Abstract
As an important insect vector, Asian citrus psyllid (ACP), Diaphorina citri Kuwayama (Hemiptera: Liviidae) transmits the pathogen ‘Candidatus Liberibacter asiaticus’ (CLas) that is associated with citrus greening also known as Huanglongbing (HLB) disease. The bacterial endosymbionts have a potential role in shaping the host range of insect herbivores and their performance on different host plants, which might affect the endosymbiont distribution in insect populations. Here, we detected and characterized Pantoea endosymbiont in nymph and adult ACP specimens collected from Citrus reticulata Blanco and Cordia myxa L. plants. The phylogenetic tree constructed using endosymbiotic bacteria 16S ribosomal RNA sequences indicated that Pantoea sp. was closely related to Mixta calida, sharing about 98% identity and was grouped with other Mixta and Pantoea endosymbionts. Our findings showed 100% and 92.3% infection of Pantoea in adults while 61.5% and 90% infection of Pantoea in nymphs collected from C. reticulata and C. myxa plants, respectively. Understanding the interaction of endosymbiotic bacteria with ACP associated with host plants could be useful for developing an effective management strategy for both ACP and HLB disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Asian citrus psyllid (ACP), Diaphorina citri Kuwayama (Hemiptera: Liviidae), is an economically important insect pest of citrus crop and vectors the pathogen ‘Candidatus Liberibacter asiaticus’ (CLas). CLas is a gram-negative, phloem-limited, fastidious bacterium, which is associated with the most threatening disease, citrus greening, also known as huanglongbing (HLB) disease. ACP and HLB have been spread in almost all citrus-growing regions worldwide. HLB has resulted in tremendous losses and the death of millions of trees worldwide [1]. There is no cure or effective control method for this disease [2].
The host range of ACP includes more than 25 species in the Rutaceae family [3, 4]. The population of ACP is well established on almost all citrus cultivars in Pakistan [5]. Besides Rutaceae, ACP may feed on a wide range of alternative host plants, which are important in allowing the insects to survive in the absence of suitable hosts. Recently, Cordia myxa L. (Family: Boraginaceae) has been reported as a new host for ACP in the south region of the state of Punjab, Pakistan [6]. However, bacterial endosymbionts associated with ACP feeding on new detected host plant have not been reported yet.
An understanding of the interactions between insects and symbionts can shed light on a new perspective for pest prevention and control technology. Symbiotic bacteria play different roles in insect biology which might affect insect growth and reproduction [7], surpass insects’ immune defenses [8], affect the tolerance to heat stress [9], alter resistance level in insect against parasitism [10], and amend the attractiveness of insects toward their host plants by changing in phloem composition [11]. Furthermore, the microbial symbionts associated with insect herbivores may allow their insect hosts to feed on different plant species [12, 13].
Generally, ACP harbors various endosymbiotic bacteria including Wolbachia, Arsenophonus spp., Candidatus Carsonella ruddii, mycetocyte symbiont, syncytium symbiont, Liberibacter sp., and Candidatus Profftella armatura [14, 15]. The genus Pantoea comprise of free-living and both pathogenic and non-pathogenic host-associating species [16]. The prevalence of different Pantoea isolates has been reported in a wide variety of insect species [17,18,19,20]. In most of them, the association has been termed as mutualistic, wherein the bacteria inhabit the insect intercellularly, and in few cases intracellularly within specialized cells of the host [21]. Recent studies related to Pantoea showed that while the bacteria contribute insect nutrition and digestion [20] insect gets to benefit from bacteria to the breakdown of toxic substances [22].
In this study, we identified for the first time Pantoea sp. associated with ACP feeding on two different host plants by molecular-phylogenetic sequence analysis based on 16S ribosomal RNA sequences. We provided data on the prevalence of endosymbiont, Pantoea with natural populations of ACP collected from C. reticulata (known host plants) and C. myxa (alternative new host plant). Our research objective was to assess whether there is a host-related differentiation in Pantoea harbored ACP nymphs and adult populations associated with C. reticulata and C. myxa plants.
Materials and Methods
Insect Samples
The nymphs and adult ACP samples were collected on 15 June 2018 from C. myxa plants at Multan (29° 35′ 20.3″ N 71° 10′ 06.5″ E) and additional location Muzaffargarh district (30° 03′ 11.5″ N 71° 10′ 16.1″ E); 472.3 m west of the initial site. The plants were about 3 to 4 years old, and the closest citrus orchards from both locations were 1.04–1.20 km northeast. For comparison, the ACP population was collected from C. reticulata orchard in Sahiwal (31° 58′ 48.5″ N 72° 18′ 37.5″ E) and Bhalwal (32° 16′ 34.7″ N 72° 55′ 32.8″ E) tehsils of Sargodha, most citrus growing district in Pakistan. The insect specimens were collected using aspirator and preserved in 96% ethanol. The preserved specimens in 1.5 ml tubes were analyzed in Molecular Entomology Laboratory, Department of Plant Protection, Ankara University, Turkey.
DNA Isolation
A total of 100 ACP individuals from two different host plants were used in this study. Only fifth nymphal instars and adults were used in molecular experiments, which were separated using morphological characters [23]. Genomic DNA was extracted from single nymph and adult of ACP. The whole insect samples were homogenized in lysis buffer (100 mM Tris, 50 mM EDTA, 1.4 M NaCl, 2% CTAB) using a sterile pestle and incubated at 65 °C for 12 h. DNA was extracted using chloroform-isoamyl alcohol (24:1) followed by isopropanol precipitation. The pellet was washed with 70% ethanol, centrifuged for 5 min at 7500 rpm, dried and eluted in sterile water [24]. DNA samples were electrophoresed in 1% agarose gel containing Pronasafe Nucleic Acid Staining Solution (Laboratorios, CONDA, S.A.), and gel images were obtained using GelCapture Software (DNR Bio-Imaging Systems, Jerusalem, Israel). The concentration of DNA samples was analyzed using the NanoDrop2000 spectrophotometer (Thermo Scientific, USA).
Polymerase Chain Reaction (PCR) Amplification and Sequence Analysis
PCRs were carried out in a 50-µl reaction volume using an equal nanogram of DNA template and GoTaq Flexi DNA polymerase (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Primers set for identification of Pantoea (5′-ACGGAGGGTGCAAGCGTTAAT-3′ as forward primer, 5′-AGGTAAGGTTCTTCGCGTTGCA-3′ as reverse primer) were designed using the sequences of the bacteria identified in ACP given in GenBank including the species Pantoea ananatis, P. cypripedii, P. agglomerans (GenBank accession numbers: KC153128.1; KC153127.1; KC153126.1). Pantoea sp. was tested for the presence of the bacteria with the expected product size of approximately 450 bp. A second primer set for mitochondrial cytochrome oxidase I (mtCOI) gene coding region, COI-F (5′-AGGAGGTGGAGACCCAATCT-3′) and COI-R (5′-TCAATTGGGGGAGAGTTTTG-3′) [25], was used to assess the quality of the template DNA with the expected product size of approximately 821 bp. Amplification conditions were as follows for Pantoea sp.; 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 90 s and a final step at 72 °C for 10 min. Amplification conditions for COI primers consisted of an initial denaturation at 94 °C for 3 min, followed by amplification for 35 cycles at 94 °C for 1 min, 53 °C for 30 s, 72 °C for 2 min, and a final step at 72 °C for 10 min. PCR products were visualized using gel electrophoresis on a 1.5% agarose gel containing Pronasafe Nucleic Acid Staining Solution (Laboratorios, Conda, S.A.), and gel images were obtained using GelCapture Software (DNR Bio-Imaging Systems, Jerusalem, Israel).
Pantoea-specific PCR products were purified with Wizard SV Gel and PCR Clean-up System (Promega) according to the manufacturer’s instructions. Sequencing reactions were carried out using CEQ 8800 Genetic Analysis System (Beckman Coulter). The sequenced rRNA sequence was submitted to BLAST against the NCBI 16S ribosomal RNA sequences (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and to the Ribosomal Database Project II server (https://rdp.cme.msu.edu). The amplified ACP Pantoea rRNA sequence was submitted to GenBank under accession number MN251864.1.
Phylogenetic Analysis
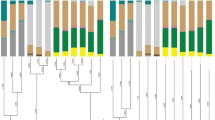
16S rRNA nucleotide sequences of Pantoea and different bacteria genus from ACP were collected from the NCBI database and aligned with MEGAX [26] suit using ClustalW [27]. The evolutionary history was inferred using the Neighbor-Joining method [28]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method [29] and are in the units of the number of base substitutions per site. All ambiguous positions were removed for each sequence pair (pairwise deletion option). Evolutionary analyses were conducted in MEGA X [26].
Results
In this study, an endosymbiotic bacterium belonging to ACP was identified as Pantoea sp. for the first time by sequencing and phylogenetic analysis based on 16S ribosomal RNA sequences. 16S rRNA BLAST analysis revealed that sequence of Pantoea 16S rRNA from ACP displayed a high sequence similarity among Pantoea and Mixta species with the highest possible scores including Mixta calida (homotypic synonym Pantoea calida) and M. gaviniae (homotypic synonym Pantoea gaviniae) (Table 1).
The Pantoea 16S rRNA gene sequence obtained from ACP was used for phylogenetic analyses. The phylogenetic tree was constructed using endosymbiotic bacteria showing highest identity to Pantoea 16S rRNA from ACP, wherein Syncytium placed in ingroup. Additionally, Wolbachia and C. Liberibacter asiaticus from D. citri were used as an outgroup. Figure 1 depicts the tree constructed by Neighbor-joining analysis. According to the phylogenetic tree, Pantoea sp. was closely related to M. calida, sharing an identity of 98% (on average). The phylogenetic tree grouped Pantoea sp. according to its expected classification within the Erwiniaceae family clade, and separated this node from the other endosymbionts of ACP. Three major clades were produced that included taxa belonging to the γ-, β-, and α-Proteobacteria, respectively, and the separation of each major eubacterial division was strongly supported by the bootstrap method. Pantoea and Mixta species were grouped in the γ-Proteobacteria in a clade. While Syncytium endosymbiont of ACP was grouped in β-Proteobacteria; Wolbachia endosymbiont of ACP and C. Liberibacter asiaticus were grouped in α-Proteobacteria.
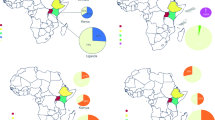
Pantoea primer set was used to detect the presence and prevalence of Pantoea infection in ACP by observing an approximately 450 bp PCR fragment. Among 100 adult and nymph ACP samples, 91 were found positive for Pantoea (91%). Out of 67 adult specimens, 65 were positive for Pantoea (97.01%), while 26 specimens out of 33 nymphs were found to be positive (78.8%) from four locations including Sahiwal, Bhalwal, Muzaffargarh, and Multan locations. As presented in Table 2, the numbers of adult ACP samples infected with Pantoea (95.3%) were higher than nymphal stages (78.8%) collected from three different locations.
Discussion
Pantoea species has been characterized by molecular phylogenetic approaches in a variety of insects such as bark beetles [30], fruit fly [31], flesh flies [32], wild mosquito [33], and termites [34]. The most abundant bacterial community genus across the different life stages of ACP samples collected from a navel orange orchard in China was detected as Profftella, Wolbachia, and Pantoea through Illumina MiSeq sequencing [35]. As far as we know, this is the first molecular phylogenetic analysis report of ACP harboring Pantoea associated with two different host plants in Pakistan. Proteobacteria was attributed to the most prevalent bacterial phylum across all many insects [33, 36, 37] due to insects actively recruiting Proteobacteria or due to proteobacterial taxa being more effective than other bacterial groups at invading and proliferating within new insect hosts [38]. As in many other insects, ACP microbiota was mainly dominated by species of the Proteobacteria phylum [35], our result also confirms the classification of Pantoea sp occurring in the ACP in γ-Proteobacteria class. It has been recently reported that Pantoea calida, Pantoea gaviniae, Pantoea theicola, and Pantoea intestinalis were moved to Mixta genus [39]. Placing of both Pantoea and Mixta genus belonging to Erwiniaceae family in the same clade underlined very high similarity of these two genera in the phylogenetic tree.
The present study concerns the presence of Pantoea endosymbiotic bacteria associated with the natural populations of ACP collected from C. reticulata and C. myxa plants. An investigation from both host plants revealed uniformity in the distribution frequency of Pantoea. We detected high prevalence with 91% of Pantoea infection rate among 100 adult and nymph ACP samples, which are consistent with 93.0% infection rate by P. agglomerans in blueberry maggot fly [20]. Furthermore, high-throughput sequencing of bacterial DNAs of gut microbiota of an invasive pest, Agrilus mali Matsumara (Coleoptera: Buprestidae) resulted in 98.8% Pantoea infection [40].
In this study, there was a significant difference detected in the infection rate of Pantoea in the nymphal and adult populations of ACP feeding on C. reticulata plants. However, no difference was found in terms of infection rate among different life stages collected from C. myxa plants that could be closely associated with the development and insect feeding habits, e.g., nymphs and adults feeding exclusively on the phloem from leaves of their host plants. A recent study reported that Pantoea was found to be more abundant during the nymph 2–5 stages among ACP samples collected from a navel orange orchard in China [35]. This study suggests that there may be a difference associated with the bacterial abundance depending on hosts and life stages. However, infection frequency by Pantoea between different instars of ACP required detailed investigation with more samples to make clear that it is more closely related to developmental stages.
Previously, a broad host plant range within the Rutaceae family has been reported for ACP [41, 42]. Development and oviposition of ACP are similar on almost all citrus cultivars and additionally on orange jasmine [43]. ACP also lays eggs on fig Ficus carica L. (Moraceae) and feeds on potatoes Solanum tuberosum L.; hackberry [44] and C. myxa, as well [6]. In comparison with two different host plants, 100% adults and 61.5% nymphs collected from C. reticulata were found to be infected with Pantoea. While the rate of Pantoea infection was 92.3% adults and 90% nymphs collected from C. myxa plants. Differences in leaf surface, wax composition, availability of sugars, and interactions with other bacterial species are notable parameters that can alter the bacterial community [45]. Furthermore, the concentrations of plant secondary compounds can also affect the bacterial composition in insect guts [46]. Further genome sequencing analysis will provide data on Pantoea metabolism and putative role in ACP physiology. Also understanding the interaction of ACP with Pantoea might be an important aspect of the integrated management of HLB disease.
The prevalence of Pantoea with symbiotic nature in insect hosts or inside the plants affected by arthropods pests originates the promising possibilities to counter the arthropods pests or pathogens by using the paratransgenesis technique. The genetically modified strains of Pantoea might have a greater chance to develop in the arthropod’s body compared to exogenous strains commonly used to kill the pest. Therefore, paratransgenesis might be also a promising tool for the management of ACP and HLB disease, as it has been identified as an antagonist of various plant pathogens including bacteria and fungi, which is related to the production of antibiotics or any other mechanisms. This technique could be used as a biocontrol agent, and being an environment-friendly procedure, which might be helpful in minimizing chemical control [47].
Conclusion
This study contributes to our understanding of the prevalence of Pantoea endosymbionts in a natural population of ACP from two host plants, C. reticulata and C. myxa. By using metagenomics libraries and genome sequencing, it is possible to identify reluctant microbes and mechanisms in order to develop evolutionary models explaining the changes undergone by endosymbiotic bacteria in their adaptation to the intracellular host environment. Vectoring of beneficial bacteria is an important concern that could replace the other methods for ACP and HLB control. Further study should be conducted to investigate whether this endosymbiotic bacterium offers any kind of physiological advantage to ACP and the fitness costs for ACP harboring this bacterium on different host plants.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Hall DG, Richardson ML, Ammar ED, Halbert SE (2013) Asian citrus psyllid, Diaphorina citri, vector of citrus Huanglongbing disease. Entomol Exp Appl 146:207–223. https://doi.org/10.1111/eea.12025
Chin EM, Mishchuk D, Bruce J, Cilia M, Coaker G, Davis C, et al. (2014) An interdisciplinary approach to combat HLB. Citrograph Magazine 2014:28–34. https://www.citrusresearch.org/citrograph-winter-2014
Paris TM, Croxton SD, Stansly PA, Allan SA (2015) Temporal response and attraction of Diaphorina citri to visual stimuli. Entomol Exp Appl 155:137–147. https://doi.org/10.1111/eea.12294
Hall DG, Hentz MG (2016) An evaluation of nine plant genotypes for rearing Asian citrus psyliid, Diaphorina citri (Hemiptera: Liviidae). Fla Entomol 99:471–480. https://doi.org/10.1653/024.099.0320
Fiaz M, Afzal M, Majeed MZ (2018) Influence of abiotic weather factors on population dynamics of Asian Citrus Psyllid, Diaphorina citri Kuwayama (Hemiptera: Psyllidae) in central Punjab. Pak J Agric Res 56(1):35–40
Arshad M, Ullah MI, Çağatay NS, Dikmen F, Abdullah A, Afzal M (2019) Cordia myxa L., a new host plant record for Asian citrus psyllid Diaphorina citri Kuwayama. Southwest Entomol 44(1):331–334. https://doi.org/10.3958/059.044.0137
Chen DQ, Montllor CB, Purcell AH (2000) Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol Exp Appl 95:315–323. https://doi.org/10.1046/j.1570-7458.2000.00670.x
De Souza DJ, Bezier A, Depoix D, Drezen JM, Lenoir A (2009) Blochmannia endosymbionts improve colony growth and immune defence in the ant Camponotus fellah. BMC Microbiol 9:29. https://doi.org/10.1186/1471-2180-9-29
Montllor CB, Maxmen A, Purcell AH (2002) Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27:189–195. https://doi.org/10.1046/j.1365-2311.2002.00393.x
Oliver KM, Campos J, Moran NA, Hunter MS (2008) Population dynamics of defensive symbionts in aphids. Proc Biol Sci 275:293–299. https://doi.org/10.1098/rspb.2007.1192
Meyer JM, Hoy MA (2008) Molecular survey of endosymbionts in Florida populations of Diaphorina citri (Hemiptera: Psyllidae) and its parasitoids Tamarixia radiata (Hymenoptera: Eulophidae) and Diaphorencyrtus aligarhensis (Hymenoptera: Encyrtidae). Fla Entomol 91:294–304. https://doi.org/10.1653/0015-4040(2008)91[294:MSOEIF]2.0.CO;2
Tsuchida T, Koga R, Fukatsu T (2004) Host plant specialization governed by facultative symbiont. Science 303:1989. https://doi.org/10.1126/science.1094611
Ferrari J, Scarborough CL, Godfray HCJ (2007) Genetic variation in the effect of a facultative symbiont on host-plant use by pea aphids. Oecologia 153:323–329. https://doi.org/10.1007/s00442-007-0730-2
Hoffmann M, Coy MR, Gibbard HNK, Pelz-Stelinski KS (2014) Wolbachia infection density in populations of the Asian citrus psyllid (Hemiptera: Liviidae). Environ Entomol 43:1215–1222. https://doi.org/10.1603/EN14193
Hall AA, Morrow JL, Fromont C, Steinbauer MJ, Taylor GS, Johnson SN, Cook JM, Riegler M (2016) Codivergence of the primary bacterial endosymbiont of psyllids versus host switches and replacement of their secondary bacterial endosymbionts. Environ Microbiol 18:2591–2603. https://doi.org/10.1111/1462-2920.13351
Nadarasah G, Stavrinides J (2014) Quantitative evaluation of the host-colonizing capabilities of the enteric bacterium Pantoea using plant and insect hosts. Microbiology 160:602–615. https://doi.org/10.1099/mic.0.073452-0
Roper M (2011) Pantoea stewartii subsp. stewartii: lessons learned from a xylem-dwelling pathogen of sweet corn. Mol Plant Pathol 12:628–637. https://doi.org/10.1111/j.1364-3703.2010.00698.x
Lauzon C, McCombs SD, Potter S, Peabody NC (2009) Establishment and vertical passage of Enterobacter (Pantoea) agglomerans and Klebsiella pneumoniae through all life stages of the Mediterranean Fruit Fly (Diptera: Tephritidae). Ann Entomol Soc Am 102:85–95. https://doi.org/10.1603/008.102.0109
Wells M, Gitaitis R, Sanders F (2002) Association of tobacco thrips, Frankliniella fusca (Thysanoptera: Thripidae) with two species of bacteria of the genus Pantoea. Ann Entomol Soc Am 95:719–723. https://doi.org/10.1603/0013-8746(2002)095[0719:AOTTFF]2.0.CO;2
Maccollom GB, Lauzon CR, Sjogren RE, Meyer WL, Olday F (2009) Association and attraction of blueberry maggot fly Curran (Diptera: Tephritidae) to Pantoea (Enterobacter) agglomerans. Environ Entomol 38:116–120. https://doi.org/10.1603/022.038.0114
Vorwerk S, Blaich R, Forneck A (2007) Pantoea sp.—an associated bacteria common in grape Phylloxera (Daktulosphaira vitifoliae Fitch). Acta Hortic 733:47–51
Sood P, Nath A (2002) Bacteria associated with Bactrocera sp. (Diptera: Tephritidae)-isolation and identification. Pest Manag Econ Zool 10:1–9
Tsai JH, Liu YH (2000) Biology of Diaphorina citri (Homoptera: Psyllidae) on four host plants. J Econ Entomol 96:1721–1725. https://doi.org/10.1603/0022-0493-93.6.1721
Guz N, Kocak E, Kilincer N (2013) Molecular phylogeny of Trissolcus species (Hymenoptera: Scelionidae). Biochemi Syst Ecol 48:85–91. https://doi.org/10.1016/j.bse.2012.12.010
Boykin LM, De Barro P, Hall DG, Hunter WB, McKenzie CL, Powell CA, Shatters RG (2012) Overview of worldwide diversity of Diaphorina citri Kuwayama mitochondrial cytochrome oxidase 1 haplotypes: two Old World lineages and a New World invasion. Bull Entomol Res 102(5):573–582. https://doi.org/10.1017/S0007485312000181
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Higgins DG, Sharp PM (1989) Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci 5:151–153. https://doi.org/10.1093/bioinformatics/5.2.151
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101:11030–11035. https://doi.org/10.1073/pnas.0404206101
Cardoza YJ, Vasanthakumar A, Suazo A, Raffa KF (2009) Survey and phylogenetic analysis of culturable microbes in the oral secretions of three bark beetle species. Entomol Exp Appl 131(2):138–147. https://doi.org/10.1111/j.1570-7458.2009.00844.x
Prabhakar CS, Sood P, Kapoor V, Kanwar SS, Mehta PK, Sharma PN (2009) Molecular and biochemical characterization of three bacterial symbionts of fruit fly, Bactrocera tau (Tephritidae: Diptera). J Gen Appl Microbiol 55(6):479–487. https://doi.org/10.2323/jgam.55.479
Gupta AK, Rastogi G, Nayduch D, Sawant SS, Bhonde RR, Shouche YS (2014) Molecular phylogenetic profiling of gut-associated bacteria in larvae and adults of flesh flies. Med Vet Entomol 28(4):345–354. https://doi.org/10.1111/mve.12054
Moro CV, Tran FH, Raharimalala FN, Ravelonandro P, Mavingui P (2013) Diversity of culturable bacteria including Pantoea in wild mosquito Aedes albopictus. BMC Microbiol 13(1):70. https://doi.org/10.1186/1471-2180-13-70
Palmer M, De Maayer P, Poulsen M, Steenkamp ET, Van Zyl E, Coutinho TA, Venter SN (2016) Draft genome sequences of Pantoea agglomerans and Pantoea vagans isolates associated with termites. Stand Genomic Sci 11(1):23. https://doi.org/10.1186/s40793-016-0144-z
Meng L, Cheng X, Zhang H (2019) 16S rRNA gene sequencing reveals a shift in the microbiota of Diaphorina citri during the psyllid life cycle. Front Microbiol 10:1948. https://doi.org/10.3389/fmicb.2019.01948
Suen G, Scott JJ, Aylward FO, Adams SM, Tringe SG et al (2010) An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet 6(9):e1001129. https://doi.org/10.1371/journal.pgen.1001129
Ravenscraft A, Berry M, Hammer T, Peay K, Boggs C (2019) Structure and function of the bacterial and fungal gut microbiota of Neotropical butterflies. Ecol Monogr 89(2):e01346. https://doi.org/10.1002/ecm.1346
Jones RT, Sanchez LG, Fierer N (2013) A cross-taxon analysis of insect-associated bacterial diversity. PLoS ONE 8(4):e61218. https://doi.org/10.1371/journal.pone.0061218
Palmer M, Steenkamp ET, Coetzee MPA, Avontuur JR, Chan WY, Van Zyl E et al (2018) Mixta gen. nov., a new genus in the Erwiniaceae. Int J Syst Evol Microbiol 68:1396–1407. https://doi.org/10.1099/ijsem.0.002540
Bozorov TA, Rasulov BA, Zhang D (2019) Characterization of the gut microbiota of invasive Agrilus mali Matsumara (Coleoptera: Buprestidae) using high-throughput sequencing: uncovering plant cell-wall degrading bacteria. Sci Rep 9(1):4923. https://doi.org/10.1038/s41598-019-41368-x
Halbert SE, Manjunath KL (2004) Asian citrus psyllids (Sternorryncha: Psyllidae) and greening disease of citrus: A literature review and assessment of risk in Florida. Fla Entomol 87(3):330–353. https://doi.org/10.1653/0015-4040(2004)087[0330:ACPSPA]2.0.CO;2
Yang Y, Huang M, Beattie GAC, Xia Y, Ouyang G, Xiong J (2006) Distribution, biology, ecology, and control of the psyllid Diaphorina citri Kuwayama, a major pest of citrus: a status report for China. Int J Pest Manage 2:343–435. https://doi.org/10.1080/09670870600872994
Nava DE, Torres ML, Rodrigues MLD, Bento MS, Parra JRP (2007) Biology of Diaphorina citri (Hem., Psyllidae) on different host plants at different temperatures. J Appl Entomol 131:709–715. https://doi.org/10.1111/j.1439-0418.2007.01230.x
Thomas DB, De Leon JH (2011) Is the old world fig, Ficus carica L. (Moraceae), an alternative host for the asian citrus psyllid, Diaphorina citri (Kuwayama) (Homoptera: Psyllidae)? Fla Entomol 94:1081–1083. https://doi.org/10.1653/024.094.0455
Lindow SE, Brandl MT (2003) Microbiology of the phyllosphere. Appl Environ Microbiol 69(4):1875–1883. https://doi.org/10.1128/aem.69.4.1875-1883.2003
Mason CJ, Rubert-Nason KF, Lindroth RL, Raffa KF (2015) Aspen defense chemicals influence midgut bacterial community composition of gypsy moth. J Chem Ecol 41(1):75–84. https://doi.org/10.1007/s10886-014-0530-1
Morales H, Sanchis V, Usall J, Ramos AJ, Marín S (2008) Effect of biocontrol agents Candida sake and Pantoea agglomerans on Penicillium expansum growth and patulin accumulation in apples. Int J Food Microbiol 122(1–2):61–67. https://doi.org/10.1016/j.ijfoodmicro.2007.11.056
Acknowledgements
We would like to express our sincere thanks to Türkiye Scholarships for supporting Muhammad Arshad during his research in Turkey.
Author information
Authors and Affiliations
Contributions
Muhammad Arshad contributed in conceptualization, methodology, investigation, original draft, data curation, and analysis; Nurper Guz contributed in conceptualization, methodology, supervision, editing, and molecular analysis; Naciye S. Cagatay contributed in DNA isolation, PCR, and editing; Asli Dageri contributed in conceptualization, methodology, supervision, writing, and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Guz, N., Arshad, M., Cagatay, N.S. et al. High Prevalence of Pantoea in Diaphorina citri (Hemiptera: Liviidae): Vector of Citrus Huanglongbing Disease. Curr Microbiol 77, 1525–1531 (2020). https://doi.org/10.1007/s00284-020-01969-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01969-6