Abstract
Key message
The study of insect-resistant transgenic tobacco provides a good foundation for the further application of the cry1Ah gene in other important crops.
Abstract
To improve transgene expression levels and insect resistance, the coding sequence of the novel Bacillus thuringiensis insecticidal gene cry1Ah (truncated cry1Ah) was modified according to the codon bias of the plant by increasing its GC content from the original 37 % to 48, 55, and 63 % (designated m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah, respectively). In addition, the m3-cry1Ah gene was linked with a transit peptide sequence for chloroplast-targeted expression (designated ctp-m3-cry1Ah). Four plant expression vectors were constructed harboring m1-cry1Ah, m2-cry1Ah, m3-cry1Ah, or ctp-m3-cry1Ah. A total of 23 transgenic tobacco lines were produced with the four constructs by Agrobacterium tumefaciens-mediated transformation. PCR, Southern hybridization, quantitative RT-PCR and ELISA indicated that the cry1Ah gene was not only integrated into the tobacco genome, but was also successfully expressed at the mRNA and protein levels. The Cry1Ah protein level in ctp-m3-cry1Ah plants reached 4.42 μg/g fresh weight, which was a 2- to 10-fold increase over the levels observed in m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah plants and resulted in the highest resistance to Helicoverpa armigera based on bioassays. Our results demonstrated that combining the codon optimization of cry1Ah gene with the targeting of Cry1Ah protein to the chloroplasts conferred a high level of protection against insects. The results of our experiments in tobacco, an important model system, provide a good foundation for enhancing the insecticidal efficacy of staple crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Insect pests cause huge crop losses and necessitate the extensive use of insecticides; thus, the development of insect-resistant transgenic crops is necessary. Until recently, only members of the Bacillus sp., particularly Bacillus thuringiensis (Bt), have been commonly used (Kumar et al. 2008). The Bt proteins cause insect death by disrupting midgut cells (Bravo et al. 2007); however, these proteins are not toxic to non-target insects, birds, or mammals (including humans) because of their high specificity for certain species of insects (Kim et al. 2009). Bt crops have increased yields and reduced the use of insecticides, thus, providing a variety of benefits for human health and the environment (Schwember 2008). The use of Bt genes in insect-resistant genetically modified crops has been a major biotechnological breakthrough in the protection of economically important crops from infestation (Gatehouse 2008).
The cry1Ah gene is a novel insecticidal gene that was cloned from B. thuringiensis isolate BT8 (Xue et al. 2008). The Cry1Ah protein is highly toxic to lepidopteran insects: this protein is more toxic than Cry1Ac to Helicoverpa armigera, Ostrinia furnacalis, and Chilo suppressalis, and it is more toxic than Cry1Ab to O. furnacalis. The truncated active Cry1Ah toxin with a molecular weight of 65 kDa has a toxicity that is similar to that of the full-length Cry1Ah toxin.
One of the key factors that affect exogenous gene expression is the GC content or codon usage pattern. The GC content of bacterial endotoxin genes is low, whereas that of plant genes tends to be high (Campbell and Gowri 1990; Murray et al. 1989). Vaeck et al. (1987) showed an expression of the cry1Ab gene in plant cells for the first time and found that exogenous protein expression accounted for only 0.0002–0.02 % of the total soluble protein. A low level of Cry1Ab protein accumulated in transgenic sugarcane plants (Arencibia et al. 1997). Increasing the GC content of the cry1Ac gene and transforming it into sugarcane plants led to increased protein expression (Weng et al. 2006). Thus, codon preference is a major barrier to the expression of bacterial genes in plants. Different species have different preferences for synonymous codons of a given amino acid. Genes from bacteria should be optimized for their codon usage before they are transformed into plants. Targeting foreign proteins to specific subcellular locations, such as the chloroplasts or endoplasmic reticulum, is another strategy to enhance the expression of the transgene. Use of the rice rbcS-tp sequence to target the foreign gene products effectively increased foreign protein production in transgenic rice plants (Jang et al. 2002).
In this study, we constructed three alleles of the truncated cry1Ah gene with optimized coding sequences (designated m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah). In addition, the m3-cry1Ah allele was linked with a chloroplast transit peptide sequence (designated ctp-m3-cry1Ah) for chloroplast-targeted expression. Next, we generated transgenic tobacco lines with the m1-cry1Ah, m2-cry1Ah, m3-cry1Ah, and ctp-m3-cry1Ah alleles. Analysis of the transgenic tobacco plants showed that the ctp-m3-cry1Ah plants had the highest Cry1Ah protein expression levels and exhibited a higher level of resistance to H. armigera compared with the other transgenic tobacco plants. Our results indicated that combining codon optimization with the use of a transit peptide sequence improved the expression levels of the transgene and conferred a high level of resistance to the insect pests.
Materials and methods
Plant materials
Tobacco seeds (wild-type NC89) were sown on Murashige and Skoog (MS) medium (Gamborg et al. 1976) and stored in the dark at 28 °C for 5 days. The plants were then transferred to light conditions with 16-h light/8-h dark photoperiods at 28 °C under illumination of 300 μmol m−2 s−1.
Modification of the cry1Ah gene and construction of transformation vectors
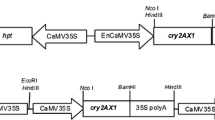
We constructed three alleles of the truncated cry1Ah insecticidal gene with varying GC content and codon usage pattern. These alleles were designated m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah. The GC content and variations in codon usage in the modified cry1Ah genes are shown in Table 1. The modified cry1Ah genes were synthesized by the GenScript Corporation (Nanjing, China). In addition, the m3-cry1Ah allele was linked with a chloroplast transit peptide sequence from the maize RuBP carboxylase, and this gene was designated ctp-m3-cry1Ah. Four expression vectors (pMhGM, pMAhb, pMhNK, and pMhCTP) containing the maize ubiquitin promoter to drive expression of m1-cry1Ah, m2-cry1Ah, m3-cry1Ah, and ctp-m3-cry1Ah were utilized in this study. These vectors also contain the bar gene driven by the cauliflower mosaic virus 35S (CaMV35S) promoter for herbicide-based selection. In addition, both sides of the gene expression cassette in pMhGM are linked with the sequences encoding maize alcohol dehydrogenase matrix attachment regions (madMars) (Fig. 1).
Maps of the expression vectors. RB right border, Mar maize alcohol dehydrogenase matrix attachment regions, Ubi ubiquitin promoter, ctp transit peptide sequence for chloroplast-targeted expression, m1-cry1Ah, m2-cry1Ah, m3-cry1Ah modified cry1Ah alleles, nos nopaline synthase gene terminator, 35S cauliflower mosaic virus 35S promoter, bar phosphinothricin acetyltransferase gene, polyA polyA terminator, LB left border, a 603-bp fragment of the bar gene was used as probe for Southern blotting
Tobacco transformation
The plant expression vectors were transformed into tobacco using Agrobacterium tumefaciens strain EHA105 (Horsch et al. 1985). A total of 320 tobacco leaf explants were infected, and the infected leaves were then selected on MS regeneration medium (Murashige and Skoog 1962) containing 2 mg/L bialaphos, 3 mg/L 6-benzyl amino purine, 0.2 mg/L naphthalene acetic acid, 400 mg/L carbenicillin, 3 % (w/v) sucrose and 0.3 % (w/v) gelrite and grown on MS regeneration medium containing 400 mg/L carbenicillin, 3 % (w/v) sucrose and 0.3 % (w/v) gelrite. The plant tissue culture reagents were obtained from PhytoTechnology Laboratories (Shawnee Mission, KS, USA).
Polymerase chain reaction (PCR) analysis
The presence of the foreign gene in the tobacco genome was verified using PCR analysis. Genomic DNA was isolated from the leaves of regenerated tobacco plants by using the plant genomic mini preparation kit (Newprobe, Beijing, China). Specific primers were designed to amplify the different cry1Ah alleles (Table 2): primer pairs F1/R1, F2/R2, and F3/R3 were used to amplify m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah, respectively. Each PCR reaction was performed in a volume of 25 μl containing 100 ng of DNA template, dNTPs (0.2 mM each), forward and reverse primers (0.1 μM each), 2.5 μl of 10× PCR buffer and 2.5 U of Taq DNA polymerase (TaKaRa, Dalian, China). All PCR reactions included an initial denaturation for 5 min at 94 °C followed by 33 cycles of denaturation (50 s at 94 °C), annealing and extension steps. For m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah, the annealing steps were 40 s at 58 °C, 50 s at 57 °C and 50 s at 60 °C, respectively. The extension temperature was always 72 °C, and the extension times were 50 s, 1 min 30 s, and 1 min 20 s for m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah, respectively. A final extension step (10 min at 72 °C) was added to all reactions. The PCR products were resolved by electrophoresis using 1 % (w/v) agarose gels, and the expected fragment lengths are shown in Table 2.
Southern blot analysis
Genomic DNA was extracted from transgenic tobacco plants using the CTAB method (Porebski et al. 1997). More than 40 μg of genomic DNA isolated from each transgenic plant was digested with HindIII. The digested samples were electrophoresed on a 0.7 % (w/v) agarose gel and blotted on Hybond™-XL nylon membranes (GE Healthcare, UK). To detect the integration of the exogenous genes in the tobacco genome, a 603-bp fragment of the bar gene was PCR amplified from pMhNK using the primers F4/R4 (Table 2) and used as probe in Southern blot analysis. The probe DNA was labeled with α-[32P]-dCTP using the Prime-a-Gene Labeling System (Promega, Madison, WI, USA) and hybridized for 20 h at 65 °C with agitation in a hybridization oven. After hybridization and washing, the blotted membranes were subjected to autoradiography for 48 h at −80 °C.
Quantitative RT-PCR analysis
The leaves of 1-month-old transgenic tobacco plants grown in the greenhouse were used for quantitative RT-PCR analysis. Total RNA was isolated from 100 mg of transgenic tobacco leaves using Trizol reagents (Tiangen, Beijing, China). Approximately 1 μg of total RNA was used as a template for reverse transcription using RevertAid™ First Strand cDNA Synthesis Kit (MBI Fermentas, EU) according to the manufacturer’s instructions. Quantitative RT-PCR was performed using an ABI PRISM 7500 instrument (Applied Biosystems). The actin gene was used as a reference gene. All reactions were run as duplicates in 96-well plates. The 25-μl reaction mixtures contained 12.5 μl of SYBR Green Real-time PCR Master Mix (TransGen, Beijing, China), 0.2 μM of each primer and 1 μl of cDNA template. The reaction conditions were as follows: an initiation step for 30 s at 95 °C followed by 45 cycles of 15 s at 95 °C and 15 s at 55 °C. The primers were designed with Primer 5.0 software. Primer pairs F5/R5, F6/R6, and F7/R7 (Table 2) were used to detect m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah transcript, respectively, and primers F8/R8 (Table 2) were used to detect the actin transcript.
Enzyme-linked immunosorbent assay (ELISA) and immunoblot analysis
The leaves of 1-month-old transgenic tobacco plants grown in the greenhouse were used for ELISA analysis. Total soluble proteins were extracted from the leaf tissues of transgenic plants. Approximately, 100 mg of tobacco leaves were homogenized in 0.5 ml of PBST extraction buffer (0.55 % Tween) provided by the manufacturer (EnviroLogix, Portland, ME, USA) and centrifuged at 10,000×g for 20 min at 4 °C. The amount of total soluble protein was determined by the Bradford assay (Bradford 1976) using the Bio-Rad protein dye concentrate and a standard curve derived from bovine serum albumin (BSA). The amount of Cry1Ah protein in transgenic plants leaves was determined using the QualiPlate Kit for Cry1Ab/Ac (EnviroLogix). Immunoassays were carried out in 96-well ELISA microplates according to the protocols provided by the manufacturers.
For immunoblot experiments, total soluble proteins were extracted following the method described above. Chloroplasts were isolated from tobacco leaves following the protocol described by Fan et al. (2009) and chloroplast proteins were extracted from the chloroplasts according to the protocol described by Phee et al. (2004). Protein extracts were separated on 10 % SDS–polyacrylamide gels and blotted onto a polyvinylidene difluoride (PVDF) membrane (Millipore Corporation, Billerica, MA, USA) using a semi-dry transfer apparatus (Bio-Rad). The primary antibody anti-Cry1Ah was used at a 1:10,000 dilution. The membrane was then washed three times prior to incubation with alkaline phosphatase-labeled IgG secondary antibody (Sigma, St. Louis, MO, USA) at a 1:10,000 dilution. The bound antibodies were detected using the NPT/BCIP kit (CWBIO, Beijing, China) as described by the manufacturer.
Insect bioassays
The transgenic tobacco plants were tested for resistance to H. armigera. The first instar neonatal larvae of H. armigera were used in the bioassay. Approximately 1,000 mg of fresh leaves were placed onto moist filter paper in plastic petri dishes (90 × 14.5 mm). The dishes were sealed with parafilm to prevent desiccation and kept in the insect rearing room at 25–28 °C. Each sample was infested with 12 neonate insect larvae, and the numbers of both the living and dead larvae were recorded twice a day with 3-day interval between measurements. We performed three biological replicates with each treatment, and an identical bioassay was also carried out with control plants. Mortality rates were presented as the proportion of dead larvae to total larvae applied (%). Leaf damage was estimated through visual inspection after 3 days of infestation, and damage was classified using a severity scale of 1–4 according to the Technical Specification for Evaluating Resistance of Cotton to Disease and Insect Pests (Part 1: Cotton Bollworm; GB/T 22101.1-2008).
The rating levels were as follows:
Level 1: no visible leaf feeding or individual shot-hole injuries on leaves.
Level 2: leaves with a small amount of shot-hole injuries but no large lesions.
Level 3: leaves with a large amount of shot-hole injuries but no large lesions.
Level 4: large lesions occupying a large amount of leaf area.
Results
Identification of the transgenic tobacco plants
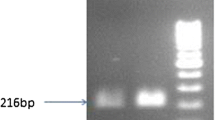
A total of 84 regenerated tobacco plants were obtained (Table 3), and we performed PCR analysis of these plants using sets of primers designed to amplify m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah. The results showed amplification of the expected products (865, 1456, and 1369 bp), which were similar to the products obtained from positive control DNA (Fig. 2a–d). In total, 23 PCR-positive lines were obtained (Table 3). No products were amplified from non-transformed tobacco plants using identical PCR conditions.
PCR analysis of the modified cry1Ah alleles and Southern blot analysis of the bar gene in the transgenic tobacco plants. a PCR analysis of the cry1Ah gene in m1-cry1Ah plants, b PCR analysis of the cry1Ah gene in m2-cry1Ah plants, c PCR analysis of the cry1Ah in m3-cry1Ah plants, d PCR analysis of the cry1Ah in ctp-m3-cry1Ah plants. All the PCR products were amplified using genomic DNA as a template. M DNA marker, P positive control, N wild-type tobacco plants and e Southern blot analysis of eight transgenic and wild-type tobacco plants. P positive control, N non-transgenic plants, all other lanes represent eight transgenic tobacco plants
Genomic DNA from plants expressing m1-cry1Ah, m2-cry1Ah, m3-cry1Ah, and ctp-m3-cry1Ah was digested with HindIII. Both the cry1Ah and bar genes were present in the T-DNA regions of the four binary vectors. To conveniently detect the integration of exogenous genes in the tobacco genome, a 603-bp fragment of the bar gene was used as a probe in Southern blot analysis. The results of Southern blot analysis indicated that a single copy (Fig. 2e, A3, A17, B1 and C18) or multiple copies (Fig. 2e, B12, C9, D11 and D12) of the transgene were present at different sites in the genome of the transgenic tobacco plants. No hybridization signal was detected in the untransformed tobacco plants (Fig. 2e, lane 4). These results confirmed that the exogenous genes were successfully integrated into the tobacco genome.
Analysis of cry1Ah transcript levels
The differences in cry1Ah transcript levels were confirmed by quantitative RT-PCR. We used the 2−ΔΔCT method according to ABI PRISM® 7700 Sequence Detection System User Bulletin #2. The above-mentioned 23 PCR-positive transgenic lines were detected individually. The actin gene was used as a reference gene. For each treatment sample, three technical replicates were set up in 96-well plates. The mean values of ΔCT of the different transgenic plants transformed with the same vector were obtained, respectively, and the mean value of ΔCT of the seven m1-cry1Ah plants was used as a calibrator. The quantitative RT-PCR results demonstrated that the ctp-m3-cry1Ah plants showed the highest cry1Ah transcript level of the four different transgenic constructs, approximately 12.5-fold compared with the level in m1-cry1Ah plants. The m3-cry1Ah plants had higher cry1Ah transcript level than m2-cry1Ah plants, approximately 9.5- and 4-fold, respectively, compared with the level in m1-cry1Ah plants (Fig. 3).
Quantitative RT-PCR analysis of quantitative changes in cry1Ah transcript levels. a The mean quantity of cry1Ah transcript in seven m1-cry1Ah plants (used as a calibrator). b The mean quantity of cry1Ah transcript in five m2-cry1Ah plants. c The mean quantity of cry1Ah transcript in six m3-cry1Ah plants. d The mean quantity of cry1Ah transcript in five ctp-m3-cry1Ah plants. The error bars represent the standard deviations of biological replicates from different transgenic lines transformed with the same vector
Analysis of Cry1Ah protein levels
A quantitative assessment of Cry1Ah endotoxin in transgenic tobacco plants was achieved by ELISA. The above-mentioned 23 PCR-positive transgenic lines were analyzed. For each treatment sample, three technical replicates were set up in 96-well plates. Quantitative analysis revealed that the protein expression levels in the modified cry1Ah lines were highest in plants expressing ctp-m3-cry1Ah followed by those expressing m3-cry1Ah, m2-cry1Ah, and m1-cry1Ah (in descending order) (Fig. 4). The quantity of Cry1Ah endotoxin in transgenic plants developed with the different constructs varied from 0.46 to 4.42 μg/g fresh weight. The protein levels in plants expressing m2-cry1Ah, m3-cry1Ah, and ctp-m3-cry1Ah were higher than the level in m1-cry1Ah plants by approximately four-, six-, and tenfold, respectively (Table 4d).
Enzyme-linked immunosorbent assay of Cry1Ah expression in different transgenic tobacco plants. A1, A3, A12, A14, A15, A17, and A20 represent m1-cry1Ah plants; B1, B3, B7, B12, and B17 represent m2-cry1Ah plants; C3, C6, C9, C12, C18, and C20 represent m3-cry1Ah plants; D4, D6, D8, D11, and D12 represent ctp-m3-cry1Ah plants; WT represents wild-type tobacco plants. The error bars represent the standard deviations of three technical replicates from each transgenic line
Based on the results of statistical analysis, the Cry1Ah expression levels in m1-cry1Ah, m2-cry1Ah, m3-cry1Ah, and ctp-m3-cry1Ah plants were significantly different (Table 5). The ELISA results showed that codon optimization and chloroplast-targeted expression of the cry1Ah gene significantly increased its protein expression levels.
To investigate whether the Cry1Ah protein expressed in ctp-m3-cry1Ah plants was targeted to the chloroplasts, we compared Cry1Ah levels in chloroplast proteins isolated from transgenic tobacco leaves with Cry1Ah levels in total soluble proteins by immunoblot analysis, which was performed with 15 μg of protein. In the ctp-m3-cry1Ah plants, Cry1Ah was more abundant in the chloroplast protein fraction than in the total soluble proteins. These results demonstrated that the Cry1Ah protein was targeted to the chloroplasts, and the Cry1Ah protein was most abundant in ctp-m3-cry1Ah plants followed by m3-cry1Ah, m2-cry1Ah, and m1-cry1Ah plants, respectively (Fig. 5). These observations strongly suggested that ctp-m3-cry1Ah plants had the highest protein expression levels, and the Cry1Ah protein was successfully targeted to the chloroplasts.
Cry1Ah protein levels in transgenic plants. Total (T) soluble proteins and chloroplast proteins (Chl) were isolated from transgenic tobacco leaves. 15 μg of protein were used for immunoblotting with anti-Cry1Ah antibody. The arrow on the right side of the panel indicates the position of the Cry1Ah protein (65 kDa). M prestained protein marker, P purified Cry1Ah protein (positive control), N wild-type tobacco plants; A3, B12, C9, and D11 represent m1-cry1Ah, m2-cry1Ah, m3-cry1Ah, and ctp-m3-cry1Ah plants, respectively
Analysis of the insect resistance of the transformed tobacco plants
All the confirmed transgenic plants were subjected to feeding first instar neonatal larvae of H. armigera. A total of 23 transgenic tobacco lines were assessed along with two wild-type plants for comparison. Photographs of leaf damage were taken after 3 days of infestation. Transgenic plants showed significant resistance to H. armigera infestation compared to wild-type plants. Variable degrees of leaf injury were observed, which might be due to the differences in the expression level of the cry1Ah gene. Three days after infestation with the neonatal larvae, wild-type leaves were badly damaged (Fig. 6, WT1 and WT2). However, the leaves of all transgenic plants showed resistance to H. armigera larvae (Fig. 6). The m1-cry1Ah plants (Fig. 6, A3 and A17) showed a higher injury rate after larval feeding than the m2-cry1Ah plants (Fig. 6, B1 and B12), which exhibited a small amount of leaf damage. No leaf damage was visible on m3-cry1Ah or ctp-m3-cry1Ah plants (Fig. 6, C12, C20, D8 and D12). These results clearly indicated that the expression of the cry1Ah gene effectively controlled H. armigera larvae. The mortality of the H. armigera larvae was recorded after 3 days of feeding to assess the effect of the Cry1Ah protein on the larvae. When feeding on control tobacco, no larval mortality was noted after 3 days of infestation (Table 4, WT1 and WT2). However, the mortality rates of larvae feeding on m1-cry1Ah, m2-cry1Ah, m3-cry1Ah, and ctp-m3-cry1Ah plants were 63, 82, 93, and 100 %, respectively (Table 4b), and the resistance rating levels were 2.57, 1.8, 1.33, and 1, respectively (Table 4c). The statistical analysis indicated that the mortality rates of larvae feeding on m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah plants were significantly different. However, the mortality rates of larvae feeding on m3-cry1Ah and ctp-m3-cry1Ah plants were not significantly different (Table 5). The results of the insect bioassays showed that these transgenic tobacco plants were toxic to H. armigera, and the insect resistance efficiency was highest in ctp-m3-cry1Ah plants followed by m3-cry1Ah, m2-cry1Ah, and m1-cry1Ah plants, respectively. The ctp-m3-cry1Ah plants exhibited the highest resistance to H. armigera.
The appearance of control (WT1 and WT2) and transgenic tobacco leaves after a larval feeding assay with H. armigera. Leaves A3 and A17 were from m1-cry1Ah plants, leaves B1 and B12 were from m2-cry1Ah plants, leaves C12 and C20 were from m3-cry1Ah plants, leaves D8 and D12 were from ctp-m3-cry1Ah plants. Photographs were taken after 3 days of infestation
Discussion
Most insect-resistant transgenic plants have been developed using Bt δ-endotoxin genes. Cry1Ab, cry1Ac and cry1Ab/cry1Ac hybrid genes are now widely used in commercial GMO crops. The toxic fragment of Cry1Ah has 72 and 82 % similarity to Cry1Ab and Cry1Ac, respectively. The results of bioassays indicated that Cry1Ah was highly toxic to four important agricultural pests, including O. furnacalis, H. armigera, C. suppressalis, and Plutella xylostella. This gene’s high level of toxicity to lepidopteran larvae makes it potentially useful for insect biocontrol (Xue et al. 2008). However, codon preference is a major barrier to the expression of bacterial genes in plants. Optimizing the codon usage patterns of exogenous genes improves the transgene expression in tobacco and tomato (Perlak et al. 1991), as well as potato (Perlak et al. 1993). Modifications in the cry1Ab gene allowed for efficient expression in plant protoplasts, and expression levels increased from four- to sixfold (Jabeen et al. 2010). Weng et al. (2011) optimized the cry1Ac gene for efficient, high level expression in sugarcane plants.
To improve transgene expression levels in plants, we modified the coding sequence of the truncated cry1Ah gene and created three alleles with increasing GC content to study the effect of GC content on transgene expression levels. We generated transgenic tobacco lines harboring the m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah alleles via Agrobacterium-mediated transformation. Quantitative analyses of the transgenic tobacco plants demonstrated that the expression levels of m2-cry1Ah and m3-cry1Ah were higher than that of m1-cry1Ah by approximately 4- and 9.5-fold in transcript levels and 4- and 6-fold in protein levels. In addition, m3-cry1Ah conferred a higher level of insect resistance compared with m2-cry1Ah and m1-cry1Ah. Vinogradov (2003) suggested that GC-rich regions were more transcriptionally active in plants and animals. Kudla et al. (2006) suggested that both RNA synthesis and degradation could potentially be affected by GC content, and the increased mRNA levels of GC-rich genes could be the results of two mechanisms: increased mRNA synthesis or decreased mRNA degradation. It, thus, appeared that the enhanced transcript levels of cry1Ah alleles with increased GC content might result from the efficient production of polyadenylated mRNA either through increased transcription or RNA processing efficiency.
Based on the results of statistical analysis, the expression levels of Cry1Ah and the mortality rates of larvae feeding on m1-cry1Ah, m2-cry1Ah, and m3-cry1Ah plants were significantly different, indicating that codon optimization of the cry1Ah gene caused an increase in protein expression levels and insect resistance efficiency. Previously, we observed that protein expression of the m1-cry1Ah gene was increased two- to fourfold with madMars on both sides of the gene expression cassette compared to the same gene without madMars in transgenic maize plants (unpublished work). The findings of our current study confirmed that codon optimization of the truncated cry1Ah gene could enhance its expression levels further. Codon optimization is a promising way to improve transgene expression levels in crops.
To date, a number of studies have suggested that chloroplasts are ideal hosts for the expression of transgenes (Jang et al. 1999; Kim et al. 2009; Verma and Daniell 2007; Wong et al. 1992). The transit peptides that act as chloroplast targeting sequences also facilitate interaction with envelope lipids, chloroplast receptors and the stromal processing peptidase (Bruce 2000). These transit peptides are cleaved from the mature protein during or after translocation into chloroplasts (Schmidt et al. 1981). In the present study, we linked a chloroplast transit peptide sequence to the N-terminus of the m3-Cry1Ah protein. Next, we generated H. armigera-resistant transgenic tobacco lines with the ctp-m3-cry1Ah gene. The ctp-m3-cry1Ah plants showed the highest Cry1Ah protein levels and a 100 % mortality rate of larvae feeding. It implied that the Cry1Ah protein might be more stable in chloroplasts than in cytosolic compartments. Based on the results of statistical analysis, Cry1Ah expression levels in m3-cry1Ah and ctp-m3-cry1Ah plants were significantly different, and the mortality rates of larvae feeding on m3-cry1Ah and ctp-m3-cry1Ah plants were 93 and 100 %, respectively. There was no significant difference on the mortality rates of larvae between them, because both m3-cry1Ah and ctp-m3-cry1Ah plants showed high mortality rates of larvae. Cry1Ah is very effective on larva mortality even at moderate levels of expression; to detect a significant difference with the mortality rates of larvae would require a moderately toxic Cry protein. Quantitative analysis showed that the transcript level of ctp-m3-cry1Ah was higher than those of m3-cry1Ah, and the difference of the two constructs is simply the presence or absence of the transit peptide. It is unlikely that the transit peptide has a significant effect on increasing transcription. Wong et al. (1992) and Kim et al. (2009) both suggested that the presence of the transit peptide led to higher transcript levels, which may be due to increased mRNA stability.
Several researchers have reported that single copy transgenes exhibited higher expression levels than multiple T-DNA insertions (Hobbs et al. 1990; Jones et al. 1987). However, we noted little difference in transgene expression in m3-cry1Ah plants, even though there were multiple copies in line C9. It was somewhat interesting that the transgene expression levels in the multiple copy lines B12, D11, and D12 were higher than the expression levels in other transgenic lines in relevant groups. The co-suppression is an interaction of homologous sequence that occurs in trans over great genetics distance (Napoli et al. 1990). In the current study, the multicopy lines may be luckily integrated in the same or close sites, and co-suppression of homologous genes was avoided.
Overall, the combined effects of exogenous gene modification and the use of a transit peptide sequence on transgene expression were shown to be significant. Further applications of this method and use of the cry1Ah gene in other important crops will be valuable in the future.
References
Arencibia A, Vázquez RI, Prieto D, Téllez P, Carmona ER, Coego A, Hernández L, De la Riva GA, Selman-Housein G (1997) Transgenic sugarcane plants resistant to stem borer attack. Mol Breed 3:247–255
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bravo A, Gill SS, Soberón M (2007) Mode of action of Bacillus thuringiensis cry and cyt toxins and their potential for insect control. Toxicon 49:423–435
Bruce BD (2000) Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol 10:440–447
Campbell WH, Gowri G (1990) Codon usage in higher plants, green algae, and cyanobacteria. Plant Physiol 92:1–11
Fan PX, Wang XC, Kuang TY, Li YX (2009) An efficient method for the extraction of chloroplast proteins compatible for 2-DE and MS analysis. Electrophore 30:3024–3033
Gamborg OL, Murashige T, Thorpe TA, Vasil IK (1976) Plant tissue culture media. In Vitr Cell Dev Biol Plant 12:473–478
Gatehouse JA (2008) Biotechnological prospects for engineering insect-resistant plants. Plant Physiol 146:881–887
Hobbs SLA, Kpodar P, DeLong CMO (1990) The effect of T-DNA copy number, position and methylation on reporter gene expression in tobacco transformants. Plant Mol Biol 15:851–864
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Jabeen R, Khan MS, Zafar Y, Anjum T (2010) Codon optimization of cry1Ab gene for hyper expression in plant organelles. Mol Biol Rep 37:1011–1017
Jang I-C, Nahm BH, Kim J-K (1999) Subcellular targeting of green fluorescent protein to plastids in transgenic rice plants provides a high-level expression system. Mol Breed 5:453–461
Jang I-C, Lee K-H, Nahm BH, Kim J-K (2002) Chloroplast targeting signal of a rice rbcS gene enhances transgene expression. Mol Breed 9:81–91
Jones JDG, Gilbert DE, Grady KL, Jorgensen RA (1987) T-DNA structure and gene expression in petunia plants transformed by Agrobacterium tumefaciens C58 derivatives. Mol Gen Genet 207:478–485
Kim EH, Suh SC, Park BS, Shin KS, Kweon SJ, Han EJ, Park S-H, Kim YS, Kim J-K (2009) Chloroplast-targeted expression of synthetic cry1Ac in transgenic rice as an alternative strategy for increased pest protection. Planta 230:397–405
Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M (2006) High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol 4:933–942
Kumar S, Chandra A, Pandey KC (2008) Bacillus thuringiensis (Bt) transgenic crop: an environment friendly insect-pest management strategy. J Environ Biol 29:641–653
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Murray EE, Lotzer J, Eberle M (1989) Codon usage in plant genes. Nucleic Acids Res 17:477–498
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279–289
Perlak FJ, Fuchs RL, Dean DA, McPherson SL, Fischhoff DA (1991) Modification of the coding sequence enhances plant expression of insect control protein genes. Proc Natl Acad Sci USA 88:3324–3328
Perlak FJ, Stone TB, Muskopf YM, Petersen LJ, Parker GB, McPherson SA, Wyman J, Love S, Reed G, Biever D, Fischhoff DA (1993) Genetically improved potatoes: protection from damage by Colorado potato beetles. Plant Mol Biol 22:313–321
Phee B-K, Cho J-H, Park S, Jung JH, Lee Y-H, Jeon J-S, Bhoo SH, Hahn T-R (2004) Proteomic analysis of the response of Arabidopsis chloroplast proteins to high light stress. Proteomics 4:3560–3568
Porebski S, Bailey LG, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15:8–15
Schmidt GW, Bartlett SG, Grossman AR, Cashmore AR, Chua N-H (1981) Biosynthetic pathways of two polypeptide subunits of the light-harvesting chlorophyll a/b protein complex. J Cell Biol 91:468–478
Schwember AR (2008) An update on genetically modified crops. Cienc Investig Agrar 35:231–250
Vaeck M, Reynaerts A, Höfte H, Jansens S, De Beuckeleer M, Dean C, Zabeau M, Montagu MV, Leemans J (1987) Transgenic plants protected from insect attack. Nature 328:33–37
Verma D, Daniell H (2007) Chloroplast vector systems for biotechnology applications. Plant Physiol 145:1129–1143
Vinogradov AE (2003) DNA helix: the importance of being GC-rich. Nucleic Acids Res 31:1838–1844
Weng LX, Deng HH, Xu JL, Li Q, Wang LH, Jiang ZD, Zhang HB, Li QW, Zhang LH (2006) Regeneration of sugarcane elite breeding lines and engineering of stem borer resistance. Pest Manag Sci 62:178–187
Weng LX, Deng HH, Xu JL, Li Q, Zhang YQ, Jiang ZD, Li QW, Chen JW, Zhang LH (2011) Transgenic sugarcane plants expressing high levels of modified cry1Ac provide effective control against stem borers in field trials. Transgenic Res 20:759–772
Wong EY, Hironaka CM, Fischhoff DA (1992) Arabidopsis thaliana small subunit leader and transit peptide enhance the expression of Bacillus thuringiensis proteins in transgenic plants. Plant Mol Biol 20:81–93
Xue J, Liang GM, Crickmore N, Li HT, He KL, Song FP, Feng X, Huang DF, Zhang J (2008) Cloning and characterization of a novel cry1A toxin from Bacillus thuringiensis with high toxicity to the Asian corn borer and other lepidopteran insects. FEMS Microbiol Lett 280:95–101
Acknowledgments
The authors gratefully acknowledge the financial support of Genetically Modified Organisms Breeding Major Projects (No. 2011ZX08003-001), the National Basic Research Program of China (973 Program) (No. 2009CB118902) and the National Natural Science Foundation of China (No. 30970231). In addition, the authors thank Dr. Liang Gemei at the Institute of Plant Protection of the Chinese Academy of Agricultural Sciences for supplying the test larvae.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by K. Kamo.
Rights and permissions
About this article
Cite this article
Li, X., Li, S., Lang, Z. et al. Chloroplast-targeted expression of the codon-optimized truncated cry1Ah gene in transgenic tobacco confers a high level of protection against insects. Plant Cell Rep 32, 1299–1308 (2013). https://doi.org/10.1007/s00299-013-1444-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-013-1444-z