Abstract
Purpose
Toxicity of 6-Mercaptopurine (6MP) is related to single nucleotide polymorphism (SNP) in genes coding for metabolizing enzymes, with TPMT analysis being recommended prior to maintenance therapy. However, ITPA and NUDT15 polymorphisms appear more important in the Asian population.
Method
In this study 63 consecutive patients with ALL, entering maintenance phase of therapy, were evaluated for TPMT, ITPA and NUDT15 polymorphisms by PCR RFLP and confirmed by sequencing. Hematological and hepatic toxicities were monitored for 36 weeks. The groups with and without any of the three studied polymorphisms (Risk SNP + and Risk SNP-) were compared.
Results
Eighteen (28.6%) patients had major polymorphisms, 17 being heterozygous. ITPA(198CA): 11(17.5%); NUDT (415CT): 6(9.5%) and TPMT*3C: in 2(3.1%). Mean cumulative dose of 6MP was lower: 10927 mg/m2 in group with one of the polymorphisms compared to 12533 mg/m2 in the group without a polymorphism (p = 0.009). The group with Risk SNP + tolerated lesser weeks of full-dose 6MP chemotherapy (20.81 vs 30.40 weeks; p = 0.001). Risk of neutropenia > 3 weeks was pronounced in Risk SNP + group. The individual TPMT, ITPA and NUDT15 polymorphism subgroups had similar cumulative 6MP dose and chemotherapy interruptions. There was no difference in the average cumulative dose of methotrexate in the two groups. No significant hepatotoxicity was noted.
Conclusion
Polymorphisms in ITPA and NUDT15 have a greater prevalence in the north Indian population. Patients with these SNPs tolerate lower doses of 6MP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thiopurine drugs like 6-Mercaptopurine (6MP) are the cornerstone of maintenance therapy in acute lymphoblastic leukaemia (ALL). However being cytotoxic, these drugs are associated with haematological and hepatic toxicities requiring dose modifications and treatment interruptions [1].
Germ line genetic variations determine the inter-individual variability in drug dosing, tolerance, side effect profile and susceptibility of therapeutics commonly used in ALL [2, 3]. Pharmacogenomics when applied in childhood ALL have implicated toxicity of 6MP to single nucleotide polymorphisms (SNP’s) in genes encoding 6MP metabolizing enzymes with thiopurine methyltransferase (TPMT) being the most researched polymorphism [4]. TPMT is inherited as an autosomal co-dominant trait. Individuals homozygous for alleles are TPMT deficient while heterozygotes have intermediate enzyme activity, leading to unexpected myelosuppression when patients are treated with recommended doses of 6MP [4]. TPMT*3A SNPs have been observed to have a greater prevalence in the Caucasian population [5] with their frequencies being less than 5% in populations of Asian ethnicity [6, 7]. The application of TPMT pre-emptive testing in patients with ALL as recommended in European and North American population is conceivably of limited clinical utility in population with Asian ancestry. Recently, SNPs of inosine triphosphate pyrophosphatase (ITPA198 CA) and nucleoside diphosphate linked moiety-type motif 15 (NUDT15415 CT) have been linked with myelosuppression and possibly are one of the unidentified reasons of high toxicity despite the low prevalence of polymorphism in TPMT genes in the Asian population [8,9,10].
Extrapolating pharmacogenomics into clinical decision making may prove to be a cost-effective way of pre-emptively genotyping all patients who need the drug. Guidelines have already been framed for pre-emptive testing of TPMT genotype and their clinical utility in tailoring 6MP doses at start of maintenance therapy in patients with ALL, which will minimize the therapeutic interruptions and subsequent chances of relapse with a resultant influence on the outcome of childhood ALL [5].
This study was performed to evaluate the prevalence of TPMT, ITPA and NUDT15 gene polymorphism in children with ALL in a tertiary care hospital in north India and assess the relationship of these polymorphisms with the doses and subsequently incurred toxicities of 6MP.
Materials and methods
This was aprospective Mendelian randomisation study conducted in a tertiary care hospital in north India from 01 January 2016 to 30 April 2017.
Patients and treatment
All children treated for ALL, entering the maintenance therapy phase of the Indian Childhood Collaborative Leukemia Group 2015 (ICICLE) protocol (the protocol for ALL being followed in the unit) from 01 January 2016 to 30 April 2017, were consecutively enrolled in the study. The patients who were CNS positive and being treated for a relapse were excluded. Informed consent was obtained from all individual participants (or their guardians) included in the study.
As per protocol, maintenance phase includes daily 6MP, weekly oral methotrexate (MTX) along with an intrathecal MTX at 3-monthly intervals. Vincristine and dexamethasone pulses are not given during maintenance therapy. All children were started at a 6MP dose of approximately 60 mg/m2/day and MTX dose of 20 mg/m2/week (adjusted to the tablet strength, liquid preparation not being available in the country). A blood sample for genotyping for TPMT, ITPA and NUDT15 was drawn prior to starting maintenance therapy. The enrolled patients were followed with a complete blood count done every 2 weeks and liver function tests done monthly. Side effect profile included; (1) myelosuppression: absolute neutrophil count (ANC) and platelet count, (2) occurrence of febrile neutropenia, (3) hepatotoxicity, (4) cumulative dose of 6MP and (5) duration of treatment interruptions; were recorded for 36 weeks.
Dose adjustment, if any, was done as per routine in the pediatric oncology clinic to maintain an ANC between 750–1500/cumm. The patients, treating clinicians and observers were blinded to the results of gene polymorphism.
Definitions
Myelosuppression ANC < 750/cumm, and/or thrombocytopenia (platelets count < 75,000/cumm).
Hepatotoxicity An increase of the liver enzyme alanine transferase (ALT) to at least 5 times(> 44 U/L) and /or rise of serum bilirubin beyond 3 times the upper limit of normal.
Febrile Neutropenia (FN) Temperature higher than 38 °C with ANC < 500/cumm or ANC showing a falling trend to reach < 500/cumm.
Genotyping
Approximately 2 ml of peripheral blood was collected from study participants in tubes containing sodium EDTA. DNA was extracted using the QIAmp DNA blood kit (Qiagen Inc.) as per the manufacturer’s instructions.
-
(i)
TPMT: Three sites of known TPMT gene polymorphisms[c.238 G > C(TPMT*2), c.460 G > A(TPMT*3A & 3B), and c.719A > G (TPMT*3A & *3C)] causing TPMT deficiency were determined according to the method described by Yates [11]. Briefly, allele-specific PCR amplification was used to detect the c.238 G > C transversion in exon 5 using primer P2W& P2C for wild-type and P2M & P2C for mutant variant (Supplementary table S1). PCR amplification and restriction enzyme digestion (PCR-RFLP) were used to detect the c.460 G > A and c.719 A > GSNPs in exon 7 and 10 respectively, using the primers listed in Supplementary table I. RFLP for the variants c.460 G > A and c.719 A > G were detected using enzymes MwoI (Thermo Fisher Sci.) and ACCI(Thermo Fisher Sci.), respectively. The condition of RFLP enzymes is detailed in Supplementary table S2.
-
(ii)
ITPA: The c.198 CA (P32T)SNP was genotyped as described previously [12]. Briefly, the genomic DNA was amplified using the primers for exon 2 and 3 (Supplementary table S1). For the 198 CA SNP, the 328-bp product was digested with enzyme NspI(Thermo Fisher Sci.).
-
(iii)
NUDT15: The c.415 CT (R139C) SNP was genotyped by amplifying genomic DNA using the primers shown in Supplementary table S1. The endonuclease TaaI (Thermo Fisher Sci.) was used to cut the mutant C > T allele (Supplementary table S2).
The SNP positive cases were further confirmed by Sanger’s sequencing using ABI 3130 genetic analyzer (Applied Biosystems) and classified as heterozygous or homozygous.
Author SK recorded the doses received, treatment interruptions and other morbidities in the cohort under study. The study was approved by the hospital ethics committee and departmental review board. A written consent was obtained from parents or guardians.
Statistical analysis
Statistical analysis was performed using IBM SPSS software version 21.0. Allelic and genotypic frequencies, disequilibrium coefficients and the associated standard error for co-dominant traits were calculated. Hardy Weinberg equilibrium (HWE) was calculated for each polymorphism studied. The association between 6MP dose along with adverse effects and gene polymorphisms was tested using Fisher’s exact test for comparison between groups and ANOVA for comparison between the subgroups. Odds ratios and 95% confidence intervals were calculated. P value < 0.05 was considered as statistically significant.
Results
A total of 67 children with ALL entering the maintenance phase of therapy were enrolled. Four children were excluded (as per criteria) and 63 underwent analysis for gene polymorphisms. Fifty-eight children had a complete follow-up for a period of 36 weeks of maintenance chemotherapy (Fig. 1).
Prevalence of studied gene polymorphisms
Sixty-three children underwent genotyping. Nineteen SNPs were identified in 18(28.5%) children. ITPA(198CA) polymorphism was, seen in 11(17.5%) cases, with 10 being heterozygous and one having homozygosity. NUDT15(415CT) polymorphism was noted in 6 (9.5%) children, all being heterozygous. TPMT polymorphism was seen in only two cases (3.1%). Both TPMT polymorphisms were TPMT*3C(c.719A > G) and were heterozygous. One case had co-existing ITPA and TPMT*3C SNP’s (Figs. 2, 3).
Screening of genetic variants in TPMT, ITPA and NUDT15 in representative patient samples (Lane 1–4, M-50 bp marker). a Allele-specific PCR showing wild-type TPMT*2 gene (exon 5); b PCR (upper panel) followed by restriction digestion (lower panel) using MwoI showing wild-type TPMT*3B (exon 7); c Wild-type TPMT*3C (exon 10) variants in different patient samples following PCR-RFLP using enzyme AccI; d Restriction digestion using NspI of PCR product of ITPA (exon 2,3) showing homozygous mutation at c.198 CA (P32T) in lane 2; e Restriction digestion using TaaI of PCR product of NUDT15 (exon 1) showing heterozygous mutation at c.415 CT (R139C) in lane 2
Hardy Weinberg equilibrium analysis was applied for the three polymorphisms to ensure that the study sample did not violate the assumption of HWE and target population for current study was in HWE at all three loci p value [TPMT (p = 0.89), ITPA (p = 0.53) and NUDT (p = 0.69)].
The 58 patients who completed the study were divided into two groups for analysis (Risk SNP + and Risk SNP-). Risk SNP+: group with any of the three studied gene polymorphism being positive (n = 16); Risk SNP-group: group with all the three studied gene polymorphism being negative (n = 42) (Fig. 1).
Comparison of two groups (Risk SNP + and Risk SNP −)
The baseline characters in the two groups including the demographic characters, along with the mean 6MP and MTX dose at start of maintenance were comparable (Table 1).
Cumulative doses of 6MP
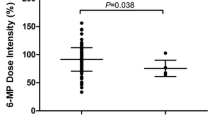
The mean cumulative dose of 6MP was 10,927 mg/m2 (9857–11,997), equivalent to 43 mg/m2/day in the Risk SNP + group and 12,533 mg/m2 (11,874–13,191) equivalent to 50 mg/m2/day in the Risk SNP- group (P = 0.011) (Fig. 4). The mean cumulative dose of 6MP in the three polymorphisms identified was (1) NUDT(415CT) (n = 6): 10,690 mg/m2 (7722–13,658), (2) ITPA(198CA) (n = 10) : 10,840 mg/m2(9569–12,211) and (3) TPMT*3C (n = 2) 10,962 mg/m2 (8946–12,978), respectively, (Supplementary table S3). We analyzed cumulative doses of 6 MP per pair of loci. It was observed that, combined NUDT + ITPA SNP group had significantly lower tolerated dose (10790 mg/m2) (p = 0.01), while other groups like NUDT + TPMT, ITPA + TPMT had tolerated 11,072 mg/m2 and 11,035 mg/m2, respectively (Supplementary table S3).The mean cumulative dose of MTX was higher in the Risk SNP-group 480 mg/m2 (451–510) compared to Risk SNP + group 425 mg/m2 (377–473) (p = 0.051).
Chemotherapy interruptions
Children with polymorphisms tolerated significantly fewer mean weeks of full-dose 6MP therapy (20.81 vs 30.40 weeks; p = 0.001). The Risk SNP + group tolerated full-dose chemotherapy for 57% of the time as compared to Risk SNP- group who received full dose for 85% of time in the 36-week follow-up period of the study. On individual SNP analysis, patients with ITPA(198CA) tolerated full dose of the drug for 58% of the time and those with NUDT(415CT) polymorphism tolerated the drug for 47% time (Supplementary table S3). The Risk SNP + group had significantly higher mean weeks of 6MP therapy interruptions (4.75 vs 2.79 weeks; p = 0.016) compared to the Risk SNP- group (Table 2).
The Risk SNP + group had significantly higher mean weeks of neutropenia (4.63 vs 2.69 weeks; p = 0.024). The risk of prolonged neutropenia (> 3 weeks) was pronounced in children with polymorphisms with an odds ratio of 6. FN was encountered more frequently (62.5% vs 47.6%) in Risk SNP + group as compared to the Risk SNP- group. However, there was no difference in the mean weeks of FN and thrombocytopenia between the two groups (p = 0.1) (Table 2).
Other side effects Hepatotoxicity was evident by increased transaminases in 18.9% of our cohort with the median alanine transferase (ALT) in the cohort being 66.5 IU/L (17–712 IU/L). Serum bilirubin was abnormal in only three cases. However, no case required maintenance therapy to be paused as a result of hepatotoxicity.
Discussion
Inter–patient variability in the doses of 6MP and MTX required to maintain the TLC and ANC in the desired range makes the maintenance phase of therapy in ALL a challenging exercise. It is estimated that 20–25% of children who are treated for a malignancy experience severe drug-related adverse effects [13]. Research in heritable genetic variations in drug metabolism are giving rise to precision medicine/individualized therapy and currently thiopurines are the only drug where there is sufficient evidence for pharmacogenomics to be incorporated into clinical practice with pre-emptive testing for the TPMT polymorphism being recommended to avoid serious adverse effects [5].
Polymorphisms in the TPMT, NUDT 15 and ITPA enzymes which metabolize 6MP have been associated with a decreased tolerance to 6MP and these polymorphisms have been noted to have a varying prevalence across different ethnic populations across the globe. TPMT(*2, *3A and *3C) is to date the most widely studied SNP for 6MP metabolism. The frequency and pattern of various TPMT alleles vary with ethnicity. The incidence is around 10–15% in the Caucasian population (commonly TPMT*3A) where currently pre-emptive testing is recommended, with the homozygous variants tolerating only 10% of the recommended doses while the heterozygotes have been observed to be having up to four times risk of neutropenia necessitating a 50% decrease in dose [5]. However, in people of south east Asian ethnicity, literature uniformly reports that the TPMT allele(commonly TPMT*3C) accounts for less than 5% prevalence, thereby having a limited role in 6MP toxicity. In the data published from the Indian sub-continent, TPMT polymorphisms was seen in < 5% of patients, with TPMT *3C being the predominant allele [14,15,16].
ITPA & NUDT15 polymorphisms are the other enzymes involved in the metabolism of 6MP. Sanitization of the cellular nucleotide pools from mutagenic base analogues is necessary and ITPA/NUDT help by removing oxidised deoxyneucleoside triphosphates (dNTP’s) responsible for 6MP toxicity [17]. South east Asia reports prominence of ITPA polymorphism. The ITPA94CA & 198CA are the most relevant and widely studied polymorphisms. In south-east Asia, quoted ITPA polymorphism prevalence ranges from 20 to 40%. A prevalence of 11–22% has been reported in patients of Indian origin [18]. This polymorphism has been seen in a bare 2% population of Hispanic and European origin, with a 3–5% prevalence reported from South America. Of late, NUDT15 polymorphisms have also been identified to be associated with intolerance to 6MP. NUDT15(415CT) frequency varied from 15% in patients with Thai ancestry to 23% in the Korean population [19]. The prevalence of NUDT15 polymorphisms is reported to be less than 1% in Europe. Studies from the Middle East and Central America also report a frequency of 1–3% while South America has reported 9% prevalence. Most of these SNPs were heterozygous barring a few homozygous mutations which were seen in less than 5% of cases. In our cohort the prevalence of TPMT*3C (c.719A > G), ITPA (198 CA) and NUDT15 (415 CT) was observed to be 3.1%, 17.5% and 9.5%. All polymorphisms except one were heterozygous. This distribution is similar to what has been observed from other south east Asian countries.
Germ line genetic variations are important determinants of inter-patient variability in susceptibility, drug response and toxicities of ALL therapy and affect survival and relapse rates [5]. The level of erythrocyte-6TGN is responsible for the tolerated doses and subsequent side effect profile [20]. In our study the mean cumulative dose of 6MP which was tolerated was significantly lower in cases with a detectable polymorphism as compared to wild type (10,927 mg/m2equivalent to 43 mg/m2/day vs 12,533 mg/m2 equivalent to 50 mg/m2/day). Independently, the mean cumulative dose of 6MP in children with the ITPA and NUDT15 polymorphism was lower, though not statistically significant. On analysing various SNP group combinations, NUDT + ITPA was found to tolerate even lower doses of 6MP compared to risk SNP + group. There is conclusive evidence of the relationship of the dose of 6MP and toxicity in individuals with the NUDT415CT variant allele with a number of studies, done mostly in Asian countries, establishing these SNP’s as significant in the Asian population [3, 8, 10, 21]. There are conflicting reports regarding the effect of ITPA198CA SNP on 6MP toxicity (hepatic & myelosuppression) implying the need for more research on the toxicity profile of this allele with the dose of 6MP [10, 18, 22, 23]. We have observed patients with this allele to receive lower mean doses (10,840 mg/m2 vs 12,533 mg/m2; p = 0.05) of 6MP. No difference was observed with MTX dosing, confirming that the 6MP metabolizing enzymes contribute to differential tolerated dose.
Pauses in therapy during the maintenance phase in ALL have an impact on the outcome of therapy. In our study, patients with any of the polymorphisms tolerated significantly fewer weeks of full-dose 6MP chemotherapy (20.81 vs 30.40 weeks; p = 0.001) and had significantly higher mean weeks of chemotherapy interruptions (4.75 vs 2.29 weeks; p = 0.01). Prolonged neutropenia (> 3 weeks) was significantly higher in Risk SNP + groups. Our findings are consistent with reports from literature where polymorphisms of ITPA & NUDT15 were associated with increased neutropenic events with resultant therapy interruptions [3, 8, 10, 17, 19, 21, 23].
There is a definite role of polymorphisms (TPMT & NUDT15) in the pharmacogenetics and pharmacokinetics of 6MP. It is clear from literature that polymorphisms differ with ancestry. Research is required for studying promoter polymorphism, respective weightage of each polymorphism in light of racial differences, role of environmental factors and standardizing a method for genotype assessment. The effect of ITPA on toxicity associated with 6MP as yet needs elucidation. Studies with larger sample size and across various ethnicities are required to define the correlation if any, between the variants of ITPA/NUDT15 gene and the long-term survival in ALL, their effect at the molecular level and whether it can be used in prediction of relapse; as seen in case of TPMT.
This was a prospective Mendelian randomization study. There are very few prospective blinded studies in literature where all the three prominent gene polymorphisms implicated in 6MP metabolism have been tested with a 6-month follow-up period. The SNP’s in gene polymorphisms detected by PCR-RFLP were confirmed with gene sequencing. Our shortcoming is that this is a single centre study with a limited number of patients which precludes adequate statistical inference. Further large geographic area based pharmacogenetic studies will help establish a robust association with pharmacokinetic effects on 6MP before pre-emptive testing of these polymorphisms may be advocated across various ethnic groups.
Abbreviations
- 6 MP:
-
6-Mercaptopurine
- SNP:
-
Single nucleotide polymorphism
- ALL:
-
Acute lymphoblastic leukaemia
- TPMT:
-
Thiopurine methyltransferase
- ITPA:
-
Inosine triphosphate pyrophosphatase
- NUDT15:
-
Nucleoside diphosphate linked moiety-type motif 15
- ICICLE:
-
Indian Childhood Collaborative Leukaemia Group 2015
- MTX:
-
Methotrexate
- PCR-RFLP:
-
PCR amplification and restriction enzyme digestion
- ALT:
-
Alanine transferase
- FN:
-
Febrile neutropenia
- ANC:
-
Absolute neutrophil count
- HWE:
-
Hardy Weinberg equilibrium
- ANOVA:
-
Analysis of variance
- 6TGN:
-
6-Thioguanine
- TLC:
-
Total leucocyte count
- CBC:
-
Complete blood count
- PCR:
-
Polymerized chain reaction
References
Wall AM, Rubnitz JE (2003) Pharmacogenomic effects on therapy for acute lymphoblastic leukemia in children. Pharmacogenomics J, 3:128
Evans WE et al (2001) Preponderance of thiopurine S-methyltransferase deficiency and heterozygosity among patients intolerant to mercaptopurine or azathioprine. J Clin Oncol 19(8):2293–2301
Moriyama T, Relling MV, Yang JJ (2015) Inherited genetic variation in childhood acute lymphoblastic leukemia. Blood 125(26):3988–3995
Relling MV et al (1999) Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst 91(23):2001–2008
Relling MV et al (2013) Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin Pharmacol Ther 93(4):324–325
Chang JG et al (2002) Molecular analysis of thiopurine S-methyltransferase alleles in South-east Asian populations. Pharmacogenetics 12(3):191–195
Srimartpirom S et al (2004) Thiopurine S-methyltransferase genetic polymorphism in the Thai population. Br J Clin Pharmacol 58(1):66–70
Tanaka Y et al (2012) The activity of the inosine triphosphate pyrophosphatase affects toxicity of 6-mercaptopurine during maintenance therapy for acute lymphoblastic leukemia in Japanese children. Leuk Res 36(5):560–564
Yang SK et al (2014) A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat Genet 46(9):1017–1020
Chiengthong K et al (2016) NUDT15 c.415C> T increases risk of 6-mercaptopurine induced myelosuppression during maintenance therapy in children with acute lymphoblastic leukemia. Haematologica 101(1):e24–e26
Yates CR et al. (1997) Molecular diagnosis of thiopurine S-methyltransferase deficiency: genetic basis for azathioprine and mercaptopurine intolerance. Ann Intern Med 126:608–614
Cao H, Hegele RA (2002) DNA polymorphisms in ITPA including basis of inosine triphosphatase deficiency. J Hum Genet 47:620
Schmiegelow K et al (2014) Mercaptopurine/methotrexate maintenance therapy of childhood acute lymphoblastic leukemia: clinical facts and fiction. J Pediatr Hematol Oncol 36(7):503–517
Kapoor G et al. (2010) Thiopurine S-methyltransferase gene polymorphism and 6-mercaptopurine dose intensity in Indian children with acute lymphoblastic leukemia. Leuk Res 34:1023–1026
Davavala SK et al (2014) Prevalence of TPMT polymorphism in Indian patients requiring immunomodulator therapy and its clinical significance. Indian J Gastroenterol 33(1):41–45
Desire S et al (2010) Frequency of TPMT alleles in Indian patients with acute lymphatic leukemia and effect on the dose of 6-mercaptopurine. Med Oncol 27(4):1046–1049
Soler AM et al. (2017) TPMT and NUDT15 genes are both related to mercaptopurine intolerance in acute lymphoblastic leukaemia patients from Uruguay. Br J Haematol 181:252–255
Wan Rosalina WR et al (2012) Polymorphism of ITPA 94C> A and risk of adverse effects among patients with acute lymphoblastic leukaemia treated with 6-mercaptopurine. J Clin Pharm Ther 37(2):237–241
Kim HT et al (2017) NUDT15 genotype distributions in the Korean population. Pharmacogenet Genom 27(5):197–200
Schmiegelow K, Bruunshuus I (1990) 6-Thioguanine nucleotide accumulation in red blood cells during maintenance chemotherapy for childhood acute lymphoblastic leukemia, and its relation to leukopenia. Cancer Chemother Pharmacol 26(4):288–292
Yang JJ et al (2015) Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol 33(11):1235–1242
Farfan MJ et al (2014) Prevalence of TPMT and ITPA gene polymorphisms and effect on mercaptopurine dosage in Chilean children with acute lymphoblastic leukemia. BMC Cancer 14(1):299
Ma X et al (2014) Inosine triphosphate pyrophosphohydrolase (ITPA) polymorphic sequence variants in Chinese ALL children and possible association with mercaptopurine related toxicity. Int J Clin Exp Pathol 7(7):4552–4556
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Khera, S., Trehan, A., Bhatia, P. et al. Prevalence of TPMT, ITPA and NUDT 15 genetic polymorphisms and their relation to 6MP toxicity in north Indian children with acute lymphoblastic leukemia. Cancer Chemother Pharmacol 83, 341–348 (2019). https://doi.org/10.1007/s00280-018-3732-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-018-3732-3