Abstract
Grain hardness of wheat is determined by the hardness (Ha)-locus region, which contains three friabilin-related genes: puroindoline-a (Pina), puroindoline-b (Pinb) and GSP-1. In our previous study, we produced the transgenic rice plants harboring the large genomic fragment of the Ha-locus region of Aegilops tauschii containing Pina and GSP-1 genes by Agrobacterium-mediated transformation. To examine the effects of the transgenes in the rice endosperms, we firstly confirmed the homozygosity of the T-DNAs in four independent T2 lines by using fluorescence in situ hybridization (FISH) and DNA gel blot analyses. The transgenes, Pina and GSP-1, were stably expressed in endosperms of the T3 and T4 seeds at RNA and protein levels, indicating that the promoters and other regulatory elements on the wheat Ha-locus region function in rice, and that multigene transformation using a large genomic fragment is a useful strategy. The functional contribution of the transgene-derived friabilins to the rice endosperm structure was considered as an increase of spaces between compound starch granules, resulting in a high proportion of white turbidity seeds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grain hardness is an important agricultural trait in wheat. After flour milling, powder particles of soft wheat are finer than those of hard wheat, influencing differences between cake flour and baking flour. The grain hardness of wheat is regulated by the hardness (Ha)-locus region, which is located on the short arm of chromosome 5D in common wheat (Triticum aestivum), and contains three linked genes: puroindoline-a (Pina), puroindoline-b (Pinb) and Grain Softness Protein-1 (GSP-1). The Pina, Pinb and GSP-1 encode ca. 15-kDa storage protein, termed friabilin (or GSP), which is present in seeds of soft wheat but absent in those of hard wheat, suggesting that these friabilin-related genes cause grain softness. These friabilins are lipid-binding proteins, which are considered to associate with starch granules in seeds, resulting in the soft kernel texture of wheat, while their original biological functions may be related to antimicrobial activities (reviewed in Bhave and Morris 2008). Puroindoline genes are expressed in starchy endosperm cells, and their products are developmentally accumulated in the endosperm (Digeon et al. 1999; Turnbull et al. 2003a; Wiley et al. 2007). Molecular genetic studies of the Pina, Pinb and GSP-1 genes in different types of wheat suggest that PINA is a major contributor of grain softness (Giroux et al. 2000; Chen et al. 2006) and functions with PINB (Amoroso et al. 2004). Although GSP-1 is apparently not a central player in the softness texture (Giroux and Morris 1998), several recent pieces of circumstantial evidence show a possible relationship between GSP-1 and grain hardness (Clarke and Rahman 2005; Ikeda et al. 2005; Morris and Bhave 2008) and propose the necessity of further experiments on the whole Ha-locus region including GSP-1 (Bhave and Morris 2008).
Rice (Oryza sativa) with hard grain texture is best used as a host cereal plant for transgenic experiments with the friabilin-related genes for the following reasons: (1) transformation of rice is more efficient than that of wheat; and (2) puroindoline genes are absent in the rice genome, while genes at the boundaries of the Ha-locus are conserved between rice and wheat (Chantret et al. 2005). Krishnamurthy and Giroux (2001) introduced puroindoline cDNAs into rice under the control of a ubiquitin promoter and showed that expression of Pina and/or Pinb in the transgenic rice seeds reduced grain hardness, demonstrating that both Pina and Pinb are involved in the soft kernel texture phenotype. However, it is still important for the overall understanding of friabilins to characterize transgenic rice plants harboring the wheat Ha-locus region including GSP-1 as a set of genes and their flanking and promoter regions.
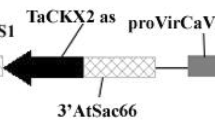
In our previous report, we produced the transgenic rice plants harboring the large genomic fragments of the wheat Ha-locus region through Agrobacterium-mediated transformation (Nakano et al. 2005). A BAC 8 binary cosmid clone (Turnbull et al. 2003b; Fig. 1) contains a 92-kb Ha-locus region with Pina and GSP-1 genes of the D genome donor of wheat, Aegilops tauschii, which is an evolutionally important species as a provider of softness texture in wheat (Chantret et al. 2005). We introduced the BAC 8 clone into rice (Oryza sativa L. cv. Yamahoushi), and characterized the genomic organization of the integrated T-DNAs in four independent transgenic lines by Southern blot and fiber FISH analyses, showing that T-DNA was integrated as 1, 1, 1 and 2 copies for lines S, N, 11 and 12, respectively (Nakano et al. 2005). The existence of Pina and GSP-1 genes in the transformants was confirmed by PCR, although rearranged T-DNAs were observed in the four transgenic lines, which could have been due to instability of the large T-DNA integration. In the present study, we further analyzed gene expression, protein accumulation and functional contribution of Pina and GSP-1 by using the transgenic rice plants.
Genomic organization of the hardness (Ha) locus region and the BAC 8 construct used in this study. Three friabilin-related genes are linked in the Ha-locus of Aegilops tauschii. The 92-kb genomic fragment containing Pina and GSP-1 genes cloned in a binary cosmid vector, pCLD04541, was used for the Agrobacterium-mediated transformation of rice
Materials and methods
Plant materials
Four lines (S, N, 11 and 12) of transgenic rice (Nakano et al. 2005) harboring a 92-kb wheat Ha-locus region (the BAC 8 construct, Turnbull et al. 2003b; Fig. 1) were used in this study. A cultivar of japonica rice, Oryza sativa L. Yamahoushi was used as an untransformant control.
Southern blot analysis
Total DNA was extracted from young leaf tissue by the procedure of Murray and Thompson (1980). Total DNA (about 1 μg) was digested with HindIII and separated on 0.8% agarose gel. After electrophoresis, DNA was transferred to nylon membranes (Roche Diagnostics). Hybridization was carried out in 5× SSC, 0.5% blocking reagent (Roche Diagnostics), 0.1% sodium N-lauroyl sarcosinate and 0.02% SDS at 65°C. The membrane was washed twice in 0.1× SSC, 0.1% SDS at 65°C for 20 min each. The digoxigenin-labeled nptII was prepared by PCR reaction using PCR DIG Labeling Mix (Roche Diagnostics). The detection of the hybridized probe was carried out according to the instructions of the hybridized manual of the DIG Luminescent Detection Kit (Roche Diagnostics) with CSPD as the substrate. Detected band(s) indicated the copy number of nptII in transgenic plants as described in Nakano et al. (2005) and Imazawa et al. (2009). Homozygosity of the T-DNA locus was estimated from signal intensity of the band(s).
FISH analysis
After pretreatment with 2 mM 8-hydroxyquinoline for 4 h at room temperature, the root tips were fixed with Farmer’s fixative (1:3 glacial acetic acid and ethanol) for several days. The FISH slides with squashed root-tip cells were prepared as described by Mukai et al. (1990). For a probe for FISH analysis, the whole BAC 8 DNA was purified by QIAGEN Plasmid Midi kit (QIAGEN) and labeled with dig-11-dUTP using a Dig-Nick Translation Mix kit (Roche Diagnostics). FISH hybridization and detection of the BAC probe were performed according to standard protocol as described in Suzuki et al. (2001) by using rhodamine-conjugated anti-dig (Roche Diagnostics).
Reverse transcription (RT)-PCR
Poly(A)+ RNA was isolated from the developing endosperm of the transgenic rice by using the FastTrack mRNA isolation kit (Invitrogen). The RNA samples were reverse-transcribed to synthesize first-strand cDNA by using the First-Strand cDNA synthesis kit (GE Healthcare). The cDNA was then used as a template for PCR amplification with specific primers; PA-F (5′-ATGAAGGCCCTCTTCCTCATAGG-3′) and PA-R (5′-TCACCAGTAATAGGCAATAGTGCC-3′) were used to amplify the Pina sequence, and GSP-F (5′-GCGATCTAAGTGGCTTCAAG-3′) and GSP-R (5′-GCTAGTGATGGGGATGTTGC-3′) were used to amplify the GSP-1 sequence. PCR was performed with ExTaq polymerase (Takara) for 35 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 60°C, and extension for 1 min at 72°C followed by a final extension for 5 min, by using DNA thermal cycler PERSONAL (Takara). The rice alpha-tubulin gene was used as a control.
Western blot analysis
Two to five seeds were crushed and homogenized with 50 mM Tris–HCl (pH7.5). After vortex mixing and leaving at room temperature for 15 min, the sample was centrifuged at 8,000g for 10 min and any supernatant liquid was removed. After the pellet was washed once more with 50 mM Tris–HCl, and once with 0.1 M NaCl and 50 mM Tris–HCl, the protein was extracted with 1% TritonX-114, 50 mM Tris–HCl by vortex mixing and leaving for 15 min, followed by centrifugation at 8,000g for 10 min. Subsequently, ca. 10 μg of the protein extract was subjected to SDS polyacrylamide-gel electrophoresis (SDS-PAGE) in the usual manner using the 15% PAGE gel (e-PAGEL, ATTO). The fractionated protein in the PAGE gel was electroblotted and Western blot detection was conducted by using ECL Western Blotting Analysis System (GE Healthcare). The anti-friabilin polyclonal antibody (Jolly et al. 1993) and the HRP-labeled anti-rabbit antibody (GE Healthcare) were used as primary and secondary antibodies, respectively.
Scanning electron microscope (SEM) analysis
Dried seeds were cut by a blade, coated with gold and subjected to a scanning electron microscope (JXA-840A, JEOL) as described in Turnbull et al. (2003a).
Results
Confirmation of homozygous plants for T-DNAs
In the four transgenic rice lines with the BAC 8 construct, both Pina and GSP-1 were expected to be expressed in the endosperm of the seed (having genotype of the next generation). Therefore, the progenies possessing homozygous T-DNAs, which could produce seeds with the same genotype, were useful for analysis of expression and function of the transgenes using a number of seeds. The T0 plants (S, N, 11, and 12) were all hemizygous, and T1 seeds (e.g., S-18, N-3, 11-6, and 12-21) were segregated. Our previous study showed that one-copy large T-DNA was integrated in the lines S, N and 11, and two copies in the lines 12 (Nakano et al. 2005). Because rearrangements that occurred in the T-DNA locus of the line 11 have resulted in the two-linked nptII genes, HindIII-digestion produced two unsegregated fragments in Southern blot analysis with the nptII probe. On the other hand, each or both the two unlinked nptII fragments were detected in progenies of the line 12. It was difficult to discriminate heterozygotes and homozygotes in the segregated progenies and identify homozygous T1 plants only by the Southern blot analysis with an nptII probe. Additionally, in the case of line 12, no homozygous plant for both the independent T-DNA loci was found in the T1 generation, owing to a small number of the T1 seeds. Therefore, we determined the homozygosity of the T-DNA loci in the T2 population by using both Southern blot and FISH analyses (Fig. 2); candidates of homozygous plants estimated by Southern blot analysis with the nptII probe (Fig. 2a) were confirmed by FISH analysis with the whole BAC 8 probe (Fig. 2b).
Confirmation of homozygosity of the T-DNAs by Southern blot and FISH analyses. a Genomic DNA of the T2 plants and an untransformant was digested with HindIII, and applied to Southern blot analysis with the nptII probe. As the DNA amount control, separated total DNAs stained with ethidium bromide before blotting are indicated in Online Resource 1. Homozygosity of the T-DNA locus was estimated from signal intensity of the band(s). Note that the two bands obtained in the line 11 (11-6-1, 11-20-6, 11-20-8, and 11-22-10) are linked in the large T-DNA locus. b Representative FISH results of mitotic chromosomes of the four independent T2 lines detected with the BAC 8 probe (signals are indicated by arrows). Homozygosity was confirmed by two signals obtained for one-locus T-DNA insertion (S-18-2, N-3-1, 11-6-1), and four signals for two-loci T-DNA insertion (12-21-5). The bar represents 10 μm
Figure 2b shows FISH detection of homozygous T-DNA loci in the four lines of T2 plants (S-18-2, N-3-1, 11-6-1 and 12-21-5). Since lines S, N and 11 contained a single T-DNA locus, two distinct FISH signals were observed in each nucleus or prophase chromosome set. Such homozygosity was also observed in three other T2 plants of the line S (S-4-1, S-15-1 and S-18-1) and three other T2 plants of the line 11 (11-20-6, 11-20-8 and 11-22-10). In the case of line 12, four distinct FISH signals (2 strong and 2 weak) were observed in a 12-21-5 T2 plant (Fig. 2b), indicating that this plant was obviously homozygous against two independent T-DNA loci. We also obtained three T2 plants (12-19-2, 12-19-6 and 12-19-9), which were homozygous only for the single T-DNA locus. The progenies of these T2 plants, whose homozygosity of T-DNA was efficiently determined by a combination of Southern blot and FISH analyses, were then subjected to the expression analysis for transgenes.
Gene expression and protein accumulation of PINA and GSP-1 in transgenic rice
To determine the expression of the transgenes at an RNA level in the BAC 8-introduced rice, RT-PCR was conducted using T3 seeds obtained from the homozygous T2 plants (S-18-2, N-3-1, 11-6-1 and 12-21-5). All four of the lines showed expression of Pina and GSP-1 in developing endosperms at 1, 2 and 3 weeks after flowering (Fig. 3). Absence of Pina and GSP-1 genes in untransformed rice was confirmed by PCR with genomic DNA templates (Fig. 3), and no or low RT-PCR product of Pina and GSP-1 was obtained in leaves (data not shown). Successful detection of transcription of Pina and GSP-1 indicated that wheat friabilin promoters are also active in rice endosperms, although these RT-PCR results did not show the quantity of the expression.
Reverse transcription (RT)-PCR analysis of T3 homozygous seeds. RT-PCR was performed with transgene-specific primers by using endosperm cDNA (1–3 weeks after flowering) of the four transgenic lines as templates. Plus and minus symbols represent existence and absence of reverse transcriptase, respectively. No band in the absence of reverse transcriptase indicates the samples to be free of genomic DNA contamination. α-tubulin was used as positive control. PCR results using the wheat and rice genomic DNA (untransformants) are also shown as a size standard for the amplified RT-PCR bands, which also demonstrate that Pina and GSP-1 are in wheat but not in rice
Subsequently, Western blot analysis was performed to analyze protein-level expression of PINA and GSP-1 in the T3 and T4 seeds. Powder of the rice endosperm was washed by 50 mM Tris and 0.1 M NaCl, and then starch-binding proteins were extracted with 1% TritonX-114, 50 mM Tris from the remaining precipitation. The extracted starch-binding proteins were subjected to SDS-PAGE followed by Western blot analysis with the anti-friabilin polyclonal antibody (Jolly et al. 1993). As a result, two bands around 15 kDa were detected in mature T3 seeds of the seven homozygous transgenic plants (S-4-1, S-18-2, N-3-1, 11-6-1, 11-20-6, 12-19-6 and 12-21-5), which were absent in the untransformed rice (Fig. 4a). Molecular weight values estimated from deduced amino acids sequences of PINA and GSP-1 of Ae. tauschii were 14.9 and 16.6 kDa, respectively (information from nucleotide DATA of CR626926, Chantret et al. 2005). Therefore, in the two ca. 15-kDa bands in our Western blot analysis, a band with a greater molecular weight might be GSP-1, and that with a lesser weight might correspond to PINA. Thus, exogenous friabilins were successfully accumulated in endosperms of the transgenic rice plants, although their amount was less than those of soft wheat (Fig. 4a). These two bands were detected from samples extracted with the TritonX-114 buffer and not with the Tris and NaCl buffers (data not shown), indicating that PINA and GSP-1 might be associated with starch granules in endosperms of the transgenic rice, just as in the case of the starch-binding friabilins in the soft wheat. No significant difference was observed in the amount of accumulated PINA and GSP-1 between seeds of 12-21-5 with two T-DNA copies and those of other plants with one T-DNA copy. Western blot analysis was also conducted with developing T4 seeds and it showed that PINA and GSP-1 proteins were accumulated in endosperms from 1 week after flowering (Fig. 4b).
Western blot analysis of T3 and T4 homozygous seeds. a Accumulation of friabilin in the T3 seeds of seven transgenic rice lines analyzed by Western blot analysis. Seeds of the untransformant rice and soft wheat were also used as negative and positive controls, respectively. Crushed mature endosperms were washed by Tris and NaCl and extracted with TritonX-114. Extracted starch-binding proteins were subjected to SDS-PAGE (the CBB-stained gel is indicated in Online Resource 2), electroblotted and detected with anti-friabilin antibody (Jolly et al. 1993). Two bands (ca. 15 kDa) were obtained in the transgenic plants. Asterisk indicates a putative GSP-1 protein estimated from the size. b Developmental regulation of protein expression of friabilin. Proteins extracted from immature endosperms (1–3 weeks after flowering) of T4 plants were used to perform Western blot analysis to detect the friabilin. Asterisk indicates the putative GSP-1 protein estimated from the size. The CBB-stained gel is indicated in Online Resource 2
Structural characterization of seeds in transgenic rice
We further characterized homozygous T4 rice seeds by structural comparison with untransformed rice seeds to understand the functional contribution of the wheat friabilins in the Pina/GSP-1-introduced transgenic rice plants. There was no difference in appearance of rice seeds with palea and lemma between the transgenic plants and untransformants (Fig. 5a). However, a high rate (ca. 75–95%) of white turbidity seeds was observed in brown rice seeds obtained from the progenies of S-4-1, S-18-2, 11-20-6, 11-6-1 and 12-21-5 plants (Fig. 5a, b). Although the lines N-3-1 showed only 10–20% proportions of the white turbidity seeds, the presence of such white seeds is a characteristic structural alternation of the Pina/GSP-1-introduced transgenic rice plants, because no white turbidity seeds were observed in the untransformant controls (Fig. 5b). All the T3 plants and untransformants were simultaneously and normally grown in a closed glasshouse under the same conditions, and the appearance of the white turbidity seeds was different from that of the milky white rice seeds, the occurrence of which was due to environmental stress during development. A similar occurrence of the turbidity seeds was also observed in the T3 seeds, which were obtained from T2 plants grown in a different year.
The T4 seeds contain white turbidity seeds. a Rice seeds with and without palea/lemma. Representative T4 seeds (11-20-6-1) and untransformant seeds are indicated. The bars represent 5 mm. b Ratio (%) of white turbidity seeds was calculated in the T4 and untransformant seeds. Two to four independent T3 plants for each homozygous line were used for analysis. All the T3 plants and untransformants were simultaneously grown in a closed glasshouse under the same conditions
We characterized endosperms of these rice grains in detail by using scanning electron microscopy (SEM). Because Turnbull et al. (2003a) examined the cut surface of the endosperms of wheat seeds using SEM and distinguished clearly between soft and hard lines (cellular components of the hard line are more tightly compacted together than those of the soft line), we applied this method to rice seeds to better understand the effects of the PINA and/or GSP-1 on the structural property of the endosperm (Fig. 6). On whole views of the cut surface, radial orientations of endosperm cells were observed in the normal perfect rice seed of the untransformant (Fig. 6a), whereas these normal orientations were lost in the transgenic plants (Fig. 6d). In the high magnification views, compound starch granules were tightly packed with each other on the smooth cut surface of the untransformant control (Fig. 6b, c), while individual compound starch granules were easy to recognize as scale-like organizations in the transgenic lines (Fig. 6e–i), indicating more spaces between the compound starch granules. In some views of the cut surface, independent starch granules, which were normal in appearance, were exposed in the transgenic seeds (data not shown).
Cut surface of the rice endosperms observed by scanning electron microscope (SEM). Dried mature seeds of untransformant (a–c) and transgenic plants (d–i) were cut by blade, and the surfaces were analyzed by SEM. b, e Close-up pictures of white square frame regions in a and d, respectively. c, f Close up pictures of black square frame regions in a and d, respectively. Bars in a and d represent 1 mm. Bars in b, c, e–i represent 20 μm
Discussion
Introduction of the Ha-locus region as multigene transformation
In the present study, we introduced a large genomic fragment including two wheat genes into rice. The approach of introducing a set of linked genes as a genomic region is useful for multigene engineering (Naqvi et al. 2010) if exogenous promoters are normally active in a recipient. The wheat Ha-locus region encompassing approximately a 100-kb region contains three endosperm-expressing genes (Pina, Pinb, and GSP-1) related to friabilin (Chantret et al. 2005); this size of the genomic region is nearly a limitation of the introducible size via an Agrobacterium-mediated transformation. Genes for friabilins are small, and the artificially fused construct including both Pina and Pinb genes with endosperm-specific promoters was successfully used for maize transformation (Zhang et al. 2009). However, multigene transformation with a large genomic fragment still has an advantage, such as co-transformation of the original regulatory sequences intricately embedded within flanking regions of the genes, from the view of stability of transgenes for generations and avoidance of gene silencing. Our transgenic plants harboring the Ha-locus region with the Pina and GSP-1 genes demonstrated that the wheat regulatory sequences in the Ha-locus region were functional in rice. Rearrangements observed in the integrated large T-DNAs (Nakano et al. 2005) might have no negative effects for transgene expression. Furthermore, Western blot analysis of the transgenic plants showed that the exogenous PINA and GSP-1 proteins were successfully accumulated in the rice endosperm and might be associated with starches as expected. Thus, the multigene transformation of a large genomic DNA has potential to be a powerful tool for plant engineering.
Functional contribution of friabilins in transgenic rice
The structure of the rice endosperm is fundamentally different from that of wheat. In the rice endosperm cell, dozens of starch granules with a polyhedral shape are tightly packed in the amyloplast to form an ellipsoidal compound starch granule, while starch granules with a spherical shape occur within the protein matrix in the wheat endosperm cell. As for localization of friabilins in the wheat endosperm, PINA and PINB are co-localized to the starch granule surface, possibly due to the association of positively charged friabilins with polar lipids on the surface of starch (Feiz et al. 2009). The friabilins on the starch granule surface might eventually prevent tight interaction between the starch and protein matrix, leading to grain softness in wheat. On the other hand, there are no friabilin-related genes in rice (Chantret et al. 2005). Krishnamurthy and Giroux (2001) reported significant reduction of grain hardness in transgenic rice plants harboring the Pina and/or Pinb cDNA, although its precise mechanism is unclear. Unlike the previous report, we introduced the genomic fragment including Pina and GSP-1 genes into rice. Although we were not able to find additive effects of GSP-1 to grain texture, GSP-1 was apparently accumulated in our transgenic rice seeds as well as PINA. Both Krishnamurthy and Giroux (2001) and this study showed significant modification of rice grain texture, indicating that friabilins actually have a potential to alternate the endosperm structure in rice. To compare the effects of friabilins in these transgenic rice plants, the same experimental evaluation of grain hardness under the same condition would be indispensable.
Considering the results of Western blot and SEM analyses in the present study, transgenes-derived PINA and GSP-1 might be localized between compound starch granules and make more spaces between them. A similar observation of the spaces surrounding the compound starch granules was recently reported in Wada et al. (2010), although differences of endosperm structure between the transgenic and non-transgenic rice plants were less obvious than those reported between hard and soft wheat plants (Turnbull et al. 2003a). The membrane surface of the amyloplast might be negatively charged as a common feature of the biomembrane structure and could associate with positively charged friabilin proteins. The possible hypothesis explaining frequent observation of the white turbidity seeds in our transgenic plants is as follows: during the drying process of the Pina/GSP-1-introduced transgenic rice seeds, air might enter into the spaces between the compound starch granules, resulting in the white turbidity seeds. A lower ratio of the white turbidity seeds in the N-3-1 lines might be due to lower expression of Pina and GSP-1 genes than in the other transgenic lines, although further detailed analysis of their expression (e.g., quantitative RT-PCR analysis) is necessary to demonstrate it in future.
This study suggests the possibility of engineering the glutinous rice-like new cultivar by introducing the friabilin-related genes from wheat. Additional introduction of the Pinb gene to the transgenic line S-4-1, S-18-2, 11-20-6, 11-6-1 or 12-21-5 would lead to a more stable production of the white turbidity seeds. Determination of the precise physicochemical properties of these transgenic rice seeds, combined with the detailed molecular analysis of the exogenous friabilins, will more accurately dissect the molecular functions of puroindolines in grain hardness.
Abbreviations
- FISH:
-
Fluorescence in situ hybridization
- GSP-1 :
-
Grain Softness Protein-1
- Ha :
-
Hardness
- Pina :
-
Puroindoline-a
- Pinb :
-
Puroindoline-b
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- SEM:
-
Scanning electron microscope
References
Amoroso MG, Longobardo L, Capparelli R (2004) Real time PCR and flow cytometry to investigate wheat kernel hardness: role of puroindoline genes and proteins. Biotechnol Lett 26:1731–1737
Bhave M, Morris CF (2008) Molecular genetics of puroindolines and related genes: allelic diversity in wheat and other grasses. Plant Mol Biol 66:205–219
Chantret N, Salse J, Sabot F, Rahman S, Bellec A, Laubin B, Dubois I, Dossat C, Sourdille P, Joudrier P, Gautier MF, Cattolico L, Beckert M, Aubourg S, Weissenbach J, Caboche M, Bernard M, Leroy P, Chalhoub B (2005) Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17:1033–1045
Chen F, He Z, Xia XC, Xia LQ, Zhang XY, Lillemo M, Morris CF (2006) Molecular and biochemical characterisation of puroindoline a and b alleles in Chinese landraces and historical cultivars. Theor Appl Genet 112:400–409
Clarke B, Rahman S (2005) A microarray analysis of wheat grain hardness. Theor Appl Genet 110:1259–1267
Digeon JF, Guiderdoni E, Alary R, Michaux-Ferrière N, Joudrier P, Gautier MF (1999) Cloning of a wheat puroindoline gene promoter by IPCR and analysis of promoter regions required for tissue-specific expression in transgenic rice seeds. Plant Mol Biol 39:1101–1112
Feiz L, Wanjugi HW, Melnyk CW, Altosaar I, Martin JM, Giroux MJ (2009) Puroindolines co-localize to the starch granule surface and increase seed bound polar lipid content. J Cereal Sci 50:91–98
Giroux MJ, Morris CF (1998) Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline-a and -b. Proc Natl Acad Sci USA 95:6262–6266
Giroux MJ, Talbert L, Habernicht DK, Lanning S, Hemphill A, Martin JM (2000) Association of puroindoline sequence type and grain hardness in hard red spring wheat. Crop Sci 30:370–374
Ikeda TM, Ohnishi N, Nagamine T, Oda S, Hisatomi T, Yano H (2005) Identification of new puroindoline genotypes and their relationship to flour texture among wheat cultivars. J Cereal Sci 41:1–6
Imazawa T, Suzuki G, Nakano A, Yamamoto M, Mukai Y (2009) Visualization of multiple T-DNA loci by FISH on extended DNA fibers. Plant Biotech 26:421–425
Jolly CJ, Rahman S, Kortt AA, Higgins TJV (1993) Characterisation of the wheat Mr 15,000 ‘grain-softness protein’ and analysis of the relationship between its accumulation and the whole seed and grain softness. Theor Appl Genet 86:589–597
Krishnamurthy K, Giroux M (2001) Expression of wheat puroindoline genes in transgenic rice confers grain softness. Nat Biotechnol 19:162–166
Morris CF, Bhave M (2008) Reconciliation of D-genome puroindoline allele designations with current DNA sequence data. J Cereal Sci 48:277–287
Mukai Y, Endo TR, Gill BS (1990) Physical mapping of the 5S rDNA multigene family in common wheat. J Hered 81:290–295
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Nakano A, Suzuki G, Yamamoto M, Turnbull K, Rahman S, Mukai Y (2005) Rearrangements of large-insert T-DNAs in transgenic rice. Mol Genet Genomics 273:123–129
Naqvi S, Farré G, Sanahuja G, Capell T, Zhu C, Christou P (2010) When more is better: multigene engineering in plants. Trends Plant Sci 15:48–56
Suzuki G, Ura A, Saito N, Do G, So B, Yamamoto M, Mukai Y (2001) BAC FISH analysis in Allium cepa. Genes Genetic Syst 76:251–255
Turnbull K-M, Marion D, Gaborit T, Appels R, Rahman S (2003a) Early expression of grain hardness in the developing wheat endosperm. Planta 216:699–706
Turnbull K-M, Turner M, Mukai Y, Yamamoto M, Morell MK, Appels R, Rahman S (2003b) The organization of genes tightly linked to the Ha locus in Aegilops tauschii, the D-genome donor to wheat. Genome 46:330–338
Wada N, Kajiyama S, Cartagena JA, Lin L, Akiyama Y, Otani M, Suzuki G, Mukai Y, Aoki N, Fukui K (2010) The effects of puroindoline b on the ultrastructure of endosperm cells and physicochemical properties of transgenic rice plant. J Cereal Sci 51:182–188
Wiley PR, Tosi P, Evrard A, Lovegrove A, Jones HD, Shewry PR (2007) Promoter analysis and immunolocalisation show that puroindoline genes are exclusively expressed in starchy endosperm cells of wheat grain. Plant Mol Biol 64:125–136
Zhang J, Martin JM, Beecher B, Morris CF, Curtis Hannah L, Giroux MJ (2009) Seed-specific expression of the wheat puroindoline genes improves maize wet milling yields. Plant Biotechnol J 7:733–743
Acknowledgments
The authors are highly obliged to Drs. Yasunori Nakamura and Akiko Kubo of the Akita Prefectural University, Dr. Tatsuya M. Ikeda of the National Agricultural Research Center for Western Region, and Dr. Noriaki Aoki of the National Institute of Crop Science for their useful technical and scientific advice. This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) and the Japan Society for the Promotion of Science (JSPS) grants, KAKENHI (18075003 and 20780240 to G.S., 22580004 and 19380194 to Y.M.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Toriyama.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2011_1134_MOESM1_ESM.doc
Online Resource 1: Separated total DNAs (HindIII digested) stained with ethidium bromide before Southern blotting (Fig. 2a) as the DNA amount control (DOC 189 kb)
299_2011_1134_MOESM2_ESM.doc
Online Resource 2 The CBB-stained SDS-PAGE gels for the protein amount control of Western blot analysis (Fig. 4). (DOC 746 kb)
Rights and permissions
About this article
Cite this article
Suzuki, G., Wada, H., Goto, H. et al. Transgenic rice plants harboring the grain hardness-locus region of Aegilops tauschii . Plant Cell Rep 30, 2293–2301 (2011). https://doi.org/10.1007/s00299-011-1134-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1134-7