Abstract
The Asian Citrus Psyllid, Diaphorina citri Kuwayama (Hemiptera: Psyllidae), is an invasive insect pest that transmits Candidatus Liberibacter spp. This insect/pathogen system was first identified in North America in the early 2000’s and has become the top threat to the citrus industry. Limited options for management of this problem exist; therefore, innovative pest management strategies are being developed. In this study, we describe the first step toward a paratransgenic approach (also referred to symbiotic control) for control of the insect vector or the pathogen. Culturable bacteria from the gut of Asian Citrus Psyllids were identified using standard culture techniques followed by sequencing of the cultured microorganisms. Further, 454 pyrosequencing of the gut was performed to audit bacterial presence in order to begin to identify any relationship between psyllid symbionts and C. Liberibacter spp.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diaphorina citri Kuwayama (Hemiptera: Psyllidae), the Asian Citrus Psyllid, is a vector of the phloem-limited bacterium Candidatus Liberibacter asiaticus (Ca. Las). Ca. Las is considered to be the etiological agent of Huanglongbing (HLB), a devastating disease of citrus. D. citri have been shown to transmit Ca. Las in both nymphal and adult stages [18, 29] yet microorganisms are suspected to grow primarily in the adult stage [4, 42] especially in the salivary glands and alimentary canal [1]. Growth of Ca. Las in the adult stage, thus, increases the vector capacity of D. citri and concurrently, the transmission efficiency of Ca. Las.
Some aspects of the transmission efficiency of D. citri remain undefined. For example, when a microorganism is acquired by D. citri through feeding, such as Ca. Las, do indigenous microbiota affect the acceptance and/or growth of the newly acquired microorganism? Some evidence exists to support this question [14] with a strong positive correlation determined between Wolbachia and Ca. Las, for example. Several reports also exist on other host responses to the introduction of allochthonous microorganisms for both invertebrate and vertebrate organisms (i.e., [17, 38, 40]). The responses are variable and often complex (i.e., [31]); however, in some cases, they can be predicted by knowing the composition of the indigenous microbiota [34, 37]. This type of information would be useful for understanding the role of D. citri for Ca. Las transmission (i.e., [26]), and ultimately, HLB management.
The aim of the work was twofold: (1) to acquire culturable bacteria that can be used in studies to begin to understand more about the microbial ecology of D. citri particularly as it relates to Ca. Las establishment and transmission, and (2) to acquire potential candidate bacteria that can be used for paratransgenic control of Ca. Las and management of HLB disease. While numerous reports exist on bacteria that inhabit Ca. Las (i.e., [16, 24, 33, 35]), herein we report on a survey of D. citri for the presence of aerobic and anaerobic bacteria using both culture and culture-independent approaches. Our intent is to add to what is currently known about the internal microbial inhabitants of D. citri paying close attention to any variability that may exist with the gut community. While the work is basic in nature, our work directly relates to finding suitable microorganisms that could be applied in a paratransgenic strategy to control HLB.
Materials and Methods
Psyllids
Dead D. citri adults (N = 227) were shipped on ice, overnight from the USDA-ARS Horticultural Laboratory in Fort Pierce, Florida to California State University where they were processed the day of receipt, or kept at 0 °C until processed, generally within 24 h. D. citri were surface sterilized using a three-step chemical soak described in Lauzon et al. [20]. Whole abdomens or individual gut samples, fore-, mid-, and hindgut, were surveyed for bacterial inhabitants.
Culture Techniques
Abdomens and individual gut samples were individually and aseptically placed into pre-sterilized Reasoner’s 2 (R2) broth (Difco Laboratories, Detroit, MI) and incubated at 24 °C for 24 h or when turbidity was observed. Individual cultures were streaked for isolation and, isolated, pure colonies of aerobic bacteria were identified using standard biochemical and morphological tests (Bergey’s 2002) in companion with API analytical strips (BioMerieux, Durham, NC). Samples were also streaked directly onto pre-reduced anaerobically sterilized media (Hardy Diagnostics, Santa Maria, CA) and incubated in an anaerobe chamber held at 24°–26° for 48 h or when visible growth presented. Pure cultures were subsequently streaked onto R2 agar and incubated overnight under air to verify anaerobic requirement. All isolates were also identified using biochemical and molecular techniques, the latter described below.
Culture-Independent Techniques for Identification of Isolated Bacteria
Bacterial DNA was isolated from 18 h-old pure cultures from abdomens and individual gut samples using the DNeasy kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. DNA was quantified using a UV-spectrophotometer (Genesys 8, Thermo Scientific, Rochester, NY) and amplified using polymerase chain reaction under the following conditions:
Two sets of primers were used to amplify regions specific for almost all bacterial 16S sequences. A region of approximately 1,500 bp from the 16S rRNA gene was amplified using the primers B27F (5′AGAGTTTGATCMGGC TCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) [19]. Each 25-μL PCR mixture contained 10 ng DNA, 2X PCR Master Mix (Promega, Madison,WI), and primers. The amplification cycle consisted of an initial denaturation step of 2 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 55 °C and 1 min at 72 °C, and a final extension step of 7 min at 72 °C. Template DNA was omitted from the reaction mixture to serve as a negative control.
Five μL of each suspension was electrophoresed on 1 % agarose gels in 1X TBE buffer, which were then stained with ethidium bromide and examined under UV light for visualization of PCR products. Five µL of PCR products were subjected to an ExoSAP-IT kit (Affymetrix, Inc) at 37 °C for 15 min followed by 80 °C for 15 min, to cleave excess primers and inactivate free nucleotides. Cleaned PCR products were subsequently used as the template for sequencing.
DNA from all samples was subjected to a BigDye V3.1 Terminator Kit (Applied Biosystems, Foster City, CA), and the reactions were run in a PTC-200 Thermal Cycler (MJ Research, Waltham, MA). Conditions were initial denaturation at 96 °C for 1 min, followed by 25 cycles at 96 °C for 10 s, 50 °C for 5 s, 60 °C for 4 min. X-Terminator Mix (Applied Biosystems, CA, USA) was added, and DNA was sequenced using a 3130 Genetic Analyzer (Applied Biosystems, CA, USA).
Peak Scanner™ Software v1.0 (Applied Biosystem, CA, USA) was used to view the sequence peaks, and each sequence was run in a BLAST search limited to a bacterial database. Bacterial DNA from pure cultures was identified by examining the top-scoring sequences from the BLAST search results.
Culture-Independent Techniques for Determination of Bacterial Communities Using 454-Pyrosequencing
Bacterial DNA was recovered from D. citri and Bactericera cockerelli (another psyllid that is known to transmit C. Liberibacter spp.) using the Qiagen DNeasy Blood and Tissue Kit (Qiagen). B. cockerelli was used as an out-group in this study to determine the relative effect of C. Liberibacter spp. presence on the bacterial community within a host insect. This insect was included to see whether the presence of Ca. Liberibacter could have an effect on indigenous microbiota of more than just ACP. Insect DNA was particularly screened for C. Liberibacter spp. using conventional PCR protocol modified from Hung et al. [18]. This focus aimed to determine if any noticeable changes in the bacterial presence existed between C. Liberibacter spp.+ and C. Liberibacter spp.− pysllids. Twenty-five µL reactions included 12.5 µL Ampli Taq Gold 360 master mix (Applied Biosystems, Foster City, CA), 1.0 µL GC enhancer (Applied Biosystems, Foster City, CA), 1 µL of each Las-specific primer, 2 µL of extracted DNA, and 7.5 µL of nuclease-free water. PCR products were verified by agarose electrophoresis. Six samples of each species were chosen, three positive for C. Liberibacter spp. and three negative. These samples were subjected to sequencing of the bacterial 16 s small ribosomal subunit using Roche 454 amplicon pyrosequencing as described in Dowd et al. [11–13]. and Hail et al. [15]. Sequencing was performed at the Research and Testing Laboratories in Lubbock, Texas.
Bacterial sequencing data were analyzed using the QIIME Microbiome Pipeline [9]. Raw reads were screened for chimeric sequences and they were removed; remaining sequences were split by sample and the barcodes and primer sequences were removed. Operational taxonomic units (OTUs) were then chosen using the Uclust method, grouping 97 % similar sequences as one reprehensive sequence. Taxonomy was then assigned to OTUs using the ribosomal database project (RDP) 16 s database [41], and taxonomy summary plots were generated. Multiple rarefactions were used to calculate alpha diversity using the Cho1 Test, and a random sample size of 4,000 sequences was chosen. Beta diversity and Jackknifed Beta diversity were calculated from which weighted (copy number of OTU taken into account) and unweighted (copy number of OTU not taken into account) bootstrapped phylogenetic trees were generated [3, 27].
Results
Two hundred and twenty seven ACP reared previously on citrus were sampled on two different dates. Gram-positive bacteria isolated included members of Bacillales, Lysinibacillus sp., Paenibacillus sp., and Bacillus cereus, Staphylococcus saprophyticus, and, Streptomyces sp., a filamentous bacterium in the Actinomycetales. Gram-negative bacteria were identified as Enterobacteriales, Enterobacter spp., and Pantoea agglomerans, and Pseudomonales, Pseudomonas putida, Chryseomonas luteola, Alcaligenes xylosoxidans denitrificans, and Acromobacter sp. (Tables 1 and 2). The identities of pure isolates of these bacteria were confirmed using standard morphological and biochemical tests and culture-independent methods. No anaerobic bacteria were isolated from D. citri.
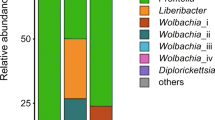
Sequencing using the 454 Pyrosequencing platform revealed the presence of the most populous bacterial genera in all ACP and B. cockerelli samples, respectively, as Wolbachia, Ralstonia, and members in the family Tremblayaceae, all of which are α-proteobacteria and β-proteobacteria. A Staphylococcus sp. was found in ACP in low numbers. Actinomycetales were not observed. Pseudomonales' one OTU was called to the Pseudomonas genus and was present in ACP. One member of the Enterobacteriales was found to be present in small numbers in ACP. In both weighted and unweighted phytogenetic analysis, insects that contained C. Liberibacter spp. had a marked impact on bacterial community analysis. More diverse microbial inhabitants were found in D. citri in the presence of Ca. Las. The amount of Wolbachia spp. in both D. citri and B. cockerelli is greater in insects infected with Ca. Las (Fig. 1).
Weighted and unweighted phylogenetic trees based on bacterial diversity of Candidatus Liberibacter infected and non-infected potato psyllid (Bactericera cockerelli) and Asian citrus psyllid (Diaphorina citri). Jackknifed trees are shown with branch distance values. Percentage of overall community composition in each sample is shown in histograms. Each color reflects an OTU, classifications are as specific as is available. Solid dots denote Liberibacter positive insects, and empty dots negative samples. Numbers in the circles denote the sample identity
Discussion
Diaphorina citri reared on citrus-harbored bacteria typically described as citrus endophytes [2, 5] and insect symbionts, such as P. agglomerans [10, 21, 22] and A. xylosoxidans denitrificans [6]. Our findings were similar to those reported by others [24, 30, 36]; however, our findings were biased due to the focus on obtaining culturable bacteria, and the limitations of basic DNA extraction, amplification, and sequencing based on 16 s rRNA. To get a more complete description of the microbiota of D. citri, Sagaram et al. [30], for example, found that PhyloChip analysis was more comprehensive in terms of microbial identifications than 16S rRNA gene clone library sequencing.
We focused our survey on culturable bacteria due to our parallel interest in finding suitable candidates for paratransgenic control of HLB. Two bacterial species, Alcaligenes xylosoxidans [6] and P. agglomerans [28, 39] have already been considered as candidates for paratransgenesis. Other microbial surveys of D. citri reported bacterial species, including intracellular [14, 24] and anaerobic bacteria [15], both groups often refractory to culture or use in the field. Our survey did not yield any anaerobic cultures which can be explained by the typical physiological state of many bacteria when removed from their habitats that make them difficult to culture [25, 32]. This may be true for other bacterial species, including aerobes and facultative anaerobes residing within D. citri. For this reason, paired studies (culture-based surveys and culture-independent surveys) are key in fully exploring bacterial communities.
As researchers begin to unravel the epidemiology of HLB disease, microbe–microbe interactions are also beginning to emerge as important areas of study and may provide opportunities for exploitation. Trivedi et al. [36] found that Ca. Las influences microbial communities in citrus and our findings are in line. Both D. citri and B. cockerelli displayed shifts in their gut microbiota in the presence of Ca. Las. For our purposes, shifts should be examined to assess the establishment and/or efficiency of an introduced bacterium (PTG agent) in the presence and absence of Ca. Las. In turn, microbes in D. citri may influence Ca. Las establishment and/or transmission. This notion is supported by the findings of Boissière et al. [7] whose findings suggested that mosquitoes harboring members of the Enterobacteriaceae are more likely to be infected by Plasmodium falciparum. Often, the focus of insect-symbiotic associations is on how the insect selects its symbionts; however, microorganisms also have a “choice” on their host [23] and influence each other in terms of growth and expression of virulence factors.
Host plant endophytic composition may be the first important factor in influencing Ca. Las transmission efficiency by indirectly seeding D. citri with bacteria that affect Ca. Las establishment and transmission. While speculative in nature, evidence does exist that support this notion (i.e., [7, 8, 30, 36]). If the case, the vector capacity and transmission efficiency for Ca. Las may be able to be manipulated artificially either using host plant endophytes or insect symbionts. An endophyte that cycles between and among D. citri and plants, such as that exhibited by Homalodisca coagulata, the glassy winged sharpshooter and A. xylosoxidans denitrificans [6], and that is associated with inhibition of Ca. Las would be ideal.
References
Ammar E-D, Shatters RG Jr, Lynch C, Hall DG (2011) Detection and relative titer of Candidatus Liberibacter asiasticus in the salivary glands and alimentary canal of Diaphorina citri (Hemiptera: Psyllidae) vector of citrus Huanglongbing disease. Annals Ent Soc Amer 104:526–533
Araújo WL, Marcon J, Maccheroni W, van Elsas JD, van Vuurde JWL, Azevedo JL (2002) Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 68:4906–4914
Arp AP, Chapman R, Crossland JM, Bextine B (2013) Low-level detection of Candidatus liberibacter Solanacearum in Bactericera cockerelli (Hemiptera: Triozidae) by 16S rRNA pyrosequencing. Environ Entomol 42:868–873. doi:10.1603/EN12260
Aubert B (1987) Epidemiological aspects of the greening (Huanglongbing) disease in Asia. Review and Abstract. 5. Workshop on citrus greening HLB disease. China
Azevedo JL, Maccheroni W, Pereira JO, de Araújo WL (2000) Endophytic microorganisms: a review on insect control and recent advances on tropical plants. Electron J Biotechnol 3:41–65
Bextine B, Lauzon C, Potter S, Lampe D, Miller TA (2004) Delivery of a genetically marked Alcaligenes sp. to the glassy winged sharpshooter for use in a paratransgenic strategy. Curr Microbiol 48:327–331
Boissière A, Tchioffo MT, Bachar D, Abate L, Marie A, Nsango SE, Shahbazkia HR, Awono-Ambene PH, Levashina EA, Christen R, Morlais I (2012) Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog 8:1–12
Buchman JL, Sengoda VG, Munyaneza JE (2011) Vector transmission efficiency of Liberibacter by Bactericera cockerelli (Hemiptera: Triozidae) in zebra chip potato disease: effects of psyllid life stage and inoculation access period. J Econ Entomol 104:1486–1495
Casporaso JG, Lauber CL, Walters WA, Berq-Lyons D, Lozupone CA, Turnbaugh PJ, Fieri N, Knight R (2010) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Nat Acad Sci 108:4516–4522
Dillon RJ, Vennard CT, Charnley AK (2002) Gut bacteria produce a locust cohesion pheromone. J Appl Microbiol 92:759–763
Dowd SE, Callaway TR, Wolcott RD, Sun Y, Mckeehan T, Hagevoort RG, Edrington TS (2008) Evaluation of the bacterial diversity in the feces of cattle using 16 S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8:125
Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA (2008) Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella infected pigs. Foodborne pathog dis 5:459–472
Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D (2008) Polymicrobial nature of chronic diabetic foot ulcer bioÞlm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 3:e3326
Fagen JR, Giongo A, Brown CT, Davis-Richardson AG, Gano KA, Triplett EW (2012) Characterization of the relative abundance of the citrus pathogen Ca. Liberibacter Asiasticus in the microbiome of its insect vector, Diaphorina citri, using high throughput 16S rRNA Sequencing. Open Microbiol J 6:29–33
Hail D, Dowd SE, Bextine BR (2012) Identification and location of symbionts associated with potato psyllid (Bactericera cockerelli) lifestages. Env Entomol 41:98–107
Hansen AK, Jeong G, Paine TD, Stouthamer R (2007) Frequency of secondary symbiont infection in an invasive psyllid relates to parasitism pressure on a geographic scale in California App. Environ Microbiol 73:7531–7535
Hao WL, Lee YK (2004) Microflora of the gastrointestinal tract: a review. Methods Mol Biol 268:491–502
Hung TH, Hung SC, Chem CN, Hsu MH, Su HJ (2004) Detection by PCR of Candidatus Liberibacter asiasticus, the bacterium causing citrus huanglongbing in vector psyllids: application to the study of vector-pathogen relationships. Plant Pathol 53:96–102
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Lauzon CR, Sjogren RE, MacCollom GB, Prokopy RJ (1998) Attraction of Rhagoletis pomonella (Diptera: Tephritidae) to odors of bacteria: apparent confinement to specialized members of enterobacteriaceae. Env Entomol 27:853–857
Lauzon CR (2003) Symbionts of tephritids. In: Bourtzis K, Miller TA (eds) Insect symbioses. CRC publication, Boca Raton, pp 115–130
Lauzon CR, McCombs SD, Potter SE (2009) Vertical passage of Pantoea (Enterobacter) agglomerans and Klebsiella pneumoniae in Ceratitis capitata Weidemann, the mediterranean fruit fly. Ann Entomol Soc Am 102(1):85–95
Mandrioli M (2009) Insect-symbiont: the key relationship to get in-depth insight on the host choice of bacteria. Info Syst J 6:98–101
Marutani-Hert M, Hunter WB, Morgan JK (2011) Associated bacteria of Asian citrus psyllid (Hemiptera: Psyllidae: Diaphorina citri). Southwest Entomol 36:322–330
Oliver JD (2005) The viable but nonculturable state in bacteria. J Microbiol 43:93–100
Pelz-Stelinski KS, Brlansky RH, Ebert TA, Rogers ME (2010) Transmission parameters for Candidatus Liberibacter asiaticus by Asian citrus psyllid (Hemiptera: Psyllidae). J Econ Entomol 103:1531–1541
Powell C, Hanson J, Bextine B (2014) Bacterial community survey of Solenopsis invicta Buren (red imported fire ant) colonies in the presence and absence of Solenopsis invicta virus (SINV). Curr Microbiol. doi:10.1007/s00284-014-0626-4
Riehle MA, Moreira CK, Lampe D, Lauzon C, Jacobs-Lorena M (2007) Using bacteria to express and display anti-Plasmodium molecules in the mosquito midgut. Int J Parasitol 37:595–603
Roistacher CN (1991) Techniques for biological detection of specific citrus graft transmissible diseases. FAO, Rome, pp 25–45
Sagaram US, DeAngelis KM, Trivedi P, Anderson GL, Lu S-E, Wang N (2009) Bacterial diversity analysis of huanglongbing pathogen-infected citrus using phylochip arrays and 16S rRNA gene clone library sequencing. Appl Environ Microbiol 75:1566–1574
Scanlan PD, Shanahan F, O’Mahony C, Marchesis JR (2006) Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol 44:3980–3988
Singh BK, Millard P, Whiteley AS, Murrell JC (2004) Unraveling rhizosphere-microbial interactions: opportunities and limitations. Trends Microbiol 12:386–393
Subandiyah S, Nikoh N, Tsuyumu S, Somowiyarjo S, Fukatsu T (2000) Complex endosymbiotic microbiota of the citrus psyllid Diaphorina citri (Homoptera: Psylloideae). Zool Sci 17:983–989
Tannock GW, Munro K, Harmsen HJ, Welling GW, Smart J, Gopal PK (2000) Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol 66:2578–2588
Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, Baumann P (2000) Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol 66:2898–2905
Trivedi P, Duan YP, Wang NA (2010) Huanglongbing, a systemic disease, restructures the bacterial community associated with citrus roots. Appl Environ Microbiol 76:3427–3436
Vanhoutte T, Huys G, Brandt E, Swings J (2004) Temporal stability analysis of the microbiota in human feces by denaturing gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol Ecol 48(3):437–446
Walter J (2008) The ecological role of lactobacilli in the gastrointestinal tract: Implications for fundamental and biomedical research. Appl Environ Microbiol. doi:10.1128/AEM.00753-08
Wang S, Ghosh AK, Bongio N, Stebbings KA, Lampe DJ, Jacobs-Lorena M (2012) Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. PNAS. doi:10.1073/pnas.1204158109
Wong CNA, Ng P, Douglas AE (2011) Low-diversity bacterial community in the gut of the fruit fly Drosophila melanogaster. Environ Microbiol. doi:10.1111/j.1462-2920.2011.02511.x
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Xu CF, Xia YH, Li KB, Ke C (1988) Further study of the transmission of citrus huanglongbing by a psyllid Diaphorina citri Kuwayama. In: Timmer LW, Garnsey SM, Navarro L (eds) Proceeding 10th Conference International Org. Citrus Virol. Nov. 17–21, 1986. pp 243–248
Acknowledgments
This work was supported in part by a California State University Center for Research Student Award. This manuscript does not contain clinical studies or patient data.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kolora, L.D., Powell, C.M., Hunter, W. et al. Internal Extracellular Bacteria of Diaphorina citri Kuwayama (Hemiptera: Psyllidae), the Asian Citrus Psyllid. Curr Microbiol 70, 710–715 (2015). https://doi.org/10.1007/s00284-015-0774-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-015-0774-1