Abstract
Seed development is an important factor in determining maize yield. The ABERRANT PANICLE ORGANIZATION 1–9 (APO1-9) gene is yield-related in maize. To study the function of ZmAPO1-9 in maize yield, Agrobacterium infection was used to transfer ZmAPO1-9 into maize and to successfully produce overexpression and gene editing in the plants. Subcellular results showed that ZmAPO1-9 proteins were localized in the nucleus. The T4 transgenic lines showed that ZmAPO1-9 genes were mainly expressed in the maize seeds at the grain filling stage, affecting the grain number per spike, row number, and 100-grain weight, and changing the starch content of seeds. Dual wavelength spectrophotometry was used to measure the amylose and amylopectin contents of the transgenic plants. The amylose content of the overexpressed plants was found to be higher than that of the wild type, and the amylopectin content of the gene-edited plants increased. Scanning electron microscopy results showed that overexpression of the ZmAPO1-9 gene can increase the diameter of starch granules. The breeding results showed that the yield of F1 generation maize can be significantly improved, indicating that ZmAPO1-9 genes play an important role in regulating yield.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize (Zea mays L.) is an annual herbaceous plant belonging to the genus Zea of the family Poaceae. It is a very important fodder, economic, and food crop worldwide. The crop has the highest total yield and the largest sown area among crops in China. Maize originated in Central and South America (White and Doebley 1998) and was widely grown in China in the mid-sixteenth century. Maize yield traits are quantitative traits, which are different from common quality traits. While qualitative traits are consistent with Mendelian inheritance and show discontinuous variation, quantitative traits display continuous variation (Zheng et al. 2021; Sharma et al. 2021), and the phenotype of the hybrid offspring has no clear correspondence with the genotype (Mackay et al. 2009). Most agronomic traits of crops are quantitative traits (Mackay 2001). Grain weight is almost entirely determined by genetic material while the ratio of infill grains is influenced by environmental factors (Sosso et al. 2015; Shen et al. 2020). Since the end of the twentieth century, breeding experts around the world have located more than 500 quantitative trait loci (QTL) related to maize yield traits, such as ear weight, grain weight, 100-kernel weight, and water content, through hybridization, mapping, and other technical means (Yang et al. 2016; Wang et al. 2020). Through QTL mapping, we can more accurately find the quantitative trait loci that can affect maize yield, screen out the genes that play a major role, and lay a theoretical foundation for subsequent functional verification.
Watkins found that recombinant APO1 from maize and Arabidopsis can bind high-affinity RNA in vitro, indicating that the unknown functional domain DUF794 constituting almost all of APO1 is an RNA domain (Watkins et al. 2011). One important way to cultivate high-yielding maize varieties using DNA marker technology is to introduce high-yield maize-related genes into excellent varieties. Yield per unit area is mainly determined by three yield components: the number of panicles per unit area, number of grains per panicle, and grain weight. The APO1 gene, an endogenous gene in maize and rice, was originally discovered by Amann et al. in a genetic screen of non-photosynthetic mutants in Arabidopsis thaliana (Amann et al. 2004). In their field experiments on three varieties of Japanese rice carrying mutants of the APO1 gene, Ikeda-Kawakatsu et al. found that the APO1 gene increased the number of spikelets per panicle, total number of spikelets per region, leaf width, and internode diameter in rice. Tiller number and panicle number per unit area were reduced (Ikeda-Kawakatsu et al. 2009). To test whether the ZmAPO1-9 gene has an effect on the development of maize kernels, overexpression and CRISPR/Cas9 technology was used in this study to transfer the ZmAPO1-9 gene into GSH9901 maize for functional verification. GSH9901 is an excellent inbred line with high yield potential. The results of this study showed that the 100-kernel weight, number of grains per ear, and grain quality of GSH9901 were significantly modified in the overexpressed and edited plants.
Materials and methods
Plant materials
Laboratory preserved callus from maize. Culture conditions were 25 °C/30 °C, 16 h/8 h light/dark cycle, and 65–75% relative humidity in a dark culture environment. Transgenic seedlings were grown in soil containing the same amount of nutrients and cultured in a culture chamber with a light intensity of 600 mol − 2 s − 1, with a temperature range of 25–35 °C, and relative humidity of 60–70%. The transgenic maize was then transplanted to the transgenic experimental base, with a normal field management.
Sequence analysis of ZmAPO1-9 from Zea mays L.
The NCBI ORF Finder tool (https://www.ncbi.nlm.nih.gov/orffinder/) was used to determine the open reading frame of the gene, and the Conserved Domain tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to predict the conserved domains of its encoded proteins. Homologous protein sequences from other species were identified using NCBI’s BLAST tool (http://blast.ncbi.nlm.nih.gov/). The Molecular Evolutionary Genetics Analysis software (MEGA7) was used to perform multiple sequence alignment and construct a phylogenetic tree.
Subcellular localization
The full-length CDS (Coding sequence) of ZmAPO1-9 was inserted into the pCAMBIA1302-YFP (modified from pCAMBIA1302-GFP) vector to construct the pCAMBIA1302-ZmAPO1-9-YFP vector by infecting tobacco leaves with Agrobacterium. The pCAMBIA1302-YFP vector was infected with Agrobacterium, followed by inoculation into Nicotiana benthamiana leaves as a control. Then, the target leaves and control leaves after Agrobacterium infection for 36 h were imaged with a laser confocal fluorescence microscope. The excitation wavelength was 488 nm, and the position of the yellow fluorescence was observed.
ZmAPO1-9 and ZmAPO1-6 gene expression pattern analysis
The roots, stems, leaves, and seeds of wild maize at the seedling stage, jointing stage, powder-scattering stage, and grain-filling stage were collected; the tissues were wrapped with tin foil, put into liquid nitrogen, and stored at − 80 °C. RNA was extracted and reverse-transcribed into cDNA for gene expression analysis.
Construction of the ZmAPO1-9 expression vector
RNA was extracted from leaves using the TRIZOL method. RNA was reverse-transcribed into cDNA and stored at − 80 °C. Primers were designed using Primer 5.0, and the primer sequences are as shown in Table S1. cDNA was used as a template for PCR amplification. The target fragment was recovered by electrophoresis and linked to the pCAMBIA3301 plasmid. Through double digestion and sequencing verification, the overexpression vector pCAMBIA3301-ZmAPO1-9 was obtained (Fig. S1A) and the plasmid transferred into Agrobacterium EHA105.
A CRISPR-Cas9 kit was acquired and the instructions carefully followed. The results are as shown in Fig. S1-b. The recombinant plasmid CRISPR-Cas9-APO1-9 was verified by sequencing, and the positive plasmid was obtained. Afterward, the plasmid was transferred into Agrobacterium EHA105.
Establishment and validation of ZmAPO1-9 transgenic maize
The recombinant plasmid pCAMBIA3301-ZmAPO1-9 was transferred into the callus of maize variety GSH9901 by Agrobacterium transference. Subsequently, T0 transgenic lines were selected with herbicides. The T0 generation plants were amplified by PCR with bar gene primers; the positive plants were identified, and the T4 generation positive plants were obtained by continuous selfing selection.
The recombinant plasmid CRISPR-Cas9-APO1-9 was transferred into the callus of maize variety GSH9901 by Agrobacterium transference. Subsequently, T0 transgenic lines were selected with herbicides. The T0 generation plants were amplified by PCR with bar gene primers, the positive plants were identified, and the T4 generation positive plants were obtained by continuous selfing selection (Fig. S4).
Gene expression analysis of ZmAPO1-9 and related genes in transgenic maize
Total RNA was extracted from overexpressed plants, gene-edited plants, and wild-type roots, stems, leaves, and seeds after four days of pollination by the TRIZOL method, and reverse-transcribed into cDNA for quantitative fluorescence analysis. Total RNA was extracted from the overexpressed plants, gene-edited plants, and wild-type seeds at 4, 8, 12, 16, and 20 days after pollination, and reverse-transcribed into cDNA for quantitative fluorescence analysis. Glucose transporter genes ZmSWEET1 (Zm00001d000222) and ZmSWEET15 (Zm00001d050577), yield-related gene ZmAPO2 (Zm00001d051790), and cell wall invertase gene ZmINCW2 (Zm00001d003776) were subjected to gene expression analysis on day 12 after pollination.
Measurement of agronomic characteristics
Transgenic plants and wild-type plants were grown at Jilin Agricultural University (Longitude: 125.410385, Latitude: 43.810433). This experiment was conducted in the summer of 2021 when the rainfall was 80.14 mm, average humidity was 55.10%, and average temperature was 20.8 °C. Plants received only natural precipitation. A randomized complete three-replicate block design was used for all field trials. Each transgenic maize variety was planted in a test area of about 3 m × 5 m = 15 m2, with 300 plants in each area, a plant spacing of 15 cm, and a row spacing of 25 cm. Plant height and stem thickness were measured with a tape measure. Ears of transgenic lines and wild-type plants were harvested at maturity, and kernel length was measured with vernier calipers. Ear length and shaft thickness were measured with a ruler. The grain weight and 100-kernel weight of the whole ear were measured with a weighing balance. The number of grains in each ear and the number of ears in each row were counted. Finally, significance analysis was performed.
Determination of amylose and amylopectin content
Preparation of the standard solutions
0.1, 0.3, 0.5, 0.7, 0.9, 1.1, and 1.3 mL respectively of amylose standard working solution were put into a 100-mL beaker, and 25 mL of purified water was added. Subsequently, 0.1 mol/L hydrochloric acid solution was used to adjust the pH value to 3.0. Then, 0.5 mL of iodine reagent was added, and the solution was diluted to 50 mL with purified water. This process produced amylose standard linear solution. In addition, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, and 5.0 mL of amylopectin standard working solution respectively were taken and used to prepare amylopectin standard linear solution, using the same method as that used to prepare the amylose standard linear solution.
Preparation of the test solutions
Dried constant weight maize starch sample (0.1 g) was weighed and added to 10 mL of 1 mol/L sodium hydroxide. The beaker was put into a water bath (85 ± 1) ℃, and the solution was stirred well until the solute was completely dissolved. After cooling, purified water was used to dilute the solution to 50 mL. Precisely, 5 mL of the sample solution was measured into a 100-mL beaker, and 25 mL of purified water was added. The pH was adjusted to 3.0 with 0.1 mol/L hydrochloric acid solution, and 0.5-mL of iodine reagent was added, bringing the solution to 50 mL with purified water.
Calculation of amylose and amylopectin content in the samples
According to the linear regression equation of amylose and amylopectin, the amylose concentration Yamylose and amylopectin concentration Yamylopectin in the test solution were calculated thus:
Determination of grain filling rate of transgenic lines
Transgenic T4 generation lines OE-2, KO-7, and wild-type material GSH9901 samples were taken after pollination; 100 grains in the middle of the ear of 4DAP, 8DAP, 12DAP, 16DAP, and 20DAP were taken, and the fresh grain was weighed. After drying in an oven at 80th℃, the dry maize was reweighed, and the difference curves of dry weight and fresh weight at the different periods were drawn.
Scanning electron microscope observation of grain starch morphology
The mature grains of the transgenic T4 generation lines OE-2, KO-7, and wild-type materials were selected, and the seeds were fixed. Then, the top of the grains were tapped with a scalpel to break them naturally, keeping the starch granules intact. The seeds were placed in a receptacle and sprayed with gold for 5 min. The material was then observed with a scanning electron microscope from three perspectives in the silty endosperm areas. The starch granule diameter of the maize endosperm was measured with ImageJ.
Evaluation of breeding effects of transgenic APO1-9 gene lines
The transgenic APO1-9 gene lines OE-2, KO-7, and the non-transgenic inbred line GSH9901 were used as the male parents; Lines Zheng 58, Chang 7-2, and Huangzao 4 were used as female parents, and crosses were performed to construct F1 hybrid combinations and to investigate the yield-related traits of F1.
Statistical analysis
Origin2021 was used for statistical analysis and graphing. Three independent biological replicates were used for each experiment.
Results
Sequence analysis of maize ZmAPO1-9
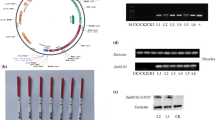
The length of the ZmAPO1-9 gene was found to be 1862 bp, the open reading frame was 1302 bp, and the encoded protein contained 433 amino acids. The conserved domain of the ZmAPO1-9 gene belongs to the F-BOX family and has high sequence homology with the F-BOX of other species (Fig. S1-C). The amino acid sequence of ZmAPO1-9 has high homology with that of Arabidopsis, rice, and tobacco. Phylogenetic analysis showed that ZmAPO1-9 has the closest evolutionary relationship with ZmAPO1-6 in maize and OsAPO1-9 in rice (Fig. 1A).
Sub-cellular localization
The fluorescence microscopy results are shown in Fig. 5. After 36 h of infection, the mesophyll nucleus showed strong fluorescence, and no yellow fluorescence was detected in the cytoplasm and the cell membrane, indicating that the ZmAPO1-9 gene was expressed normally in tobacco and localized in the nucleus (Fig. 1B).
Establishment and validation of ZmAPO1-9 transgenic maize
Five herbicide-resistant ZmAPO1-9-OE transgenic maize plants in the T0 generation were obtained (Fig. S3A). Three herbicide-resistant ZmAPO1-9-KO transgenic maize plants in the T0 generation were also obtained (Fig. S3B). After continuously selfing the plants to produce the T4 generation seeds, 4 overexpression lines and 4 gene-edited plants were obtained (Fig. S4). These were then subjected to qRT-PCR to detect the expression of ZmAPO1-9. According to the qRT-PCR results, 3 OE lines (OE-2, OE-4, and OE-5) and 3 KO lines (KO-3, KO-6, and KO-7) were selected for ZmAPO1-9 functional analysis (Fig. S5).
ZmAPO1-9 and ZmAP01-6 gene expression pattern analysis
As shown in Fig. 2A, the expression levels of ZmAPO1-9 genes in different growth stages of maize showed a gradual upward trend. The expression levels of roots, stems, and leaves did not change at the seedling and jointing stages; the expression levels of the roots and stems at the tasseling and kernel filling stages did not change, but leaf gene expression began to rise. The ZmAPO1-9 gene was expressed at a low level at the seedling, jointing, and tasseling stages, but the expression level was significantly increased in maize grains at the kernel filling stage.ZmAP01-6 and ZmAP01-9 in maize panicles 20, 16, 12, 8, 12, 4 days before pollination and 4, 8, 12, 16, 20 days after pollination respectively, it was found that ZmAP01-6 and ZmAP01-9 had small fluctuations in expression quantity before pollination, and there was an increase in expression quantity after pollination, both of which reached their peak expression quantity in 12 days, and the expression quantity of ZmAP01-9 gene exceeded that of ZmAP01-6 gene (Fig. S2). The results showed that the expression levels of the ZmAPO1-9 gene in the roots, stems, and leaves of the seedling, jointing, and tasseling stages were low, and the expression was significantly higher in the grains of maize during the kernel filling stage.
ZmAPO1-9 gene expression pattern analysis and transgenic maize kernel composition analysis and related gene expression analysis. A The expression of ZmAPO1-9 in roots, stems, leaves and grains at seeding, jointing, tasseling and grain filling stages *p < 0.05 (n = 10 for each genotype). B Expression of Wild-type and transgenic maize ZmAPO1-9 in root, stalk, leaf ear at 4 days for pollination. C Expression of Wild-type and transgenic maize ZmAPO1-9 in kernel at 4, 8, 12, 16, 20 days for pollination. D Content of Protein, starch and fat in wild-type and transgenic maize kernel. Take full, consistent size and shape kernel for experiments, n = 10 for each genotype. E Relevant gene expression in overexpression maize. F Relevant gene expression in gene editing maize *p < 0.05
Analysis of individual tissues of overexpressed and gene-edited plants
The results showed that there were significant differences in the expression levels between the edited ZmAPO1-9 maize and the ZmAPO1-9 gene kernels (Fig. 2B). In plants overexpressing the ZmAPO1-9, the gene expression levels in kernels were higher than those of the wild type. The expression levels of ZmAPO1-9 proteins in kernels were then determined at different times, that is, 8, 12, and 16 days after pollination (Fig. 2C). The results showed that ZmAPO1-9 protein expression levels remained low in the gene-edited maize but peaked 12 days after pollination and then decreased in overexpressing plants. This phenomenon showed that the ZmAPO1-9 gene played an active role in the grain development of maize at the kernel filling stage.
Agronomic trait analysis of overexpressed and gene edited plants
The results showed that the number of maize kernels and the number of rows of corn in the gene-edited plants decreased by 45.2% compared with those of the wild-type (Fig. 3). In the overexpressed maize, the number of kernels in the row increased, and the yield increased by 15.5% compared with those of the wild-type. And, the 100-kernel weight did change significantly. When the grains of the OE and KO plants were analyzed for kernel composition and a near-infrared kernel analyzer was used to analyze their protein, fat, and starch content, no difference in protein and fat content was found between the OE and KO plants; significant differences were observed in the starch content (Fig. 2D).
Agronomic trait of wild type and transgenic maize. A Ear kernel, B hundred kernel weight, C spike length, D ear diameter, E ear rows, F row kernel, G plant height, H shaft thickness, I Ear and Kernel Morphology, 10 kernels were obtained from the middle of the maize kernel, all kernels:remove all kernels from the ear and lay flat on a flat surface *p < 0.05
The expression levels of yield-related genes [ZmSWEET1, ZmSWEET15, ZmAPO2 and ZmINCW2 (Chourey et al. 2006; Bi et al. 2018)] in the OE and KO plants were measured. The results showed that the expression levels of all the genes had risen significantly (Fig. 2E). The expression levels of ZmSWEET1, ZmSWEET15, ZmAPO2, and ZmINCW2 in KO-6 and KO-7 maize kernels were measured four days after pollination, and the results showed that the expression levels of all genes had significantly decreased (Fig. 2F). Changes in the expression levels of these genes resulted in changes in the phenotype of panel A and were found to be proportional to maize yield.
Determination of grain filling rate of transgenic lines
The kernels in the middle of the ear of the transgenic lines OE-2, KO-7, and the wild-type at different periods after pollination were collected. Figure 4 shows that the dry and fresh weights of OE-2 were higher than those of the wild type at 8–20 days after pollination. As the Fig. 4 shows, overexpression of the APO1-9 gene in maize can improve the kernel filling rate, and the kernel filling rate is higher at the later stage of kernel development.
Scanning electron microscopy of the grain starch morphology
As shown in Fig. 2D, in the overexpressed material, the starch content increased by up to 2%. To directly and clearly assess the change in the size of the starch granules, the starch granules in the silty endosperm region of mature kernels in the transgenic plants were observed under a scanning electron microscope. The results showed that starch granules in the wild-type, OE-2, and KO-7 were all irregular in shape (Fig. 5A). Starch granules are categorized into three groups by diameter following the categorization of Ji et al. (2003): group A, starch granules of diameter < 9 μm; group B, diameter 9–13 μm; group C, diameter > 13 μm. Most of the starch granules of the wild-type and KO-7 fell into group B, while those of OE-2 fell into group C (Fig. 5B). This phenomenon indicates that APO1-9 genes can increase yield by affecting starch granule size.
The amylose and amylopectin contents of the OE and KO plants were determined by dual wavelength spectrophotometry. Before the experiment, the standard curves of amylose and amylopectin were obtained: amylose standard curve: y = 21.723x – 0.0969, R2 = 0.9996, and amylopectin standard curve: y = 3.2587x + 0.0213, R2 = 0.9949 (Fig. S6). As the results in Fig. 5C show, there were significant changes in amylose and amylopectin content between OE and KO. In the absence of the ZmAPO1-9 gene, the amylopectin content of maize kernels increased. In the case of overexpression of ZmAPO1-9, the amylose content significantly increased. This indicates that APO1-9 played an important role in the formation of amylose and amylopectin during the kernel filling stage.
Evaluation of breeding effects of transgenic APO1-9 gene lines
Hybrid plants containing the APO1-9 gene were identified by PCR reactions (Fig. S7). Nine hybrid combinations were phenotyped in the summer of 2021, and the results showed that the hybrid combinations containing the APO1-9 gene and the non-transgenic control combinations had significant increases in kernel width, 100-kernel weight, and grain weight (Table 1). Specifically, the grain width increased by 15%, the 100-kernel weight by 14%, and the ear weight by 15%. These results show that the overexpression of the APO1-9 gene can improve maize yield.
Discussion
Increasing grain yield is the goal of seed developers. Previous research has shown that overexpression of the APO1 gene in rice cultivars can significantly improve rice grain yield (Ikeda et al. 2005). In this study, ZmAPO1-9 was found to significantly increase the number of corn rows, 100-kernel weight, and yield per plant, a finding consistent with the results of previous studies. ZmAPO1-9 belongs to the F-BOX family and has close homology to the APO1 gene in rice. F-box proteins (FBPs) belong to one of the largest protein superfamilies found in plants, and the FBP family in plants is more diverse than that of other families (Abd-Hamid et al. 2020; Yu et al. 2007). Hua et al. reported that the number of FBP genes in 18 plants ranged from 159 to 980 (Hua et al. 2011). FBPs are characterized by the presence of a loosely conserved F-box motif consisting of approximately 40–60 amino acid residues with a few invariant positions throughout the consensus sequence (Ji et al. 2020). Most of the F-box proteins are localized in the nucleus, which is the same as the subcellular localization in the nucleus established in this experiment (Ikeda-Kawakatsu et al. 2009, 2012).
The APO1 gene was overexpressed in rice, and it was found that the shorter panicle and reduced number of spikelets in APO1 mutant rice plants were due to the early transformation of the inflorescence meristem to the spikelet meristem on the main axis and branches. Therefore, APO1 is a regulator of meristems. In maize, ZmAPO1-9 was mainly expressed in the early stage of ear development, especially within the 4–12-day period after pollination, during which the expression level gradually increased. Changes in meristem development over time are critical for the control of inflorescence architecture (McSteen et al. 2000; Bortiri et al. 2006; Wang and Li 2008).
In rice, the transition from vegetative to reproductive stages induces a transition from the shoot apical meristem (SAM) to the inflorescence meristem (IM), which initiates lateral meristem development (PBs; Ito et al. 2005). Thus, in rice, the timing of IM development determines the number of primary branches in the inflorescence (Ikeda et al. 2007). The meristems of PB produce lateral meristems and then differentiate into spikelet meristems (Irish 1997). A spikelet is a small branch containing a variable number of flowers, affecting the number of spike branches and yield in rice (Chongloi et al. 2019; Ikeda et al. 2021).
In this study, OE plants showed increased grain number in rows, while KO plants showed reduced kernel numbers in rows and severe tip baldness, indicating that the function of ZmAPO1-9 in regulating meristems was similar to that of the APO1 gene in rice. Although APO1 gene regulates the meristem and affects yield, we used a near-infrared grain analyzer to determine the starch, fat, and protein content of OE and KO grains. It was found that ZmAPO1-9 genes also affected maize kernel composition, and the starch content in OE and KO kernels changed significantly. Genes related to grain-filling in maize were successfully isolated. The SWEET gene family is involved in a variety of developmental processes and plays a key role in sugar transport. Xuan et al. found that AtSWEET1 acts as a glucose transporter in Arabidopsis (Xuan et al. 2013; Park et al. 2022). AtSWEET15 (also known as SAG29) in Arabidopsis thaliana seeds showed a specific temporal and spatial expression pattern during the development of seeds (Huang et al. 2020; Zhang et al. 2018). The knockout of this gene resulted in severe loss of nutrients in seeds, resulting in a wrinkled phenotype in seeds at maturity (Wang et al. 2022). In the early stage of grain filling, the gene INCW2 encodes a cell wall invertase required for carbon source allocation, which regulates sucrose unloading and promotes grain filling, thereby resulting in grain yield (Juárez-Colunga et al. 2018; Carlson and Chourey 1999).
We found that the INCW2 gene is a tissue-specific gene, which is specifically expressed in the maize endosperm at the grain filling stage (Chourey et al. 2006). LARGE2 encodes a hect domain E3 ubiquitin ligase OsUPL2 and regulates panicle size and grain number in rice (Kyozuka et al. 1998). LARGE2 is highly expressed in young panicles and grains. Studies have shown that LARGE2 is physically associated with abnormal ear organization (APO1) and positive ear size and grain number regulation (APO2). The gene also regulates their stability. Genetic analysis indicated that LARGE2 regulates panicle size and grain number in a manner similar to APO1 and APO2 (Huang et al. 2021). The analysis of the expression levels of these genes related to grain-filling showed that the expression levels of the genes changed significantly when the expression of ZmAPO1-9 fluctuated. These fluctuations had an impact on the transport and utilization of sugar during the grain-filling period of maize, resulting in changes in starch content as well as changes in amylose and amylopectin content. It was found that the amylose content of grains overexpressing ZmAPO1-9 increased, indicating that ZmAPO1-9 played a positive role in the formation of amylose during kernel filling.
Data availability
Datasets supporting the conclusions of this article are included within the article.
References
Abd-Hamid NA, Ahmad-Fauzi MI, Zainal Z, Ismail I (2020) Diverse and dynamic roles of F-box proteins in plant biology. Planta 251:68. https://doi.org/10.1007/s00425-020-03356-8
Amann K, Lezhneva L, Wanner G, Herrmann RG, Meurer J (2004) ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 16:3084–3097. https://doi.org/10.1105/tpc.104.024935
Bi YJ, Sun ZC, Zhang J et al (2018) Manipulating the expression of a cell wall invertase gene increases grain yield in maize. Plant Growth Regul 84:37–43. https://doi.org/10.1007/s10725-017-0319-7
Bortiri E, Chuck G, Vollbrecht E, Rocheford T, Martienssen R, Hake S (2006) ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18:574–585. https://doi.org/10.1105/tpc.105.039032
Carlson SJ, Chourey PS (1999) A re-evaluation of the relative roles of two invertases, INCW2 and IVR1, in developing maize kernels and other tissues. Plant Physiol 121:1025–1035. https://doi.org/10.1104/pp.121.3.1025
Chongloi GL, Prakash S, Vijayraghavan U (2019) Regulation of meristem maintenance and organ identity during rice reproductive development. J Exp Bot 70:1719–1736. https://doi.org/10.1093/jxb/erz046
Chourey PS, Jain M, Li QB, Carlson SJ (2006) Genetic control of cell wall invertases in developing endosperm of maize. Planta 223:159–167. https://doi.org/10.1007/s00425-005-0039-5
Hua Z, Zou C, Shiu SH, Vierstra RD (2011) Phylogenetic comparison of F-Box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS ONE 6:e16219. https://doi.org/10.1371/journal.pone.0016219
Huang C, Yu J, Cai Q, Chen Y, Li Y, Ren Y, Miao Y (2020) Triple-localized WHIRLY2 influences leaf senescence and silique development via carbon allocation. Plant Physiol 184:1348–1362. https://doi.org/10.1104/pp.20.00832
Huang L, Hua K, Xu R, Zeng D, Wang R, Dong G, Zhang G, Lu X, Fang N, Wang D, Duan P, Zhang B, Liu Z, Li N, Luo Y, Qian Q, Yao S, Li Y (2021) The LARGE2-APO1/APO2 regulatory module controls panicle size and grain number in rice. Plant Cell 33:1212–1228. https://doi.org/10.1093/plcell/koab041
Ikeda K, Nagasawa N, Nagato Y (2005) ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev Biol 282:349–360. https://doi.org/10.1016/j.ydbio.2005.03.016
Ikeda K, Ito M, Nagasawa N, Kyozuka J, Nagato Y (2007) Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J 51(6):1030–1040. https://doi.org/10.1111/j.1365-313X.2007.03200.x
Ikeda M, Takasaki H, Mitsuda N (2021) Thermomemory in shoot apical meristem: regulation of carbohydrate metabolism and stem cell identity. Mol Plant 14:1427–1429. https://doi.org/10.1016/j.molp.2021.07.003
Ikeda-Kawakatsu K, Yasuno N, Oikawa T, Iida S, Nagato Y, Maekawa M, Kyozuka J (2009) Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiol 150:736–747. https://doi.org/10.1104/pp.109.136739
Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh J, Nagato Y (2012) ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J 69:168–180. https://doi.org/10.1111/j.1365-313X.2011.04781.x
Irish EE (1997) Experimental analysis of tassel development in the maize mutant tassel seed 6. Plant Physiol 114:817–825. https://doi.org/10.1104/pp.114.3.817
Ji Y, Seetharaman K, Wong K, Pollak LM, Duvick S, Jane J, White PJ (2003) Thermal and structural properties of unusual starches from developmental corn lines. Carbohyd Polym 51:439–450
Ji H, Liu D, Zhang Z, Sun J, Han B, Li Z (2020) A bacterial F-box effector suppresses SAR immunity through mediating the proteasomal degradation of OsTrxh2 in rice. Plant J 104:1054–1072. https://doi.org/10.1111/tpj.14980
Juárez-Colunga S, López-González C, Morales-Elías NC, Massange-Sánchez JA, Trachsel S, Tiessen A (2018) Genome-wide analysis of the invertase gene family from maize. Plant Mol Biol 97:385–406. https://doi.org/10.1007/s11103-018-0746-5
Kyozuka J, Konishi S, Nemoto K, Izawa T, Shimamoto K (1998) Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc Natl Acad Sci USA 95:1979–1982. https://doi.org/10.1073/pnas.95.5.1979
Mackay TF (2001) The genetic architecture of quantitative traits. Annu Rev Genet 35:303–339. https://doi.org/10.1146/annurev.genet.35.102401.090633
Mackay TF, Stone EA, Ayroles JF (2009) The genetics of quantitative traits: challenges and prospects. Nat Rev Genet 10:565–577. https://doi.org/10.1038/nrg2612
McSteen P, Laudencia-Chingcuanco D, Colasanti J (2000) A floret by any other name: control of meristem identity in maize. Trends Plant Sci 5:61–66. https://doi.org/10.1016/s1360-1385(99)01541-1
Park J, Chavez TM, Guistwhite JA, Gwon S, Frommer WB, Cheung LS (2022) Development and quantitative analysis of a biosensor based on the Arabidopsis SWEET1 sugar transporter. Proc Natl Acad Sci USA 119:e2119183119. https://doi.org/10.1073/pnas.2119183119
Sharma M, Gangurde SS, Salgotra RK, Kumar B, Singh AK, Pandey MK (2021) Genetic mapping for grain quality and yield-attributed traits in Basmati rice using SSR-based genetic map. J Biosci 46:50
Shen S, Li BB, Deng T et al (2020) The equilibrium between sugars and ethylene is involved in shading- and drought-induced kernel abortion in maize. Plant Growth Regul 91:101–111. https://doi.org/10.1007/s10725-020-00590-8
Sosso D, Luo D, Li QB, Sasse J, Yang J, Gendrot G, Suzuki M, Koch KE, McCarty DR, Chourey PS, Rogowsky PM, Ross-Ibarra J, Yang B, Frommer WB (2015) Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet 47:1489–1493. https://doi.org/10.1038/ng.3422
Wang Y, Li J (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59:253–279. https://doi.org/10.1146/annurev.arplant.59.032607.092902
Wang G, Zhao Y, Mao W, Ma X, Su C (2020) QTL analysis and fine mapping of a major QTL conferring kernel size in maize (Zea mays). Front Genet 11:603920. https://doi.org/10.3389/fgene.2020.603920
Wang C, Chen N, Liu J et al (2022) Overexpression of ZmSAG39 in maize accelerates leaf senescence in Arabidopsis thaliana. Plant Growth Regul. https://doi.org/10.1007/s10725-022-00874-1
Watkins KP, Rojas M, Friso G, van Wijk KJ, Meurer J, Barkan A (2011) APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell 23:1082–1092. https://doi.org/10.1105/tpc.111.084335
White S, Doebley J (1998) Of genes and genomes and the origin of maize. Trends Genet 14:327–332. https://doi.org/10.1016/s0168-9525(98)01524-8
Xuan YH, Hu YB, Chen LQ, Sosso D, Ducat DC, Hou BH, Frommer WB (2013) Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc Natl Acad Sci USA 110:E3685–E3694. https://doi.org/10.1073/pnas.1311244110
Yang C, Zhang L, Jia A, Rong T (2016) Identification of QTL for maize grain yield and kernel-related traits. J Genet 95:239–247. https://doi.org/10.1007/s12041-016-0628-z
Yu H, Wu J, Xu N, Peng M (2007) Roles of F-box proteins in plant hormone responses. Acta Biochim Biophys Sin 39:915–922. https://doi.org/10.1111/j.1745-7270.2007.00358
Zhang C, Teng XD, Zheng QQ, Zhao YY, Lu JY, Wang Y, Guo H, Yang ZN (2018) Ethylene signaling is critical for synergid cell functional specification and pollen tube attraction. Plant J 96:176–187. https://doi.org/10.1111/tpj.14027
Zheng Y, Yuan F, Huang Y, Zhao Y, Jia X, Zhu L, Guo J (2021) Genome-wide association studies of grain quality traits in maize. Sci Rep 11:9797. https://doi.org/10.1038/s41598-021-89276-3
Acknowledgements
We would like to thank Dr. Peng Jiao for the discussion and suggestions to do with the work of this paper.
Funding
This work was supported by Jilin Province Science and Technology Development Plan Project [20210302003NC,20200402023NC].
Author information
Authors and Affiliations
Contributions
ZJ is responsible for writing the article, DJ and HZ are responsible for data analysis, JQ and SL are responsible for sampling, SG and YM are responsible for field management.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Dali Zeng.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, Z., Jin, D., Zhang, H. et al. Effects of overexpression of ZmAPO1-9 gene on maize yield. Plant Growth Regul 99, 493–503 (2023). https://doi.org/10.1007/s10725-022-00920-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-022-00920-y