase1) mutants, which are impaired in the MPK3-inactivation (Bartels et al. 2009).
Previously, overexpression of VIP1 in tobacco resulted in developmental retardation (Avivi et al. 2004). It is proposed that VIP1 regulates stress gene expression by VIP1-targeted tobacco promoters. However, this hypothesis does not exclude the possibility that VIP1-overexpressing tobacco might be the consequence of an imbalance of endogenous bZIP proteins. If VIP1 overexpression activated stress gene expression also in tobacco, suppression of plant growth as a consequence of activated immunity might reflect the functionality of an innate trade-off between plant immunity and growth programs.
Spatial restriction of defense regulators by the nuclear envelope and stimulus-induced nuclear translocation constitute an important level of defense-associated gene regulation (Wiermer et al. 2007; Garcia and Parker 2009). Recently, a plant F-box protein VBF was demonstrated to promote destabilization of VIP1 in plant cell nucleus (Zaltsman et al. 2010). Under normal plant-growth conditions, it is possible that VIP1 predominately localizes to the cytoplasm and the nuclear VIP1 may be continuously cleared via proteasome-mediated degradation to prevent transcription of target genes. VIP1 may be retained partially in the cytoplasm as complex with other proteins and its phosphorylation by MPK3 is responsible for release and nuclear localization. Recently, MPK6 and MPK4 were reported to regulate gene expression by releasing transcription factors upon activation (Qiu et al. 2008; Bethke et al. 2009). Whether MPK3 regulates VIP1 activity via a manner resembling that of MPK6 and MPK4 is an interesting question.
Agrobacterium-induced plant immunity signaling
In addition to a range of preformed barriers, plants possess an elaborate multi-layered defense system that relies on the intrinsic ability of plant cells to perceive the presence of pathogens and trigger local and systemic responses (Chisholm et al. 2006; Jones and Dangl 2006). Transmembrane PRRs (pattern recognition receptors) detect highly conserved microbial features, PAMPs (pathogen/microbe-associated molecular pattern), and activate signaling cascades that induce defense gene expression (Boller and Felix 2009; Nicaise et al. 2009; Pitzschke et al. 2009b). These responses are referred to as PTI (PAMP-triggered immunity). PTI occurs immediately upon host-pathogen contact and is considered as the first inducible layer of plant defense (Zipfel 2009). The unique and sophisticated mechanism of A. tumefaciens in infecting and colonizing the plant prompted us to ask how the plant perceives and responds to this pathogen.
The best known PTI paradigm in plants is the flagellin sensing 2 (FLS2)-mediated signaling pathway (Panstruga et al. 2009; Pitzschke et al. 2009b). In Arabidopsis, the leucine-rich repeat receptor kinase (LRR-RK) FLS2 senses bacterial flagellin, or derivatives of the conserved flg22 epitope (Felix et al. 1999; Gomez-Gomez and Boller 2000), and initiates immune signaling by association with another LRR-RK-like kinase, BRI1-associated kinase 1 (BAK1) (Chinchilla et al. 2007; Heese et al. 2007). More recently, botrytis-induced kinase 1 (BIK1) was demonstrated to be released from the FLS2–BAK1 complex to activate downstream intracellular signaling (Lu et al. 2010). Mitogen-activated protein kinase (MAPK, MPK) cascades, minimally composed of a MAPKKK (MAPKK kinase, MEKK), a MAPKK (MAPK kinase, MKK) and a MAPK, are signaling modules that transduce extracellular stimuli to a range of cellular responses (MAPK Group 2002; Pitzschke et al. 2009b; Andreasson and Ellis 2010). Downstreaming of flg22 perception, FLS2 activates two simultaneous MAPK cascades: one consists of an unknown MEKK (MEKK-MKK4/5-MPK3/6) and acts positively on PTI, while the other, consisting of MEKK1-MKK1/2-MPK4, acts negatively on PTI (Nicaise et al. 2009).
Most interestingly, flagellin preparations from A. tumefaciens, even at high concentrations, were completely inactive in inducing plant defense responses, e.g. alkalinization response and oxidative burst. It was found that flagellin sequences from A. tumefaciens are extremely divergent from the N-terminal domain conserved in other bacterial species (Felix et al. 1999). Whether this flagellin is originally non-detectable by FLS2 receptor or the inactive is the consequence of co-evolution of Agrobacterium-plant is unclear. It is speculated that microbes living in close association with plant may be under selective pressure to modify or lose these molecules that could potentially alarm the plant surveillance of their presence.
At present, cold shock protein (CSP) (Felix and Boller 2003), elongation factor Tu (EF-Tu) (Kunze et al. 2004; Zipfel et al. 2006) and peptidoglycan (PGN) (Erbs et al. 2008) from A. tumefaciens have been demonstrated as PAMPs. However, the only defined PRR is EF-Tu receptor (EFR) (Zipfel et al. 2006). EFR and FLS2 belong to the same subfamily of LRR-RLKs, LRRXII. Moreover, flagellin and EF-Tu activate a common set of signaling events and defense responses but without clear synergistic effects (Zipfel et al. 2006; Aslam et al. 2009).
Both A. tumefaciens and elf18 (conserved epitope of EF-Tu) can induce activities of MPK3, MPK4 and MPK6 (Zipfel et al. 2006; Djamei et al. 2007; Nekrasov et al. 2009; Saijo et al. 2009). Elf18-induced MAPK activation is EFR-dependent, since this MAPK activation was almost completely abolished in efr plants (Saijo et al. 2009). No additive effect on these kinases was detectable for co-treatment with elf18 and flg22, suggesting signaling induced by the two PAMPs appears to converge at a step upstream of MAP kinase cascades (Zipfel et al. 2006). The convergent point might be BAK1 (Chinchilla et al. 2009), albeit the direct interaction between EFR and BAK1 has not yet been reported. The weaker effect of the bak1 mutation on elf18 responses suggests that other adaptors might interact with EFR (Chinchilla et al. 2007). BIK1 is also phosphorylated by EF-Tu in addition to flg22, suggesting EFR may also use BIK1 for signal transduction (Lu et al. 2010). Thus, Agrobacterium triggers Arabidopsis PTI, at least via EFR-mediated signaling transduction.
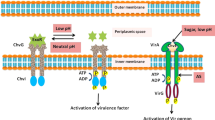
VIP1 is a transcription factor in host defense, whose subcellular localization is regulated by Agrobacterium-induced MAPK cascade (Agrobacterium-EFR-?-MEKK-MKK-MPK3-VIP1). To perform its functions, VIP1 is phosphorylated by MPK3 and transfers to the host-cell nucleus where VIP1 activates pathogenesis-related genes via VREs. Agrobacterium may use the phosphorylated VIP1 to deliver its T-complex into the host-cell nucleus, suggesting a mechanism that in addition to subvert host defense, Agrobacterium-mediated transformation is closely interlaced with host defense signaling (Fig. 1).
Conceptual model for plant immunity signaling in Agrobacterium-mediated transformation. Following vir-region activation, T-strands are processed from the Ti-plasmid and enter the plant cell via T4SS. Once inside the plant-cell cytoplasm, the T-strand is thought to be packed with numerous virE2 which separately enters the plant cell via T4SS. Then, T-complex travels through the cytoplasm and is actively imported into the host-cell nucleus, where T-DNA is stripped and integrated into the host genome. Using EFR receptor, plants are able to recognize EF-Tu of Agrobacterium. Upon recognition of EF-Tu, EFR activates downstream signaling including MPK3 activation. VIP1 is phosphorylated by MPK3 and transfers to the nucleus in which VIP1 activates pathogenesis-related genes via VREs. As one mechanism, Agrobacterium T-complex is attached to VIP1 and via backpacking enters the plant nucleus
Since Agrobacterium induces and uses the host defense signaling to deliver its T-complex into the host-cell nucleus, the question is why Arabidopsis efr mutants showed enhanced susceptibility to the A. tumefaciens, as revealed by a higher efficiency of T-DNA transformation (Zipfel et al. 2006). The explanation for the seemingly antagonistic interaction between MPK3-VIP1 signaling and EFR signaling may be that although EFR-mediated MPK3 signaling may facilitate nuclear import of T-DNA, the central function of EFR signaling is involved in Arabidopsis defense that can restrict intruding of most pathogens. Consistent with this notion, comparison of differentially expressed genes revealed most of Arabidopsis genes affected by Agrobacterium strain C58 responded in a similar manner to elf26 (conserved epitope of EF-Tu) (Lee et al. 2009).
Conclusion and perspectives
Agrobacterium tumefaciens is best known for horizontal gene transfer that plays an essential role in basic scientific research and in agricultural biotechnology. Similar to many other pathogens, Agrobacterium hijacks host cellular mechanisms and signaling pathways for infection. Recent reports support the model that T-complex is attached to VIP1 and via backpacking enters the plant nucleus (Dafny-Yelin et al. 2008) (Fig. 1). These studies highlighted the interplay between plant immunity and Agrobacterium-mediated transformation. However, since EFR is only found in the Brassicaceae and may have evolved more recently (Zipfel et al. 2006), it is not clear whether the ability of Agrobacterium-induced MAPK signaling in transformation is a common strategy.
Despite the extensive research on VIP1, several key issues are still unresolved. The role of VIP1 phosphorylation and dimerization in nuclear import and integration of T-DNA needs further to be studied. VirF is able to bind VIP1 for proteolysis and thus uncoating of T-DNA. Is there mechanism to prevent undesirable proteolysis of VIP1 in cell cytoplasm or in the nucleus before chromatin targeting?
VirF is one of the substrates of T4SS and assists in Agrobacterium infection via interacting with VIP1. It should be noted, however, that all characterized effectors identified to date participate in some way to the T-DNA transformation. Since Agrobacterium could manipulate plant hormone balance and defense gene expression, whether the armament of translocated effectors includes proteins whose functions are unrelated to T-DNA movement and instead involved in disruption of plant physiological processes to promote the overall infection process is an interesting question.
Further studies are also clearly required in order to shed more light on key issues associated with plant immunity signaling in Agrobacterium-mediated transformation. Although Agrobacterium and EF-Tu can trigger MAPK activities, the direct link between EF-Tu-triggered MPK3 and VIP1 phosphorylation is still lacking. Since Agrobacterium-triggered efr defense responses have not been assessed, whether other PAMPs from Agrobacterium also trigger signaling resembling that mediated by EFR remains open question.
Answering these questions can contribute to elucidate how Agrobacterium interacts with host immunity and contribute to our complete understanding of T-DNA transfer. In the long run, progress in this field will allow more easy engineering of agronomically important crops.
References
Anand A, Krichevsky A, Schornack S, Lahaye T, Tzfira T, Tang Y, Citovsky V, Mysore KS (2007) Arabidopsis VIRE2 INTERACTING PROTEIN2 is required for Agrobacterium T-DNA integration in plants. Plant Cell 19:1695–1708
Andreasson E, Ellis B (2010) Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci 15:106–113
Aslam SN, Erbs G, Morrissey KL, Newman MA, Chinchilla D, Boller T, Molinaro A, Jackson RW, Cooper RM (2009) Microbe-associated molecular pattern (MAMP) signatures, synergy, size and charge: influences on perception or mobility and host defence responses. Mol Plant Pathol 10:375–387
Avivi Y, Morad V, Ben-Meir H, Zhao J, Kashkush K, Tzfira T, Citovsky V, Grafi G (2004) Reorganization of specific chromosomal domains and activation of silent genes in plant cells acquiring pluripotentiality. Dev Dyn 230:12–22
Bartels S, Anderson JC, Besteiro MA, Carreri A, Hirt H, Buchala A, Metraux JP, Peck SC, Ulm R (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21:2884–2897
Baxevanis AD, Vinson CR (1993) Interactions of coiled coils in transcription factors: where is the specificity? Curr Opin Genet Dev 3:278–285
Bethke G, Unthan T, Uhrig JF, Poschl Y, Gust AA, Scheel D, Lee J (2009) Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc Natl Acad Sci USA 106:8067–8072
Bhattacharjee S, Lee LY, Oltmanns H, Cao H, Veena, Cuperus J, Gelvin SB (2008) IMPa-4, an Arabidopsis importin alpha isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell 20:2661–2680
Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406
Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G (2009) Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–1015
Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448:497–500
Chinchilla D, Shan L, He P, de Vries S, Kemmerling B (2009) One for all: the receptor-associated kinase BAK1. Trends Plant Sci 14:535–541
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124:803–814
Citovsky V, Warnick D, Zambryski P (1994) Nuclear import of Agrobacterium VirD2 and VirE2 proteins in maize and tobacco. Proc Natl Acad Sci USA 91:3210–3214
Citovsky V, Kapelnikov A, Oliel S, Zakai N, Rojas MR, Gilbertson RL, Tzfira T, Loyter A (2004) Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J Biol Chem 279:29528–29533
Dafny-Yelin M, Levy A, Tzfira T (2008) The ongoing saga of Agrobacterium-host interactions. Trends Plant Sci 13:102–105
Ditt RF, Kerr KF, de Figueiredo P, Delrow J, Comai L, Nester EW (2006) The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol Plant Microbe Interact 19:665–681
Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H (2007) Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science 318:453–456
Durrenberger F, Crameri A, Hohn B, Koukolikova-Nicola Z (1989) Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc Natl Acad Sci USA 86:9154–9158
Erbs G, Silipo A, Aslam S, De Castro C, Liparoti V, Flagiello A, Pucci P, Lanzetta R, Parrilli M, Molinaro A, Newman MA, Cooper RM (2008) Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomonas elicit plant innate immunity: structure and activity. Chem Biol 15:438–448
Escobar MA, Dandekar AM (2003) Agrobacterium tumefaciens as an agent of disease. Trends Plant Sci 8:380–386
Felix G, Boller T (2003) Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J Biol Chem 278:6201–6208
Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18:265–276
Foster R, Izawa T, Chua NH (1994) Plant bZIP proteins gather at ACGT elements. FASEB J 8:192–200
Fronzes R, Christie PJ, Waksman G (2009a) The structural biology of type IV secretion systems. Nat Rev Microbiol 7:703–714
Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G (2009b) Structure of a type IV secretion system core complex. Science 323:266–268
Garcia AV, Parker JE (2009) Heaven’s Gate: nuclear accessibility and activities of plant immune regulators. Trends Plant Sci 14:479–487
Gelvin SB (1998) Agrobacterium VirE2 proteins can form a complex with T strands in the plant cytoplasm. J Bacteriol 180:4300–4302
Gelvin SB (2003) Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67:16–37
Gelvin SB (2010a) Finding a way to the nucleus. Curr Opin Microbiol 13:53–58
Gelvin SB (2010b) Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu Rev Phytopathol 48:3.1–3.24. doi:10.1146/annurev-phyto-080508-081852
Gomez-Gomez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5:1003–1011
Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104:12217–12222
Hirooka T, Rogowsky PM, Kado CI (1987) Characterization of the virE locus of Agrobacterium tumefaciens plasmid pTiC58. J Bacteriol 169:1529–1536
Ho MS, Ou C, Chan YR, Chien CT, Pi H (2008) The utility F-box for protein destruction. Cell Mol Life Sci 65:1977–2000
Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16:3496–3507
Lacroix B, Vaidya M, Tzfira T, Citovsky V (2005) The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J 24:428–437
Lacroix B, Tzfira T, Vainstein A, Citovsky V (2006) A case of promiscuity: Agrobacterium’s endless hunt for new partners. Trends Genet 22:29–37
Lacroix B, Loyter A, Citovsky V (2008) Association of the Agrobacterium T-DNA-protein complex with plant nucleosomes. Proc Natl Acad Sci USA 105:15429–15434
Lee CW, Efetova M, Engelmann JC, Kramell R, Wasternack C, Ludwig-Muller J, Hedrich R, Deeken R (2009) Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell 21:2948–2962
Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V (2005) Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc Natl Acad Sci USA 102:5733–5738
Loyter A, Rosenbluh J, Zakai N, Li J, Kozlovsky SV, Tzfira T, Citovsky V (2005) The plant VirE2 interacting protein 1. A molecular link between the Agrobacterium T-complex and the host cell chromatin? Plant Physiol 138:1318–1321
Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107:496–501
MAPK Group (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7:301–308
Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, van Esse HP, Jorda L, Schwessinger B, Nicaise V, Thomma BP, Molina A, Jones JD, Zipfel C (2009) Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J 28:3428–3438
Nicaise V, Roux M, Zipfel C (2009) Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol 150:1638–1647
Panstruga R, Parker JE, Schulze-Lefert P (2009) SnapShot: plant immune response pathways. Cell 136:978
Pelczar P, Kalck V, Gomez D, Hohn B (2004) Agrobacterium proteins VirD2 and VirE2 mediate precise integration of synthetic T-DNA complexes in mammalian cells. EMBO Rep 5:632–637
Pemberton LF, Rosenblum JS, Blobel G (1999) Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J Cell Biol 145:1407–1417
Phair RD, Misteli T (2000) High mobility of proteins in the mammalian cell nucleus. Nature 404:604–609
Pitzschke A, Hirt H (2010) New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J 29:1021–1032
Pitzschke A, Djamei A, Teige M, Hirt H (2009a) VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc Natl Acad Sci USA 106:18414–18419
Pitzschke A, Schikora A, Hirt H (2009b) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12:421–426
Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, Brodersen P, Grasser KD, Mattsson O, Glazebrook J, Mundy J, Petersen M (2008) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J 27:2214–2221
Regensburg-Tuink AJ, Hooykaas PJ (1993) Transgenic N. glauca plants expressing bacterial virulence gene virF are converted into hosts for nopaline strains of A. tumefaciens. Nature 363:69–71
Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Haweker H, Dong X, Robatzek S, Schulze-Lefert P (2009) Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J 28:3439–3449
Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuink TJ, Crosby WL, Hooykaas PJ (2001) Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr Biol 11:258–262
Tzfira T, Citovsky V (2006) Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr Opin Biotechnol 17:147–154
Tzfira T, Vaidya M, Citovsky V (2001) VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J 20:3596–3607
Tzfira T, Vaidya M, Citovsky V (2002) Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis nuclear protein VIP1. Proc Natl Acad Sci USA 99:10435–10440
Tzfira T, Li J, Lacroix B, Citovsky V (2004a) Agrobacterium T-DNA integration: molecules and models. Trends Genet 20:375–383
Tzfira T, Vaidya M, Citovsky V (2004b) Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431:87–92
Veena, Jiang H, Doerge RW, Gelvin SB (2003) Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J 35:219–236
Wiermer M, Palma K, Zhang Y, Li X (2007) Should I stay or should I go? Nucleocytoplasmic trafficking in plant innate immunity. Cell Microbiol 9:1880–1890
Zaltsman A, Krichevsky A, Loyter A, Citovsky V (2010) Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe 7:197–209
Zhu Y, Nam J, Humara JM, Mysore KS, Lee LY, Cao H, Valentine L, Li J, Kaiser AD, Kopecky AL, Hwang HH, Bhattacharjee S, Rao PK, Tzfira T, Rajagopal J, Yi H, Veena, Yadav BS, Crane YM, Lin K, Larcher Y, Gelvin MJ, Knue M, Ramos C, Zhao X, Davis SJ, Kim SI, Ranjith-Kumar CT, Choi YJ, Hallan VK, Chattopadhyay S, Sui X, Ziemienowicz A, Matthysse AG, Citovsky V, Hohn B, Gelvin SB (2003) Identification of Arabidopsis rat mutants. Plant Physiol 132:494–505
Ziemienowicz A, Merkle T, Schoumacher F, Hohn B, Rossi L (2001) Import of Agrobacterium T-DNA into plant nuclei: two distinct functions of VirD2 and VirE2 proteins. Plant Cell 13:369–383
Ziemienowicz A, Tzfira T, Hohn B (2008) Mechanisms of T-DNA integration. In: Tzfira T, Citovsky V (eds) Agrobacterium: from biology to biotechnology. Springer, Berlin, pp 395–440
Zipfel C (2009) Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol 12:414–420
Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125:749–760
Acknowledgments
We apologize to authors whose work was not cited because of space limitations. Researches in our laboratory are supported by the National Natural Science Foundation of China (Nos. 30471052, 30871457), the State Key Basic Research and Development Plan of China (No. 2009CB118500) and the Program for Changjiang Scholars and Innovative Research Team in University (No. IRT0635).
Conflict of interest statement
The authors have no conflict of interest with the work cited in this paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Reski.
Rights and permissions
About this article
Cite this article
Liu, Y., Kong, X., Pan, J. et al. VIP1: linking Agrobacterium-mediated transformation to plant immunity?. Plant Cell Rep 29, 805–812 (2010). https://doi.org/10.1007/s00299-010-0870-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-010-0870-4