Abstract
Introduction of large-DNA fragments into cereals by Agrobacterium-mediated transformation is a useful technique for map-based cloning and molecular breeding. However, little is known about the organization and stability of large fragments of foreign DNA introduced into plant genomes. In this study, we produced transgenic rice plants by Agrobacterium-mediated transformation with a large-insert T-DNA containing a 92-kb region of the wheat genome. The structures of the T-DNA in four independent transgenic lines were visualized by fluorescence in situ hybridization on extended DNA fibers (fiber FISH). By using this cytogenetic technique, we showed that rearrangements of the large-insert T-DNA, involving duplication, deletion and insertion, had occurred in all four lines. Deletion of long stretches of the large-insert DNA was also observed in Agrobacterium.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Agrobacterium-mediated plant transformation is the most popular and reliable method for introducing foreign genes into plants. Although monocots are not natural hosts for Agrobacterium, recent production of transgenic cereals has also been achieved by Agrobacterium-mediated transformation (Chan et al. 1992; Hiei et al. 1994; Ishida et al. 1996; Cheng et al. 1997). Agrobacterium transports a single stranded T-DNA carrying foreign genes into the plant genome: the T-DNA is delimited by 25-bp direct repeats, referred to as right border (RB) and left border (LB) sequences, which flank the DNA of interest. It has generally been postulated that T-DNA integration is initiated from the RB and terminated at the LB. The structure of the junctions between T-DNA and plant DNA has been extensively studied in transgenic dicotyledonous plants (Krizkova and Hrouda 1998; Buck et al. 1999; Windels et al. 2003) and in monocot plants (Kumar and Fladung 2002; Kim et al. 2003). These analyses showed that insertions of filler DNA and small deletions can occur at the junctions. Vector backbone sequences were also frequently observed in the regions flanking the T-DNA (Martineau et al. 1994; Kononov et al. 1997). Recent studies of transgene integration and organization have been reviewed by Kohli et al. (2003).
Binary vectors related to bacterial artificial chromosome (BAC) vectors, such as the binary BAC (BIBAC) and transformation-competent artificial chromosome (TAC), which can be used in Agrobacterium-mediated plant transformation, have been developed to transfer large DNA fragments into plant chromosomes (Hamilton et al. 1996; Liu et al. 1999). In addition, Tao and Zhang (1998) reported that over 100-kb segments of DNA could be inserted in conventional binary vectors. Transformation-ready large-insert libraries have been constructed to date for Arabidopsis thaliana (Liu et al. 1999), Brassica napus (Cui et al. 2000), tomato (Hamilton et al. 1999), wheat (Moullet et al. 1999; Liu et al. 2000), and petunia (McCubbin et al. 2000). These libraries are useful resources for introducing large and complex genomic regions bearing whole sets of genes into important crops, as well as for complementation analyses during map-based cloning. The progressive introduction of large foreign genomic fragments into plant genomes, which we refer to as ‘genome fusion’, would be a fascinating strategy for creating new crops by molecular breeding.
However, little information is available regarding the chromosomal integration and organization of large-insert T-DNA in transgenic plants, especially in transgenic cereals. In this study, we introduced a 92-kb fragment of the wheat genome into rice by Agrobacterium-mediated transformation. Rearrangements of the integrated T-DNA in transgenic rice plants were successfully visualized and analyzed by fluorescence in situ hybridization on extended DNA fibers (fiber FISH).
Materials and methods
Transformation
Binary cosmid DNA containing a 92-kb segment of the wheat Ha-locus region (Turnbull et al. 2003, Fig. 1) was purified using a Plasmid Midi Kit (Qiagen) and electroporated into the Agrobacterium tumefaciens strain EHA105 (Hood et al. 1993) using the Gene Pulser II (Bio-Rad) set to 2.5 kV, 25 μF, and 100 Ω. A cultivar of japonica rice, Oryza sativa L. Yamahoushi, was used to produce transgenic plants by Agrobacterium-mediated transformation as described by Yokoi and Toriyama (1999) with some modifications. Geneticin (G418, Sigma, 70 mg/l) was used as a selection agent, as the binary cosmid vector contains the neomycin phosphotransferase gene (nptII) in the T-DNA region (Fig. 1).
Structure of the original BAC 8 construct used in this study. The relative positions of the probes employed are indicated by the open bars below the construct. A 92-kb genomic fragment from wheat containing the genes puroindoline-a and GSP-1 was cloned in the binary cosmid vector pCLD04541 (Turnbull et al. 2003)
PCR
To confirm the presence of the puroindoline-a and GSP-1 genes in Agrobacterium and transgenic plants, PCR was performed with ExTaq polymerase (Takara), for 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min, followed by a final extension for 5 min, by using the Personal DNA thermal cycler (Takara). The primers used to amplify the puroindoline-a sequence were PA-F (5′-ATGAAGGCCCTCTTCCTCATAGG-3′) and PA-R (5′-TCACCAGTAATAGGCAATAGTGCC-3′), and GSP-F (5′-GCGATCTAAGTGGCTTCAAG-3′) and GSP-R (5′-GCTAGTGATGGGGATGTTGC-3′) were used to amplify the GSP-1 sequence.
DNA gel-blot analysis
Total DNA was extracted from young leaf tissue of transgenic and non-transgenic rice plants by the procedure described by Murray and Thompson (1980). Total DNA (about 1 μg) and binary cosmid DNA (about 5 ng) isolated from Escherichia coli or A. tumefaciens were digested with HindIII, and fractionated on a 0.8% agarose gel. After electrophoresis, DNA was transferred to nylon membranes (Roche Diagnostics). Hybridization was carried out in 5×SSC, 0.5% blocking reagent (Roche), 0.1% sodium N-lauroyl sarcosinate and 0.02% SDS at 65°C. The membrane was washed twice in 0.1SSC, 0.1% SDS at 65°C for 20 min each. The digoxigenin (dig)-labeled nptII and BAC 8 probes (Fig. 1) were prepared by PCR using PCR DIG Labeling Mix (Roche) and random priming using the DIG DNA Labeling Kit (Roche), respectively. Detection of the hybridized probe was carried out according to the instructions in the manual supplied with the DIG Luminescent Detection Kit (Roche) using CSPD as the substrate.
Probes for FISH analysis
Binary cosmid DNA was purified with the Plasmid Midi Kit (Qiagen). For use as a BAC 8 probe (Fig. 1) for FISH analysis, the binary cosmid DNA was labeled with dig-11-dUTP or biotin-16-dUTP using a Nick Translation Kit (Roche). A PCR product amplified from the binary cosmid DNA was labeled with dig- or biotin-dUTP (Roche) and used as the T-DNA-LB probe (Fig. 1). The primers employed for amplification of the T-DNA-LB region were pCLD-F (5′-CCGGTACGTACCAGCTTTTG-3′) and pCLD-R (5′-AAATGTAGATGTCCGCAGCG-3′).
FISH on extended DNA fibers
Extended DNA fibers were prepared from young leaves, according to the protocol of Fransz et al. (1996a) with minor modifications. Multicolor FISH on the extended DNA fibers was performed by using the BAC 8 and T-DNA-LB probes. The dig-labeled probe was detected using an anti-Dig mouse antibody (Roche) and Cy3 conjugated anti-mouse IgG (Sigma). For detection of the biotin-labeled probe, two layers of Fluorescein Avidin DCS (Vector Laboratories) and one layer of biotinylated anti-avidin D (Vector Laboratories) were applied. Slides were counterstained with DAPI and mounted in an anti-fade solution (DABCO in 90% glycerol).
Results
Characterization of transgenic lines by PCR and Southern hybridization
We introduced a 92-kb binary cosmid clone, BAC 8 (Fig. 1), into rice by Agrobacterium-mediated transformation in order to examine structure of the integrated large T-DNA in transgenic plants. BAC 8 contains a genomic fragment from wheat that includes the puroindoline-a and GSP-1 genes, which are located at the grain hardness locus, Ha (Turnbull et al. 2003). Of the nine transgenic rice lines obtained, four (lines S, N, 11, and 12) were used to dissect the genomic organization of the integrated T-DNA. The presence of the puroindoline-a and GSP-1 genes in the transformants was confirmed by PCR (data not shown), and the copy number of the nptII gene was estimated by DNA gel-blot analysis as 1, 1, 2 and 2 copies for the lines S, N, 11, and 12, respectively (Fig. 2). DNA gel blot analysis of the segregated T1 plants showed that the two copies of nptII in each of the lines 11 and 12 were linked and unlinked, respectively (data not shown). A representative homozygous T1 plant of the lines S, N and 12 was used for subsequent DNA gel blot and FISH analyses, but a T0 plant was used for the FISH analysis of line 11.
Determination of the copy number of the nptII gene in the four transgenic lines S, N, 11, and 12. Genomic DNA (1 μg) from the transgenic rice lines (S, N, 11, and 12) and an untransformed host (U) was digested with HindIII and subjected to DNA gel-blot analysis with the nptII probe. The estimated copy number of the nptII sequence is indicated below each lane
To analyze rearrangements of the T-DNA in the Agrobacterium and transgenic plants, DNA gel blot analysis was performed with a BAC 8 probe (Fig. 3). This probe can specifically hybridize with the whole BAC 8 construct including vector DNA (Fig. 1). Several HindIII bands detected in the original BAC 8 clone in E. coli were lost from the BAC 8 clone in Agrobacterium (Fig. 3); in all, some 20 kb of the DNA was deleted in Agrobacterium. This rearrangement of the BAC 8 construct in Agrobacterium implies that actual size of the construct used for Agrobacterium-mediated gene transfer in the present experiment was approximately 72 kb. Most of the HindIII bands found in the transgenic lines S, 11 and 12, were the same as those seen in Agrobacterium, whereas changes in band lengths were observed in line N (Fig. 3). The HindIII fragments including the nptII gene (marked by asterisks in Fig. 3) might contain T-DNA/plant DNA junctions at the LB, resulting in appearance of the bands different from those seen in Agrobacterium. The presence of several other bands that differed from those in Agrobacterium suggested that rearrangements of T-DNA had also occurred in the transgenic plants.
Rearrangements of the BAC 8 construct occur both in Agrobacterium and in transgenic rice plants. Aliquots of the binary cosmid DNA (5 ng) isolated from E. coli or A. tumefaciens, and of genomic DNA (1 μg) from representative T1 plants of the four transgenic lines (S, N, 11, and 12) and an untransformed control (U) were digested with HindIII and subjected to DNA gel-blot analysis with the BAC 8 probe, which consists of the complete BAC 8 construct including vector DNA. The asterisks indicate fragments that also hybridized with the nptII probe (Fig. 2)
Analysis of the T-DNA inserts by fiber FISH
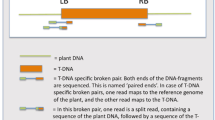
It is difficult to determine the complex structure of the large T-DNA loci by DNA gel blot analysis. Therefore, for further investigation of the genomic organization of the integrated T-DNA, we visualized the integrated regions by FISH on extended DNA fibers (fiber FISH) obtained from the transgenic rice plants. Representative results of the fiber FISH with the biotin-labeled BAC 8 probe (detected as green signals) and the Dig-labeled T-DNA-LB probe (detected as red signals) are depicted in Fig. 4. The BAC 8 probe can hybridize to all segments of the whole BAC 8 construct as in the case of DNA gel blot analysis, and the T-DNA-LB probe can detect the 6.5-kb LB region including the nptII gene (Fig. 1). The average lengths of the different signals were converted to kb lengths by using the stretching factor of 3.27 kb/μm reported by Fransz et al. (1996a). The predicted structure of the integrated region is schematically represented in Fig. 5.
Visualization of the large-insert T-DNA on extended DNA fibers from the four transgenic lines S, N, 11, and 12. Extended DNA fibers were prepared from the lines S, N, 11, and 12, and hybridized with BAC 8 (green) and T-DNA-LB (red) probes. The scale bar represents 10 μm. A schematic representation of the FISH signal for each line is shown below the hybridized preparation. The DNA fibers obtained from the lines S, N, and 12 were isolated from the same T1 plants as those used for the DNA gel blot analysis shown in Fig. 3. The DNA fibers for line 11 were isolated from a T0 plant. However, the patterns of bands detected in T0 and T1 plants by DNA gel blot analysis with the nptII and BAC 8 probes (data not shown) were found to be identical
Schematic representation of the deduced organization of the integrated T-DNAs in the four transgenic lines. The gray and black bars represent the integrated T-DNA regions that hybridized with BAC 8 and T-DNA-LB probes, respectively; a broken line between the bars denote rice genomic DNA. The average length (kb) of each region (±SD) is given in parentheses. The arrows indicate the estimated position and orientation (RB to LB) of the BAC 8 construct
In the case of the line S, a long and contiguous green signal (~50 μm) with two short red signals (approximately 2 and 4 μm) was observed on the DNA fiber (Fig. 4). The mean length of the integrated region in the line S was estimated to be 140.2 kb (SD=12.0, n=22). This total length suggested that two copies of the 72-kb construct had integrated in this line, although only one copy of the nptII gene was detected by DNA gel blot analysis (Fig. 2). This difference in the copy number estimate could be explained by assuming that the 4-μm red signal (13.8 kb, SD=4.0) might correspond to two copies of the 6.5-kb T-DNA-LB region duplicated in opposite orientation (LB to LB orientation), because the single 13-kb HindIII band was scored as one copy in the DNA gel blot analysis with the nptII probe (Fig. 2; see the construct shown in Fig. 1). We assume that the 2-μm red signal (5.2 kb, SD=2.7) does not include the nptII gene, and that this fragmented T-DNA has been copied from the neighboring construct with the short insert fragment (9.7 kb, SD=5.0). The internal green signal (77.9 kb, SD=8.5) with the two red signals on both sides might correspond to one copy of the intact BAC 8 insert, whereas the outer green signal (33.8 kb, SD=10.4) on the same side as the 4-μm red signal might correspond to another copy of the deleted BAC 8 insert. To summarize the possible integration events in the line S, two copies of the BAC 8 construct have been integrated in inverted orientation, a deletion has occurred in one of the copies, and short segments of T-DNA and insert DNA have been duplicated and transferred to the terminal (Fig. 5).
In the case of the line N, a ~10-μm green signal with a red signal and a ~20-μm green signal, which were interrupted by a gap, were observed on the same DNA fiber (Fig. 4). The total length of the integrated region in the line N (without the gap) was estimated to be 86.3 kb (SD=13.6, n=23). This length and the presence of a signal from the T-DNA-LB region at one end indicated that it consisted of one copy of the BAC 8 construct. The fragment corresponding to the gap of 32.4 kb (SD=7.9) must be derived from genomic DNA of rice inserted in one copy of the construct (Fig. 5). Additional small insertions may be present on the predicted RB-side of the construct (54.9 kb, SD=18.8). However, the small gaps could not be distinguished from the artifacts of the fiber-FISH signals which are typically detected as a ‘beads-on-a-string’ pattern rather than continuous tracks (Fransz et al. 1996a).
The fiber-FISH signals in the line 11 were detected as an approximately 40-μm contiguous green signal with two red signals (Fig. 4). The length of the integrated region in the line 11 was calculated to be 119.0 kb (SD=11.2, n=19). The presence of the two red signals on the same fiber was consistent with the detection of two linked nptII copies by DNA gel blot analysis. One explanation for this arrangement in the line 11 is that one copy of the BAC 8 construct has been interrupted by the insertion of a T-DNA-LB region (Fig. 5). Alternatively, two deleted constructs might be arranged in tandem. In both cases, the existence of another integrated region (34.2 kb, SD=9.8), detected as the green signal beyond the LB, might be due to co-integration of vector backbone sequences and/or internal duplication.
The two independent copies of the BAC 8 construct in the line 12 were detected as two different types of signals (Fig. 4): a green signal with a terminal red signal (type A) and another with a subterminal red signal (type B). The integrated regions of types A and B were, on average, 128.0 kb (SD=27.5, n=20) and 119.1 kb (SD=28.5, n=19) long, respectively, indicating that the both copies were longer than the expected insert size (Fig. 5). Internal duplication might have occurred in the unlinked two T-DNA regions of the line 12. In addition, type B showed the subterminal red signal because the integrated region possibly contained vector backbone sequences beyond the LB.
Thus, various types of rearrangements that occurred in the integrated large-insert DNA were successfully visualized in the four transgenic lines by two-color fiber FISH. The fiber FISH with a reverse combination of the probes was also performed to confirm the arrangement of the different signals (data not shown).
Discussion
FISH is an effective tool for the analysis of transgene organization. Single-copy T-DNA regions have been visualized by FISH on mitotic chromosomes (Dong et al. 2001; Khrustaleva and Kik 2001) and in interphase nuclei (ten Hoopen et al. 1999). Chromosome rearrangements in transgenic plants have also been characterized by FISH (Fransz et al. 1996b; Papp et al. 1996). Furthermore, recent improvements in the fiber FISH technique (Wolters et al. 1998) now permit detailed analysis of the rearrangement and tandem duplication of transgenes.
In this study, we successfully visualized large-insert T-DNA by fiber FISH in order to determine the length of the integrated region in transgenic rice plants. These visual data provide new and important information on the genomic organization of large transgenes in plants—an issue that has been quite difficult to analyze using traditional DNA gel-blot analysis. We were able to deduce the orientation, copy number, and extent of deletion of the large-insert T-DNA by using two-color fiber FISH with a combination of the probes for the 6.5-kb LB region (the T-DNA-LB probe) and a full-length 119-kb BAC construct (the BAC 8 probe).
The genomic organization of the integrated T-DNAs differed in the four transgenic lines analyzed in this study. All the integrated large transgenes had been rearranged. Inverted integration of two constructs and deletions in one copy were observed in the line S, an insertion of rice genomic DNA into the construct was detected in the line N, insertion of the T-DNA-LB region or the tandem duplication of deleted constructs was visualized in line 11, and two longer constructs were present in line 12. These duplication, deletion and insertion events might have occurred independently in the transgenic plants during or after the integration of the T-DNA. Our analysis suggested that the large T-DNAs integrated by Agrobacterium-mediated transformation tend to be rearranged in transgenic rice plants. Similar rearrangements were also observed in other transgenic rice plants harboring large genomic fragments from wheat (our unpublished results). The integration of intact T-DNA should therefore be routinely confirmed by cytogenetic analysis in the case of transgenic plants transformed with large transgenes using Agrobacterium.
To our surprise, the BAC 8 construct used in this study had suffered significant deletion in Agrobacterium; the 92-kb insert in the original BAC 8 construct was apparently converted into a 72-kb insert in Agrobacterium prior to infection of rice. Recently, Song et al. (2003) reported that potato BIBAC and TAC constructs containing genomic DNA fragments larger than 100 kb were unstable in Agrobacterium. Similarly, in the present investigation, deletions have been observed in the BAC 8 construct in Agrobacterium; these might be due to the presence of repetitive sequences in the 92-kb insert of wheat genomic DNA. In fact, FISH analysis with the BAC 8 probe on wheat mitotic chromosomes resulted in the strong signals over entire chromosomes, suggesting the existence of highly repetitive sequences in the BAC 8 insert (unpublished results). In transgenic experiments, it is therefore important that large-insert clones must be constructed to be as short as possible, and the constructs should be checked for rearrangements in Agrobacterium before using them for plant transformation. The initial identification of a stable clone is especially important, because clone instability in Agrobacterium may facilitate rearrangements in the transgene loci.
References
Buck SD, Jacobs A, Montagu MV, Depicker A (1999) The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J 20:295–304
Chan MT, Lee TM, Chang HH (1992) Transformation of indica rice (Oryza sativa) mediated by Agrobacterium tumefaciens. Plant Cell Physiol 33:577–583
Cheng M, Fry JE, Pang SZ, Zhou HP, Hironaka CM, Duncan DR, Conner TW, Wan YC (1997) Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol 115:971–980
Cui Y, Bi YM, Brugière N, Arnoldo M, Rothstein SJ (2000) The S locus glycoprotein and the S receptor kinase are sufficient for self-pollen rejection in Brassica. Proc Natl Acad Sci USA 97:3713–3717
Dong J, Kharb P, Teng W, Hall TC (2001) Characterization of rice transformed via an Agrobacterium-mediated inflorescence approach. Mol Breed 7:187–194
Fransz PF, Alonso-Blanco C, Liharska TB, Peeters AJM, Zabel P, de Jong JH (1996a) High-resolution physical mapping in Arabidopsis thaliana and tomato by fluorescence in situ hybridization to extended DNA fibers. Plant J 9:421–430
Fransz PF, Stam M, Montijn B, ten Hoopen R, Wiegant J, Kooter JM, Oud O, Nanninga N (1996b) Detection of single-copy genes and chromosome rearrangements in Petunia hybrida by fluorescence in situ hybridization. Plant J 9:767–774
Hamilton CM, Frary A, Lewis C, Tanksley SD (1996) Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc Natl Acad Sci USA 93:9975–9979
Hamilton CM, Frary A, Xu Y, Tanksley SD, Zhang H-B (1999) Construction of tomato genomic DNA libraries in a binary-BAC (BIBAC) vector. Plant J 18:223–229
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hood EE, Gelvin SB, Melchers S, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants (EHA105). Transgene Res 2:208–218
Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T (1996) High efficiency transformation of maize (Zea mays) mediated by Agrobacterium tumefaciens. Nat Biotechnol 14:745–750
Khrustaleva LI, Kik C (2001) Localization of single-copy T-DNA insertion in transgenic shallots (Allium cepa) by using ultra-sensitive FISH with tyramide signal amplification. Plant J 25:699–707
Kim S-R, Lee J, Jun S-H, Park S, Kang H-G, Kwon S, An G (2003) Trangene structures in T-DNA-inserted rice plants. Plant Mol Biol 52:761–773
Kohli A, Twyman RM, Abranches R, Wegel E, Stoger E, Christou P (2003) Trangene integration, organization and interaction in plants. Plant Mol Biol 52:247–258
Kononov ME, Bassuner B, Gelvin SB (1997) Integration of T-DNA binary vector ‘backbone’ sequences into the tobacco genome: evidence for multiple complex patterns of integration. Plant J 11:945–957
Krizkova L, Hrouda M (1998) Direct repeats of T-DNA integrated in tobacco chromosome: characterization of junction regions. Plant J 16:673–680
Kumar S, Fladung M (2002) Transgene integration in aspen: structures of integration sites and mechanism of T-DNA integration. Plant J 31:543–551
Liu Y-G, Shirano Y, Fukaki H, Yanai Y, Tasaka M, Tabata S, Shibata D (1999) Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc Natl Acad Sci USA 96:6535–6540
Liu Y-G, Nagaki K, Fujita M, Kawaura K, Uozumi M, Ogihara Y (2000) Development of an efficient maintenance and screening system for large-insert genomic DNA libraries of hexaploid wheat in a transformation competent artificial chromosome. Plant J 23:687–696
Martineau B, Voelker TA, Sanders RA (1994) On defining T-DNA. Plant Cell 6:1032–1033
McCubbin AG, Zuniga C, Kao T-h (2000) Construction of a binary bacterial artificial chromosome library of Petunia inflata and the isolation of large genomic fragments linked to the self-incompatibility (S-) locus. Genome 43:820–826
Moullet O, Zhang H-B, Lagudah ES (1999) Construction and characterization of a large DNA insert library from the D genome of wheat. Theor Appl Genet 99:305–313
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Papp I, Iglesias VA, Moscone EA, Michalowski S, Spiker S, Park Y-D, Matzke MA, Matzke AJM (1996) Structural instability of a transgene locus in tobacco is associated with aneuploidy. Plant J 10:469–478
Song J, Bradeen JM, Naess SK, Helgeson JP, Jiang J (2003) BIBAC and TAC clones containing potato genomic DNA fragments larger than 100 kb are not stable in Agrobacterium. Theor Appl Genet 107:958–964
Tao Q, Zhang H-B (1998) Cloning and stable maintenance of DNA fragments over 300 kb in Escherichia coli with conventional plasmid-based vectors. Nucleic Acids Res 26:4901–4909
Ten Hoopen R, Montijn BM, Veuskens JTM, Oud O, Nanninga N (1999) The spatial localization of T-DNA insertions in petunia interphase nuclei: consequences for chromosome organization and transgene insertion sites. Chromosome Res 7:611–623
Turnbull K-M, Turner M, Mukai Y, Yamamoto M, Morell MK, Appels R, Rahman S (2003) The organization of genes tightly linked to the Ha locus in Aegilops tauschii, the D-genome donor to wheat. Genome 46:330–338
Windels P, Buck SD, Bockstaele EV, Loose MD, Depicker A (2003) T-DNA integration in Arabidopsis chromosomes. Presence and origin of filler DNA sequences. Plant Physiol 133:2061–2068
Wolters A-MA, Trindade LM, Jacobsen E, Visser RGF (1998) Fluorescence in situ hybridization on extended DNA fibres as a tool to analyse complex T-DNA loci in potato. Plant J 13:837–347
Yokoi S, Toriyama K (1999) Transgenic rice (Oryza sativa L.). In: Bajaj YPS (ed) Transgenic crops I (Biotechnology in agriculture and forestry), vol 46.. Springer-Verlag, Berlin Heidelberg, ppI.1/3–13
Acknowledgments
The authors are highly obliged to Drs. Yasunori Nakamura and Akiko Kubo of Akita Prefectural University for their useful technical advice, and to Drs. Kinya Toriyama and Fumitaka Abe of Tohoku University who kindly donated rice and Agrobacterium strains.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nakano, A., Suzuki, G., Yamamoto, M. et al. Rearrangements of large-insert T-DNAs in transgenic rice. Mol Genet Genomics 273, 123–129 (2005). https://doi.org/10.1007/s00438-005-1116-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-005-1116-y