Abstract
A novel Gram-stain-positive, obligately anaerobic, spore-forming rod, designated strain ChDC B114T, was isolated from a human dental plaque of a gingivitis lesion. The strain was characterized by polyphasic taxonomic analysis to identify it at the species level. The 16S ribosomal RNA gene (16S rDNA) sequence analysis revealed that the strain belongs to the genus Lachnoanaerobaculum. The percent similarity of the 16S rDNA of the strain was closest to the homologous gene sequence of Lachnoanaerobaculum orale N1T (98.5%) and Lachnoanaerobaculum saburreum CCUG 28089T (97.6%). The major fatty acids of strain ChDC B114T were C16:0 (30.7%), C14:0 (17.7%), iso-C19:0 (14.9%), and C17:0 2OH (12.0%). The draft genome of strain ChDC B114T was 3,097,953 bp in length. The G+C content of the strain was 35.9 mol %. Average nucleotide identity values between strain ChDC B114T and L. orale N1T and L. saburreum CCUG 28089T were 83.2% and 82.0%, respectively. Genome-to-genome distance values between strain ChDC B114T and L. orale N1T and L. saburreum CCUG 28089T were 26.8% (24.5–29.3%) and 26.30% (24.0–28.8%), respectively. Based on these results, strain ChDC B114T (= KCOM 2030T = JCM 33452T) should be classified as a novel species of genus Lachnoanaerobaculum, for which the name Lachnoanaerobaculum gingivalis sp. nov. is proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Lachnoanaerobaculum is Gram-stain-positive, obligately anaerobic, spore-forming rod and is composed of Lachnoanaerobaculum orale, Lachnoanaerobaculum saburreum (formerly, Eubacterium saburreum), and Lachnoanaerobaculum umeaense, which were isolated from the saliva of a healthy young man, human dental plaque, and the small intestinal biopsy from a child with coeliac disease, respectively [4]. E. saburreum was founded in the exposed pulp space of primary endodontic infections [13]. We isolated a strain, ChDC B114T, from the subgingival dental plaque of a gingivitis lesion of a female (43 years old) in 2001 in the Republic of Korea. According to polyphasic taxonomic characterization, strain ChDC B114T represents a novel species of the genus Lachnoanaerobaculum.
Materials and Methods
Bacterial Strain and Culture Conditions
Strain ChDC B114T was grown on tryptic soy agar (TSA; BD Difco Laboratories, Franklin Lakes, NJ, USA) plate supplemented with 0.5% yeast extract, 0.05% cysteine HCl-H2O, 0.5 mg/ml hemin, and 2 μg/ml vitamin K1 (TSA-YCHVk) at 37°C in an anaerobic chamber (Bactron I, Sheldon Manufacturing Inc., Cornelius, OR, USA) under 10% H2, 5% CO2, and 85% N2 [2]. The strain was cultured in Brucella broth (BD Difco Laboratories) at 37°C for 5 days in anaerobic conditions to enable detection of the spores.
Phylogenetic Analysis
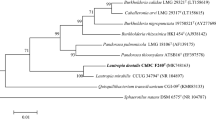
The 16S ribosomal RNA gene (16S rDNA) of strain ChDC B114T was cloned using PCR and the pGEM-T Easy Vector System and sequenced using the Sanger method as described previously [7]. The 16S rDNA sequences of the type strains of Lachnoanaerobaculum spp. were obtained from GenBank (Fig. 1, Supplementary Table S1). Multiple sequences were aligned using the CLUSTAL W algorithm, and sequence similarities were calculated using the MegAlign program (DNAStar Lasergene™ 8.0, DNAStar Inc., Madison, WI, USA) [2]. The evolutionary distance was calculated according to the Kimura two-parameter model [8], and phylogenetic trees were constructed using the neighbor-joining method [12] in MEGA 6.06 software [14]. The stability of the phylogenetic trees was assessed by the bootstrap analysis of 1000 replicates [3].
Neighbor-joining phylogenetic tree based on 16S ribosomal RNA gene (16S rDNA) sequences showing the relationships between strain ChDC B114T and type strains of closely related species. GenBank accession number of 16S rDNA of each type strain is written in parenthesis. The stability of the trees was assessed by the bootstrap analysis of 1000 replicates [3] using MEGA version 6.06 [14]. Bar indicates 0.02 changes per nucleotide position
Genome Sequence
Genomic DNAs of strain ChDC B114T and L. orale N1T were prepared using the phenol–chloroform extraction method as previously described [2].
Genomic DNAs of strain ChDC B114T and L. orale N1T were sequenced using the Illumina Hiseq 2500 platform by Macrogen Inc. (Seoul, Korea). In the case of strain ChDC B114T, three libraries of 350 bp paired-end, 5 kb mate-pair, and 8 kb mate-pair were constructed and sequenced which generated approximately 1848 Mb (596.7×) with 18,865,104 filtered subreads, 1188 Mb (383.4×) with 13,429,312 filtered subreads, and 841 Mb (271.6×) with 9,425,860 filtered subreads, respectively. In the case of L. orale N1T, two libraries of 350 bp paired-end and 5 kb mate-pair were constructed and sequenced which generated approximately 2401 Mb (857.1×) with 24,571,586 filtered subreads and 1270 Mb (453.4×) with 14,588,138 filtered subreads, respectively. The de novo assembly was performed by SPAdes (http://bioinf.spbau.ru/spades) [1]. All gaps among the scaffolds were filled by GapCloser (http://soap.genomics.org.cn/soapdenovo.html) [10]. Error correction was performed by Pilon (http://platanus.bio.titech.ac.jp/platanus-assembler) [6]. Genome annotation was conducted by the NCBI Prokaryotic Genome Annotation Pipeline [15] through the NCBI Genome Submission Portal (GenomeSubmit at http://ncbi.nlm.nih.gov). The GenBank accession numbers of the genome of strain ChDC B114T and L. orale N1T were RRCO00000000 and RRCM00000000, respectively.
Whole Genome Comparison Assay
Average nucleotide identity (ANI) and genome-to-genome distance (GGD) analyses were performed as previously described [9, 11]. Whole genome sequences of type strains for ANI and GGD analyses were downloaded from the GenBank database (https://www.ncbi.nlm.nih.gov/genome). The GenBank accession number of the whole genome sequence of L. saburreum CCUG 28089T was AEPW01000000.
Morphological and Physiological Characterization, Biochemical Analysis, and Chemotaxonomic Characteristics
Cell shape and size were investigated by scanning electron microscopy (SEM) as described previously [2]. The presence of spores was investigated using malachite green staining described previously [4].
The optimal growth conditions of strain ChDC B114T dependent on temperature, pH, and NaCl concentration were investigated as described previously [2]. Briefly, growth at different temperatures (25–45 °C at intervals of 5 and 37 °C) was determined on TSB-YCHVk agar medium for 3 days. Growth at various pHs (5–10 at intervals of 0.5) was assessed on TSB-YCHVk agar medium at 37 °C for 3 days. Growth at various NaCl concentrations was assessed on TP-YCHVk agar medium containing 0, 1, 2, or 3% (w/v) NaCl (pH 7 at 37°C) for 3 days.
API 32A and API 20A test strips (bioMerieux, Marcy-l’Etoile, France) were used to analyze the enzyme activities and sugar fermentation patterns of the strain ChDC B114T according to the manufacturer’s instructions [5].
The cellular fatty acid compositions of strain ChDC B114T were determined using the MIDI/Hewlett Packard Microbial Identification System (MIDI, Microbial ID, Newark, DE, USA) by the Korean Culture Center of Microorganisms (Seoul, Korea).
Results and Discussion
Phylogenetic analysis revealed that the strain ChDC B114T belonged to the genus Lachnoanaerobaculum (Fig. 1). The stability of the resulting tree was confirmed by the minimum evolution and maximum likelihood methods (data not shown). The 16S rDNA sequence of strain ChDC B114T was most closely related to Lachnoanaerobaculum orale N1T (98.5%) and Lachnoanaerobaculum saburreum CCUG 28089T (97.6%). The draft genome size of strain ChDC B114T was 3,097,953 bp, which was longer than those of the other Lachnoanaerobaculum spp., L. orale N1T (2,800,999 bp), L. saburreum CCUG 28089T (2,970,488 bp), and L. umeaense (2,705,257 bp). The DNA G+C content of ChDC B114T was 35.9 mol % which was derived from genome sequence. This was similar to values previously reported for Lachnoanaerobaculum species (35.0–37.8 mol %) [4]. ANI values between strain ChDC B114T and L. orale N1T and L. saburreum CCUG 28089T, and L. umeaense were 83.2% and 82.0%, respectively. GGD values between strain ChDC B114T and L. orale N1T and L. saburreum CCUG 28089T were 26.8% (24.5–29.3%) and 26.3% (24.0–28.8%), respectively. The 16S rDNA similarity between strain ChDC B114T and L. saburreum CCUG 28089T was higher than that between strain ChDC B114T and L. umeaense CD3:22T, although GGD value between strain ChDC B114T and L. saburreum CCUG 28089T was lower than that between strain ChDC B114T and L. umeaense CD3:22T. This was an unexpected result and the reason should be revealed in future studies. These results indicate that strain ChDC B114T represented a novel Lachnoanaerobaculum species.
The major cellular fatty acids of strain ChDC B114T were C16:0 (30.66%), C14:0 (17.74%), iso-C19:0 (14.94%), and C17:0 2OH (12.01%) (Table 1). C16:0 and C14:0 were the major cellular fatty acids of the analyzed strains of Lachnoanaerobaculum spp., whereas iso-C19:0 and C17:0 2OH were detected in strain ChDC B114T, but not detected in the other Lachnoanaerobaculum spp. (Table 1). In contrast, C18:1ω7c DMA and C18:1ω9c DMA were detected in the other Lachnoanaerobaculum spp. but not in strain ChDC B114T. Therefore, iso-C19:0 and C17:0 2OH are marker cellular fatty acids which discriminate the strain ChDC B114T from the other species. The major fatty acids of ChDC B114T were compared to those of type strains of Lachnoanaerobaculum spp. and are shown in Table 1.
In the API 32A system, strain ChDC B114T had positive reactions for indole production and enzyme activities of α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, and pyroglutamic acid arylamidase (Table 2, Supplementary Table S2). The strain was negative for the other biochemical reactions (Table 2, Supplementary Table S2). In API 20A system, strain ChDC B114T was positive for indole production and esculin hydrolysis but was negative for gelatin hydrolysis (Table 2 and Supplementary Table S3). The strain was able to produce acid from d-glucose, d-lactose, d-saccharose, d-maltose, d-xylose, glycerol, d-mannose, d-melezitose, d-raffinose, and d-trehalose but not from the others tested in this study (Table 2, Supplementary Table S3). The catalase reaction was negative for strain ChDC B114T. The discriminating characteristics of biochemical tests are summarized in Table 2.
Spores were detected in strain ChDC B114T in Brucella broth (BD Difco Laboratories) at 37°C for 5 days in anaerobic condition (Supplementary Fig S2).
According to the phylogenetic, morphological, physiological, and chemotaxonomical data, strain ChDC B114T represented a novel species of the genus Lachnoanaerobaculum, for which the name Lachnoanaerobaculum gingivalis is proposed.
Description of Lachnoanaerobaculum gingivalis sp. nov
Lachnoanaerobaculum gingivalis [gin.gi.val’.is. L. n. gingiva, gum; L. fem. suff. -alis, suffix denoting pertaining to; N.L. fem. adj. gingivalis, pertaining to the gums, gingival].
Colonies were circular shaped with undulate margin, shiny and transparent and spread to a diameter of approximately 1.26 ± 0.24 mm on TSA-YCHVk agar at 37°C after 2 days. Cells were Gram-stain-positive, obligately anaerobic, spore-forming, and long rod-shaped bacterium with a typical cell size of 10.32 ± 6.62 μm in length and 0.62 ± 0.07 μm in diameter. The temperature range for growth was 30–37 °C, and the optimal temperature was 35–37 °C. The pH range for growth was 5.5–9.5 (optimum 6.5). Growth occurred in the presence of 0–0.5% (w/v) NaCl with optimum growth at 0.5% NaCl. The type strain was positive for urease activity, indole formation, and esculin hydrolysis. It produced acid from d-glucose, d-lactose, d-saccharose, d-maltose, d-xylose, glycerol, d-mannose, d-melezitose, d-raffinose, and d-trehalose. Major cellular fatty acids of the type strain were C16:0, C14:0, iso-C19:0, and C17:0 2OH. The DNA G+C content of type strain was 35.9 mol %.
The type strain ChDC B114T (= KCOM 2030T = JCM 33452T) was isolated from the subgingival dental plaque of a gingivitis lesion from a female (43 years old) in 2001 in the Republic of Korea.
References
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Cho E, Park SN, Lim YK, Shin Y, Paek J, Hwang CH, Chang YH, Kook JK (2015) Fusobacterium hwasookii sp. nov., isolated from a human periodontitis lesion. Curr Microbiol 70:169–175. https://doi.org/10.1007/s00284-014-0692-7
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Hedberg ME, Moore ER, Svensson-Stadler L, Hörstedt P, Baranov V, Hernell O, Wai SN, Hammarström S, Hammarström ML (2012) Lachnoanaerobaculum gen. nov., a new genus in the Lachnospiraceae: characterization of Lachnoanaerobaculum umeaense gen. nov., sp. nov., isolated from the human small intestine, and Lachnoanaerobaculum orale sp. nov., isolated from saliva, and reclassification of Eubacterium saburreum (Prevot 1966) Holdeman and Moore 1970 as Lachnoanaerobaculum saburreum comb. nov. Int J Syst Evol Microbiol 62:2685–2690. https://doi.org/10.1099/ijs.0.033613-0
Jo E, Park SN, Lim YK, Paek J, Shin Y, Kim H, Kim SH, Shin JH, Chang YH, Kook JK (2018) Capnocytophaga endodontalis sp. nov., isolated from a human refractory periapical abscess. Curr Microbiol 75:420–425. https://doi.org/10.1007/s00284-017-1397-5
Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, Kohara Y, Fujiyama A, Hayashi T, Itoh T (2014) Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 24:1384–1395. https://doi.org/10.1101/gr.170720.113
Kim HS, Lee DS, Chang YH, Kim MJ, Koh S, Kim J, Seong JH, Song SK, Shin HS, Son JB, Jung MY, Park SN, Yoo SY, Cho KW, Kim DK, Moon S, Kim D, Choi Y, Kim BO, Jang HS, Kim CS, Kim C, Choe SJ, Kook JK (2010) Application of rpoB and zinc protease gene for use in molecular discrimination of Fusobacterium nucleatum subspecies. J Clin Microbiol 48:545–553. https://doi.org/10.1128/JCM.01631-09
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Lee I, Kim YO, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103. https://doi.org/10.1099/ijsem.0.000760
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. Erratum in: Gigascience (2015) 4:30
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. https://doi.org/10.1186/1471-2105-14-60
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sassone LM, Fidel RA, Faveri M, Figueiredo L, Fidel SR, Feres M (2012) A microbiological profile of unexposed and exposed pulp space of primary endodontic infections by checkerboard DNA-DNA hybridization. J Endod 38:889–893. https://doi.org/10.1016/j.joen.2012.03.021
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. https://doi.org/10.1093/nar/gkw569
Acknowledgments
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science and ICT (2017M3A9B8065844), in part by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2018R1A2B5002239), and in part the KRIBB Research Initiative Program funded by the Ministry of Science, ICT and Future Planning.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
DPD number: TA00879
GenBank accession number of the 16S rRNA gene for strain ChDC B114T: MK751703
GenBank accession number of the genome for strain ChDC B114T: RRCO00000000
Electronic supplementary material
Below is the link to the electronic supplementary material.
284_2019_1747_MOESM2_ESM.pptx
Supplementary Material 2 (PPTX 1611 kb). Supplementary Fig S1. Maximum likelihood (A) and the minimum evolution (B) phylogenetic tree based on 16S ribosomal RNA genes of strain ChDC B114T and type strains of closely related species. The GenBank accession number of 16S rDNA of each type strain is written in parenthesis. Stability of the phylogenetic trees was assessed by the bootstrap analysis of 1,000 replicates [3] with MEGA version 6.06 [14]. Bars indicate 0.02 (A) or 0.02 (B) changes per nucleotide position. Supplementary Fig S2. Light microscope image of strain ChDC B114T for detecting the presence of spores using malachite green staining method. The stain was cultured in Brucella broth at 37°C for five days in anaerobic condition. Original magnification: 1,000. Bar = 30 μm.
Rights and permissions
About this article
Cite this article
Lim, Y.K., Park, SN., Jo, E. et al. Lachnoanaerobaculum gingivalis sp. nov., Isolated from Human Subgingival Dental Plaque of a Gingivitis Lesion. Curr Microbiol 76, 1147–1151 (2019). https://doi.org/10.1007/s00284-019-01747-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01747-z