ABSTRACT
Purpose
Genetic factors and hereditary forms of osteonecrosis of the femoral head (ONFH) have been elucidated through genetic association studies. The significance of these cases is that they suggest an alternative hypothesis to the development of the disease. This review presents a summary of single nucleotide polymorphisms (SNPs) and other genetic mutation variations found in association with ONFH, including our recent identification of a novel mutation in the transient receptor potential vanilloid 4 (TRPV4) gene in association with inherited ONFH. The purpose of this review is to consolidate and categorize genetic linkages according to physiological pathways.
Methods
A systematic review of literature from PubMed and Google Scholar was undertaken with a focus on genetic linkages and hereditary case studies of the disease. Recent genetic analysis studies published after 2007 were the focus of genetic linkages in non-hereditary cases.
Results
The summary of these genetic findings identifies biological processes believed to be involved in the development of ONFH, which include circulation, steroid metabolism, immunity, and the regulation of bone formation.
Conclusion
Taken together, these associations may lead to new pathways of bone repair and remodeling while opening new avenues for therapeutic targets. Knowledge of genetic variations could help identify individuals considered to be at higher risk of developing ONFH and prevent the multiple hit effect.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
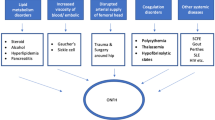
Non-hereditary osteonecrosis of the femoral head (ONFH), also known as avascular necrosis (AVN), ischemic necrosis, or aseptic necrosis, is a debilitating disease with 20,000 new cases diagnosed each year in the USA [1]. ONFH is thought to arise from a temporary or permanent loss of blood flow to the femoral head, causing bone necrosis and eventually its collapse [2]. This usually results in severe hip pain that is treated with conservative methods such as weight-bearing restriction, the use of bisphosphonates and statins, or femoral head decompression at early stage [3]. Invasive surgical procedures such as nonvascularized or vascularized bone grafting of the affected area, or total hip replacement are more often used at advanced stages [4]. Classic clinical presentations are seen in late adolescence and young to middle-aged adults; however, the disease may occur in children between four and 12 years old and is then called Legg-Calvé-Perthes Disease (LCPD), often with early asymptomatic ischemic changes [3]. There is a wide spectrum of aetiological risk factors in non-traumatic ONFH including alcohol consumption and chemotherapy; however, the most common risk factor is the use of glucocorticoids [3]. While most cases of non-traumatic ONFH are secondary, several studies have shown involvement of genetic factors, with hereditary ONFH found in Asian and Caucasian families [5, 6]. Some studies also report a genetic predisposition that explains ethnic and individual differences in ONFH incidence [7]. Here, we present a summary of recent genetic studies and review articles on ONFH pathogenesis, including 17 case-control and 7 meta-analyses which identify genetic variations thought to play a pivotal role in ONFH development (Fig. 1), as well as a summary of a recent discovery of a novel mutation in TRPV4 gene, in a family affected by hereditary ONFH [8].
Hereditary ONFH
COL2A1 was the first gene to be linked to hereditary ONFH. Chen et al. identified two four-generation Taiwanese families which showed an autosomal dominant mode of inheritance [5]. A genome wide scan for linkage analysis isolated candidate genes to a region on chromosome 12q13. Specifically, they proposed that COL2A1 and VDR could be putative causal genes. In the following year, an additional Taiwanese family with ONFH was reported with the same autosomal dominant heredity [9].
Genetic analyses were completed comparing all three families with hereditary ONFH, idiopathic cases of ONFH (sporadic cases), and wild type individuals. Candidate genes were selected following haplotype analysis of the families. Thirty-nine microsatellites repeat markers were analyzed in DNA from leukocytes and the causal gene was mapped to the same region that had been previously identified (12q13) [9].
Liu et al. identified COL2A1 mutations (NM_001844: c.3655G>A; p. Gly1170Ser and c.2149G>A; p. Gly717Ser) that were not seen in sporadic cases and controls. None of the individuals with inherited ONFH had predisposing risk factors, suggesting that the COL2A1 mutations are causal for the disease [9]. Kannu et al. identified a missense mutation of COL2A1 gene (NM_001844.3: c. 4148 C>T; p. Thr1383Met) located on the conserved C-propeptide region which is required for collagen chain trimerization [10].
Inherited forms of LCPD show a similar association with COL2A1, with Miyamoto et al. identifying a Japanese family with five members affected with LCPD [11]. They identified the same mutation (p. Gly1170Ser) as Liu et al. Su et al. also found the same mutation in a five-generation Chinese family with 16 individuals affected with some degree of ONFH [12]. Although these findings are interesting from a clinical perspective, these mutations were not characterized functionally and therefore the exact mechanism and their clinical impact remain poorly understood.
Recently, our group has identified a Canadian family of Greek origin where four of six siblings were diagnosed by x-ray and/or MR imaging to have advanced bilateral osteonecrosis [8]. The affected siblings showed no signs of known ONFH risk factors. Genetic analysis by exome sequencing showed a NM_021625.4 c.2480_2483delCCCG frameshift deletion followed by a c.2486T>A substitution of TRPV4 (transient receptor potential vanilloid 4 cation channel, subfamily V) gene. These mutations result in amino acid changes p.829V>W and p.830V>N followed by a stop codon at position 831 resulting in premature truncation of a highly conserved region [8].
TRPV4 is a non-selective cation channel involved in a broad range of physiological processes such as pain, thermoregulation, and calcium homeostasis. It is also known to regulate vascular tone and osteoclastic differentiation while also exhibiting mechanosensitive properties, proposed to play a role in the sensing of weight loading essential to bone development and mechanosensation in endothelial cells [13]. Considering that our functional studies showed impaired closure of the calcium channels, identifying specific TRPV4 pathways could have the potential to develop new targeted therapies in ONFH.
Polymorphisms in genes affecting blood circulation
The leading hypothesis for the development of ONFH is that vascular obstruction prevents blood flow to the femoral head, leading to the deterioration and collapse of the bone [14]. This hypothesis is supported by the association between sickle cell disease (SCD) and other haemoglobinopathies with ONFH. To help elucidate the pathogenesis and delineate the biological changes of the disease, several studies have focused on serological levels of factors participating in the coagulation pathway (extrinsic and intrinsic), including fibrinolysis, while others have searched for genetic variations in genes related to coagulation defects. Amongst genes involved in this process, factor V, prothrombin, and 5, 10-methylenetetrahydrofolate reductase (MTHFR) are of most interest, being known as thrombophilic risk factors. Some well-known variations in these genes have been associated with ONFH [7].
MTHFR gene encodes for an enzyme involved in the process of converting homocysteine to methionine. Several polymorphisms of the MTHFR gene affect its activity and leads to an elevated plasma homocysteine (hyperhomocysteinemia), a condition that has been associated with the development of vascular disease, including stroke, acute myocardial infarction, peripheral artery disease, and venous thrombosis especially in homozygous SNP findings [15]. The most studied polymorphisms of this gene (rs1801133 also known as c.677C>T) leads to reduced enzyme activity, with several studies exploring association of this SNP with ONFH and showing inconsistent results. Shang et al. performed a meta-analysis including data from eight studies for a total of 778 patients with ONFH and 1162 controls and found an association in non-Asian populations, but there was no clear evidence of this association across worldwide populations [16]. Another meta-analysis including all published studies (12 studies with 1181 patients and 1961 controls) found no overall association [17]. This difference is explained by differences in the design, inclusion criteria, and characteristics of the patients (age, gender, etiology) enrolled in these studies.
Factor V is a coagulation factor that binds to activated platelets and is inactivated by activated protein C. This inactivation is prevented by a change in amino acid (p. Arg506Gln, c. 1691G>A (rs6025) that is associated with venous thrombosis and known as Factor V Leiden [18]. Shang et al. performed a meta-analysis of 7 studies with 481 patients and 867 controls and concluded that patients with the A allele have a fourfold higher risk of ONFH when compared to carriers of the G allele [19]. While Gagala et al. failed to find this association in 68 patients and 100 controls in Poland, they found a statistically significant association with a genetic variation (rs1880669) in the tissue plasminogen activator (PLAT) gene [20]. tPA, the gene product of PLAT is a major component of the fibrinolytic system and the rs1880669 (TPA 25 I/D) polymorphism is an insertion/deletion of an Alu sequence on the intron 8. It affects the release of tPA after endothelial cell activation with the D allele being the slowest [21]. Gagala et al. showed higher frequency of the D allele in ONFH patients compared to controls [20].
Plasminogen activator inhibitor-1 (PAI-1) is a key regulator of the coagulation-fibrinolysis pathway, a process affected in ONFH. Several studies have shown an upregulation of the PAI-1 serum levels in ONFH patients compared to controls [20] which may be caused by a genetic difference. In fact, the PAI-1 4G/5G SNP (rs1799889) is associated with elevated PAI-1 plasma levels. 4G/4G carriers have higher PAI-1 plasma levels compared to 4G/5G which is higher than 5G/5G [22]. A meta-analysis of PAI-1 4G/5G polymorphisms (five studies with 419 cases of ONFH and 969 controls) showed that the 4G/4G genotype is a significant risk factor for predicting ONFH [23]. Li et al. analyzed two other PAI-1 SNPs (rs6092 and rs7242) in a population of 106 ONFH patients and 151 healthy controls and found no association between these two SNPs with ONFH. However, their statistical analysis showed that the haplotype G-T may be a protective factor of ONFH [24].
Tissue factor pathway inhibitor (TFPI) is an important regulator of the tissue-factor mediated blood coagulation pathway. 37339T>A, a novel SNP located in the 3’-UTR and 24999A>G (rs8176592) are related to both alcohol-induced and idiopathic ONFH in Korean individuals [25].
Blood flow may also be impeded by hyperlipidemia. PON-1 is a member of the paraoxonase gene family and is involved in lipid metabolism. The rs662 SNP affects PON-1 catalytic efficiency and has been shown to be associated with steroid-induced ONFH in Greek [26] and Han Chinese [27, 28] patients. Sterol regulatory element-binding transcription factor 1 (SREBF1) is a transcription factor involved in cholesterol and fatty acid metabolism [29]. In a study of Korean ONFH patients and controls, rs4925115, an SNP located on intron 7 of SREBP1 gene, was linked to ONFH, particularly amongst male ONFH patients. When the research participants were classified into subgroups based on etiological risk factors, the alcohol-induced group was associated with this SNP. SREBP-2 is closely related to SREBF1 and is also involved in lipid metabolism, playing a role in lipid homeostasis by stimulating gene expression of cholesterol biosynthetic pathways. The rs2267439 and rs2267443 SREBP-2 SNPs have an increased frequency in a study of 49 ONFH patients and 42 controls [30].
Another factor to consider in genes that impact blood flow is oxygen and oxidative dynamics. Nitric oxide is synthesized by endothelial nitric oxide synthase (eNOS) and has been implicated in many biological processes affected in ONFH including bone angiogenesis, thrombosis, and cellular turnover. An eNOS gene polymorphism that consists of a variable number of tandem repeats of 27 bp in intron 4 can impact nitric oxide synthesis. Two alleles have been identified, a larger form with five repeats (4b) and a smaller form with 4 repeats (4a). Subjects with the smaller form (4a) have lower levels of nitric oxide [31]. The eNOS 4a/b polymorphism has been previously associated with ONFH [32] with a subsequent examination of 68 patients and 100 controls of Polish descent supporting this finding [20]. Song et al. performed a meta-analysis of five studies (566 cases and 833 controls) and showed the same association between the eNOS 4a/b polymorphism and osteonecrosis [33].
Samples from 460 ONFH patients and 300 controls were analyzed using SNP chip array, a microarray chip used to detect SNP. Six candidate genes were found to be associated with ONFH: Transferrin (TF), Kinase insert domain receptor (KDR), Vascular endothelial growth factor C (VEGFC), Insulin-like growth factor binding protein 3 (IGFBP3), Neuropilin 1 (NRP1), and Angiotensin I converting enzyme (ACE) [34]. Interestingly, some SNPs in KDR, VEGFC, and NRP1 were linked to protection against ONFH (listed in Table 1). The protective effect of VEGF was reported by another group that investigated SNPs in the promoter and 5’ UTR regions of the gene [38]. Liu et al. performed a meta-analysis of three studies and confirmed that VEGF -634G/C SNP was significantly associated with increased risk for ONFH [47].
Sickle-cell disease (SCD) is a group of inherited blood disorders characterized by the presence of hemoglobin S (HbS), an abnormal haemoglobin caused by a mutation on chromosome 11 that results in an amino acid substitution of valine for glutamic acid at the sixth position of the beta-globin subunit of haemoglobin. This mutation causes the red blood cells to become rigid, sticky and distorted (sickled). These sickled cells can get stuck in small blood vessels, leading the blood flow to slow or stop which explains why osteonecrosis is a common complication of SCD and why several studies have reported a high prevalence (11 to 50%) of ONFH in SCD patients [48].
SCD is an autosomal recessive disease, which means that it requires the inheritance of two sickle cell genes, HbSS. The patients who are homozygous (HBSS), or the association with another abnormal hemoglobin gene, particularly with HbC (HBSC), have a high risk of osteonecrosis. Recently, Daltro et al. studied the clinical features of ON in SCD patients in Bahia, Brazil. They showed that the majority of patients (246/283) have bilateral ON; 74.6% have ON on different sites (hip, shoulder, knee, and ankle). They found no genetic effect (genotype SS vs SC) on clinical features of ON. They recommend that SCD patients should have radiological examination more frequently in order to avoid subsequent complications [49].
From a clinical and vascular bed perspective, osteonecrosis is a microvascular disease and thrombophilic markers involving the fibrinolytic pathway are probably the most relevant. Clinically, markers that usually involve macrovessels (thrombosis of lower extremities and pulmonary bed) such as Factor V Leiden and Prothrombin 20210A are probably less important in terms of risk factors for microvascular disease such as ONFH. Conversely, VEGF is more likely to have a role due to its function in microvascular disease, regeneration of blood vessels, endothelial cell differentiation, and function in bone remodeling [50]. VEGF remains a promising target in the clinical setting with some in vitro and in vivo studies having looked at the expression of VEGF in glucocorticoid-induced ONFH [51, 52].
Steroid metabolism and ONFH: a spotlight on ABCB1
Glucocorticoids are a major risk factor in the development of ONFH and are often administered at high dose for treatment of diverse disorders which may lead to the development of ONFH. Variation in the susceptibility of patients receiving high doses of glucocorticoids to ONFH strongly suggests a genetic variation that potentially involves the presence of SNPs [53, 54]. Several SNPs in adenosine triphosphate-binding cassette B1 (ABCB1), also known as MDR1, have been linked to ONFH. ABCB1 encodes the transport protein P-glycoprotein (P-gp) that plays an important role in absorption and distribution of a broad range of therapeutic compounds [55]. Higher P-gp activity was shown to be protective from ONFH development [56]. There are over 50 known SNPs in ABCB1 which may account for the variation in individual sensitivity to steroids [57]. The two most studied SNPs of this gene, rs1045642 and rs2032582, impact its activity. Studies investigating the association of these two SNPs with ONFH gave conflicting results prompting Zhou et al. to perform a meta-analysis of 7 studies and concluded that these SNPs are associated with low risk of glucocorticoid-induced ONFH [58]. Zhang et al., showed the same results for the rs1045642 SNP in a meta-analysis of 5 studies [59].
Lastly, an analysis of combinations of SNPs between genes involved in steroid metabolism show that ABCB1 and CBP, which encodes an important transcriptional co-regulator of glucocorticoid receptors, may interact with each other. Thirty-four patients diagnosed with glucocorticoid-induced ONFH and 123 controls (who were exposed to glucocorticoids but did not develop ONFH) were screened for SNPs in ABCB1 (C3435T), apoliprotein B (ApoB; c.7623C>T), and cAMP-response element binding protein (CBP; rs3751845). A synergistic index > 1.00 (1.99) was observed between ABCB1 and CBP, and the odds ratio of the presence of both SNPs in ONFH was very high (22.91). This suggests that both genes are involved in the pathology of ONFH through steroid metabolism [45]. These findings are significant because screening for the combination of SNPs in ABCB1 and CBP in patients undergoing high dose glucocorticoids could provide the identification of susceptible individuals at higher risk of developing ONFH. Susceptible individuals undergoing glucocorticoid treatment would then require close monitoring of joint symptoms and frequent radiological evaluations of their hip joints.
Immunity and ONFH
Several genes related to immunity have been associated with ONFH. Interleukin (IL)-1α is a proinflammatory cytokine that stimulates the expression of genes related to inflammation and immunity [60]. The rs1800587 SNP of IL-1α has been associated with increased risk of ONFH likely because of the stimulating role IL-1α plays in bone resorption [60]. In this study of 112 ONFH affected individuals and 438 healthy controls, the author also found an association of ONFH with IL-10, which can inhibit the synthesis of other proinflammatory cytokines [60]. The specific SNPs with the haplotype (rs1800896 G, rs1800871C, and rs1800872C) are listed in Table 1.
IL23 and IL-33 are two other interleukin genes shown to be predictive for increased risk of ONFH. The proinflammatory cytokine IL23 regulates the activity of an immune response by promoting inflammation through the IL23 receptor (IL23R). The two cytokines are primarily expressed in cells that are closely related to the immune system such as T cells, macrophages, and dendritic cells [61, 62]. The rs4655686, rs1569922, and rs7539625 SNPs on IL23R gene were found to be associated with an increased risk of ONFH in a Korean study that involved 443 ONFH patients and 272 controls [44]. However, Wang et al. showed another SNP on this gene (rs6693831) to be protective for ONFH [63].
Interleukin 33 (IL-33) is also implicated in immune responses in a similar fashion to IL23R, inducing helper T cells, mast cells, eosinophils, and basophils to produce type 2 cytokines. IL-33 is also known to be released from necrotic cells and is constitutively expressed in osteoblasts. IL-33 serum levels were significantly higher in two studies with a total of 165 ONFH patients compared to controls, with no significant differences seen across glucocorticoid-induced, alcohol-induced, or idiopathic cases [64, 65]. Zheng et al. proposed that instead of using MRI scans that are expensive and have limited availability in some countries, measuring IL-33 plasma levels could be a cost-effective and efficient method for diagnosing the early stages of ONFH [64]. While this study did not investigate specific genetic anomalies, i.e., SNPs or mutations, these findings emphasize the substantial clinical applicability of genetic research that aims to investigate the biological contributions of candidate genes to ONFH. However, before considering the use of IL-33 plasma levels as a potential biomarker of early ONFH, further clinical studies are needed to support a positive correlation of increased IL-33 plasma levels with ONFH development on MRI scans over time.
Tumor necrosis factor (TNF)-α is a potent inflammatory cytokine released by macrophages to regulate an immune response by promoting the expression of other cytokine molecules [60]. As such, TNF-α is suggested to act upon osteoblasts or bone marrow stromal cells to release cytokines that are associated with osteoclast proliferation and maturation [66].
TNF- α gene contains a large number of polymorphisms. The most important ones are located on the gene promoter which gives them the ability to alter the TNF- α gene expression and subsequently alter its physiological roles [67]. For example, the GA genotype of the -238(G/A) (rs361525) SNP site is associated with an increase in TNF-α expression, altering the osteoblast-osteoclast compositional balance [60, 68]. Peng et al. have performed a meta-analysis of 5 studies to confirm conflicting results concerning the association of 2 TNF- α SNPs (rs361525 and rs1800629) with ONFH susceptibility. Their results showed that the 2 SNPS are associated with ONFH but for the SNP rs1800629, this association is significant only in Asians [69].
Genes involved in the regulation of bone formation
Although the exact pathogenesis of ONFH is unknown, the genes mentioned to this point support that ONFH results from disrupted blood flow to the femoral head. Impeded circulation would lead to a failure to provide nutrients and immune cells necessary for the maintenance of the bone tissue, leading to its eventual collapse. However, a hypothesis for ONFH relating to the bone physiology of the hip itself is also a possible explanation as compromised bone physiology at the hip, possibly exacerbated by the weight bearing nature of the joint, could lead to a similar outcome. Therefore, genes involved in bone formation and bone structure are important to consider in the onset and progression of ONFH. This section summarizes the genes that influence processes regulating osteoblasts or osteoclasts.
As mentioned above, bone necrosis is a frequent manifestation in sickle cell disease. A case study of Indian patients with SCD revealed a linkage in the rs7170178 SNP site of the annexin A2 (ANXA2) gene [39]. Genes in the annexin family play a role in regulating cellular growth, with ANXA2 encoding an autocrine factor that heightens osteoclast formation and bone resorption. In the study, patients and control subjects were screened for five SNPs of ANXA2, of which only the rs7170718 was seen amongst the study population. The rs7170718 A/G and A/A genotypes were statistically more frequent in the patient groups indicating that detection of this SNP could be a screening tool for sickle cell patients [39]. Another study of sickle cell patients revealed a significant association of the rs3812163 SNP in bone morphogenetic protein 6 (BMP6) with ONFH [70]. Members of the BMP family induce endochondral bone formation when implanted in ectopic sites.
Insulin-like growth factors (IGFs) are the most abundant growth factors stored in bones and produced by osteoblasts [71]. IGFBP-3 is a member of the IGF binding protein family and helps regulate growth and metabolism, especially bone cell metabolism [30]. The TT and TC genotypes of IGFBP-3 rs2453839 were found to be more frequent in ONFH patients than in the control group [30].
SNPs associated with a protective effect against ONFH have also been identified at the bone level. Transforming growth factor (TGF)-β regulates the proliferation and differentiation of various cells and functions in cell growth, differentiation, apoptosis, cell migration, immune cell function, and extracellular matrix production [60]. A higher frequency of the homozygous C allele at the 25th codon (rs1800471) has been associated with a decreased level of TGF-β and linked to a protective effect [60]. The authors hypothesize that this results in the induction of BMPs, promoting bone regeneration. These findings support the hypothesis that deteriorated bone structure plays a major role in the disease and that SNPs in genes related to bone regulation provide valuable insight into the cellular mechanisms that lead to ONFH.
Joint-preserving surgical procedures are presently the most commonly used approach to prevent femoral head collapse [72]. Core decompression (CD) is the most widely accepted procedure and involves drilling a single 8–10-mm core into the necrotic lesion that could provide pain relief and reduce intraosseous pressure. Since the clinical success rate of CD was not satisfactory, CD was combined with different techniques where bone regeneration was stimulated using some bone substitutes, stem cells, or cellular growth factors (such as TGF-β, BMP, FGF-2, VEGF, and IGF) [73, 74]. Andriolo et al. have performed a meta-analysis of 41 studies where regenerative techniques were used for the treatment of ONFH. They have concluded that combination of CD with regenerative techniques increases significantly survivorship of ONFH patients compared to CD alone [75].
Karol et al. performed a genome-wide association study of 2285 patient with acute lymphoblastic leukemia treated with high doses of glucocorticoids [40]. Two hundred fifty of these patients developed ONFH, identifying a SNP near the Glutamate Receptor, Ionotropic, N-Methyl-D-Aspartate 3A (GRIN3A) gene (rs10989692) in association with glucocorticoid-induced ONFH. Glutamate signaling has been shown to regulate bone formation induced by mechanical loading which induces glutamate release from osteocytes. The released glutamate induces osteoblast receptor through activation of calcium channels [40, 76], which could implicate TRPV4 in this process, and thus provide a novel additional insight into the study of glucocorticoid-induced ONFH.
Conclusion
ONFH is a multifactorial disease representing a complex interplay of genetic anomalies and environmental factors. Knowledge of genetic variations thought to be involved in the disease could be useful in determining individuals considered at higher risk to develop ONFH with the ultimate goal of preventing the multiple hit effect. COL2A1 mutations remain one of the strongest association with hereditary ONFH while ABCB1 and GRIN3A (glutamate pathway) are gaining importance in glucocorticoid-induced ONFH. Since SNPs are identified within specific pathways, they could account for differences in disease susceptibility and responses to drug therapies; therefore, screening an individual at risk for ONFH for the presence of genetic risk factors may be beneficial in evaluating treatment options. As our society is moving towards a more preventive and personalized medicine, genetic studies will likely become of greater value and ONFH is a superb example of the potential applicability of translational research from basic science to patient care.
Abbreviations
- ABCB1:
-
adenosine triphosphate-binding cassette B1
- ACE:
-
angiotensin I converting enzyme
- ANXA2:
-
annexin A2
- ApoB:
-
apoliprotein B
- AVN:
-
avascular necrosis
- BMP:
-
Bone morphogenetic protein
- CAT:
-
catalase
- CBP:
-
CREB-binding protein
- COL2A1:
-
Collagen 2 aplha 1
- CTDP1:
-
C-terminal domain of RNA polymerase II subunit A, phosphatase of subunit 1
- CYP27C1:
-
cytochrome P450, family 27, subfamily C, polypeptide 1
- eNOS:
-
endothelial nitric oxide synthase
- GRIN3A:
-
Glutamate Receptor, Ionotropic, N-Methyl-D-Aspartate 3A
- IGF:
-
Insulin-like growth factors
- IGFBP3:
-
Insulin-like growth factor binding protein 3
- IGFBP-3:
-
Insulin-like growth factors binding protein
- IL:
-
Interleukin
- KDR:
-
Kinase insert domain receptor
- LCPD:
-
Legg-Calvé-Perthes disease
- MDR1:
-
Multidrug resistance 1
- MRI:
-
magnetic resonance imaging
- MTHFR:
-
5, 10-methylenetetrahydrofolate reductase
- NRP1:
-
Neuropilin 1
- ONFH:
-
osteonecrosis of the femoral head
- PAI-1:
-
Plasminogen activator inhibitor-1
- P-gp:
-
P-glycoprotein
- PLAT or TPA:
-
tissue plasminogen activator
- PON-1:
-
paraoxonase 1
- SNP:
-
single nucleotide polymorphism
- SREBF1:
-
Sterol regulatory element-binding transcription factor 1
- SREBF2:
-
Sterol regulatory element-binding transcription factor 2
- TF:
-
Transferrin
- TFPI:
-
Tissue factor pathway inhibitor
- TGF-β:
-
Transforming growth factor
- TNF-α:
-
Tumor necrosis factor
- TRPV4:
-
Transient receptor potential vanilloid 4
- VDR:
-
vitamin D receptor
- VEGFC:
-
Vascular endothelial growth factor C
References
Larson E, Jones LC, Goodman SB, Koo KH, Cui Q (2018) Early-stage osteonecrosis of the femoral head: where are we and where are we going in year 2018? Int Orthop. https://doi.org/10.1007/s00264-018-3917-8
Kerachian MA, Harvey EJ, Cournoyer D, Chow TY, Seguin C (2006) Avascular necrosis of the femoral head: vascular hypotheses. Endothelium 13(4):237–244. https://doi.org/10.1080/10623320600904211
Mont MA, Cherian JJ, Sierra RJ, Jones LC, Lieberman JR (2015) Nontraumatic osteonecrosis of the femoral head: where do we stand today? A ten-year update. J Bone Joint Surg Am 97(19):1604–1627. https://doi.org/10.2106/JBJS.O.00071
Zalavras CG, Lieberman JR (2014) Osteonecrosis of the femoral head: evaluation and treatment. J Am Acad Orthop Surg 22(7):455–464. https://doi.org/10.5435/JAAOS-22-07-455
Chen WM, Liu YF, Lin MW, Chen IC, Lin PY, Lin GL, Jou YS, Lin YT, Fann CS, Wu JY, Hsiao KJ, Tsai SF (2004) Autosomal dominant avascular necrosis of femoral head in two Taiwanese pedigrees and linkage to chromosome 12q13. Am J Hum Genet 75(2):310–317. https://doi.org/10.1086/422702
Su P, Li R, Liu S, Zhou Y, Wang X, Patil N, Mow CS, Mason JC, Huang D, Wang Y (2008) Age at onset-dependent presentations of premature hip osteoarthritis, avascular necrosis of the femoral head, or Legg-Calve-Perthes disease in a single family, consequent upon a p.Gly1170Ser mutation of COL2A1. Arthritis Rheum 58(6):1701–1706. https://doi.org/10.1002/art.23491
Hadjigeorgiou G, Dardiotis E, Dardioti M, Karantanas A, Dimitroulias A, Malizos K (2008) Genetic association studies in osteonecrosis of the femoral head: mini review of the literature. Skelet Radiol 37(1):1–7. https://doi.org/10.1007/s00256-007-0395-2
Mah W, Sonkusare SK, Wang T, Azeddine B, Pupavac M, Carrot-Zhang J, Hong K, Majewski J, Harvey EJ, Russell L, Chalk C, Rosenblatt DS, Nelson MT, Seguin C (2016) Gain-of-function mutation in TRPV4 identified in patients with osteonecrosis of the femoral head. J Med Genet. https://doi.org/10.1136/jmedgenet-2016-103829
Liu YF, Chen WM, Lin YF, Yang RC, Lin MW, Li LH, Chang YH, Jou YS, Lin PY, Su JS, Huang SF, Hsiao KJ, Fann CS, Hwang HW, Chen YT, Tsai SF (2005) Type II collagen gene variants and inherited osteonecrosis of the femoral head. N Engl J Med 352(22):2294–2301. https://doi.org/10.1056/NEJMoa042480
Kannu P, O'Rielly DD, Hyland JC, Kokko LA (2011) Avascular necrosis of the femoral head due to a novel C propeptide mutation in COL2A1. Am J Med Genet A 155A(7):1759–1762. https://doi.org/10.1002/ajmg.a.34056
Miyamoto Y, Matsuda T, Kitoh H, Haga N, Ohashi H, Nishimura G, Ikegawa S (2007) A recurrent mutation in type II collagen gene causes Legg-Calve-Perthes disease in a Japanese family. Hum Genet 121(5):625–629. https://doi.org/10.1007/s00439-007-0354-y
Su P, Zhang L, Peng Y, Liang A, Du K, Huang D (2010) A histological and ultrastructural study of femoral head cartilage in a new type II collagenopathy. Int Orthop 34(8):1333–1339. https://doi.org/10.1007/s00264-010-0985-9
Darby WG, Grace MS, Baratchi S, McIntyre P (2016) Modulation of TRPV4 by diverse mechanisms. Int J Biochem Cell Biol 78:217–228. https://doi.org/10.1016/j.biocel.2016.07.012
Urbaniak JR, Harvey EJ (1998) Revascularization of the femoral head in osteonecrosis. J Am Acad Orthop Surg 6(1):44–54
Seguin C, Abid MR, Spokes KC, Schoots IG, Brkovic A, Sirois MG, Aird WC (2008) Priming effect of homocysteine on inducible vascular cell adhesion molecule-1 expression in endothelial cells. Biomed Pharmacother 62(6):395–400. https://doi.org/10.1016/j.biopha.2008.02.008
Shang XF, Su H, Chang WW, Wang CC, Han Q, Xu ZW (2012) Association between MTHFR C677T polymorphism and osteonecrosis of the femoral head: a meta-analysis. Mol Biol Rep 39(6):7089–7094. https://doi.org/10.1007/s11033-012-1540-0
Chai W, Zhang Z, Ni M, Geng P, Lian Z, Zhang G, Shi LL, Chen J (2015) Genetic association between methylenetetrahydrofolate reductase gene polymorphism and risk of osteonecrosis of the femoral head. Biomed Res Int 2015:196495. https://doi.org/10.1155/2015/196495
Castoldi E, Rosing J (2004) Factor V Leiden: a disorder of factor V anticoagulant function. Curr Opin Hematol 11(3):176–181
Shang X, Luo Z, Li X, Hu F, Zhao Q, Zhang W (2013) Meta-analysis of Factor V Leiden G1691A polymorphism and osteonecrosis of femoral head susceptibility. Biomed Rep 1(4):594–598. https://doi.org/10.3892/br.2013.93
Gagala J, Buraczynska M, Mazurkiewicz T, Ksiazek A (2013) Prevalence of genetic risk factors related with thrombophilia and hypofibrinolysis in patients with osteonecrosis of the femoral head in Poland. BMC Musculoskelet Disord 14:264. https://doi.org/10.1186/1471-2474-14-264
Jern C, Ladenvall P, Wall U, Jern S (1999) Gene polymorphism of t-PA is associated with forearm vascular release rate of t-PA. Arterioscler Thromb Vasc Biol 19(2):454–459
Munoz-Valle JF, Ruiz-Quezada SL, Oregon-Romero E, Navarro-Hernandez RE, Castaneda-Saucedo E, De la Cruz-Mosso U, Illades-Aguiar B, Leyva-Vazquez MA, Castro-Alarcon N, Parra-Rojas I (2012) PAI-1 mRNA expression and plasma level in rheumatoid arthritis: relationship with 4G/5G PAI-1 polymorphism. Rheumatol Int 32(12):3951–3956. https://doi.org/10.1007/s00296-011-2279-y
Liang XN, Xie L, Cheng JW, Tan Z, Yao J, Liu Q, Su W, Qin X, Zhao JM (2013) Association between PAI-1 4G/5G polymorphisms and osteonecrosis of femoral head: a meta-analysis. Thromb Res 132(2):158–163. https://doi.org/10.1016/j.thromres.2013.06.023
Li Y, Liu FX, Yuan C, Meng L (2017) Association between plasminogen activator inhibitor gene polymorphisms and osteonecrosis of the femoral head susceptibility: a case-control study. Medicine (Baltimore) 96(42):e7047. https://doi.org/10.1097/MD.0000000000007047
Dai XL, Hong JM, Oh B, Cho YS, Lee JY, Park EK, Kim CY, Kim SY, Kim TH (2008) Association analysis of tissue factor pathway inhibitor polymorphisms and haplotypes with osteonecrosis of the femoral head in the Korean population. Mol Cells 26(5):490–495
Hadjigeorgiou GM, Malizos K, Dardiotis E, Aggelakis K, Dardioti M, Zibis A, Dimitroulias A, Scarmeas N, Tsezou A, Karantanas A (2007) Paraoxonase 1 gene polymorphisms in patients with osteonecrosis of the femoral head with and without cerebral white matter lesions. J Orthop Res 25(8):1087–1093. https://doi.org/10.1002/jor.20393
Wang Z, Zhang Y, Kong X, Li S, Hu Y, Wang R, Li Y, Lu C, Lin N, Chen W (2013) Association of a polymorphism in PON-1 gene with steroid-induced osteonecrosis of femoral head in Chinese Han population. Diagn Pathol 8:186. https://doi.org/10.1186/1746-1596-8-186
Li JM, Li Y, Wang L (2017) The genetic association between PON1 polymorphisms and osteonecrosis of femoral head: a case-control study. Medicine (Baltimore) 96(42):e8198. https://doi.org/10.1097/MD.0000000000008198
Lee HJ, Choi SJ, Hong JM, Lee WK, Baek JI, Kim SY, Park EK, Kim SY, Kim TH, Kim UK (2009) Association of a polymorphism in the intron 7 of the SREBF1 gene with osteonecrosis of the femoral head in Koreans. Ann Hum Genet 73(1):34–41. https://doi.org/10.1111/j.1469-1809.2008.00490.x
Song Y, Du ZW, Li QJ, Zhang GZ, Wang LL, Wu N, Wang JC, Gao ZL (2012) Association of sterol regulatory element binding protein 2 and insulin-like growth factor binding protein 3 genetic polymorphisms with avascular necrosis of the femoral head in the Chinese population. Chin Med J 125(22):4037–4043
Tsukada T, Yokoyama K, Arai T, Takemoto F, Hara S, Yamada A, Kawaguchi Y, Hosoya T, Igari J (1998) Evidence of association of the ecNOS gene polymorphism with plasma NO metabolite levels in humans. Biochem Biophys Res Commun 245(1):190–193. https://doi.org/10.1006/bbrc.1998.8267
Koo KH, Lee JS, Lee YJ, Kim KJ, Yoo JJ, Kim HJ (2006) Endothelial nitric oxide synthase gene polymorphisms in patients with nontraumatic femoral head osteonecrosis. J Orthop Res 24(8):1722–1728. https://doi.org/10.1002/jor.20164
Song GG, Lee YH (2017) Association of eNOS polymorphisms with susceptibility to osteonecrosis of the femur head : a meta-analysis. Z Rheumatol 76(3):267–273. https://doi.org/10.1007/s00393-016-0093-3
Hong JM, Kim TH, Kim HJ, Park EK, Yang EK, Kim SY (2010) Genetic association of angiogenesis- and hypoxia-related gene polymorphisms with osteonecrosis of the femoral head. Exp Mol Med 42(5):376–385. https://doi.org/10.3858/emm.2010.42.5.039
Kannu P, Irving M, Aftimos S, Savarirayan R (2011) Two novel COL2A1 mutations associated with a Legg-Calve-Perthes disease-like presentation. Clin Orthop Relat Res 469(6):1785–1790. https://doi.org/10.1007/s11999-011-1850-x
Chang JD, Hur M, Lee SS, Yoo JH, Lee KM (2008) Genetic background of nontraumatic osteonecrosis of the femoral head in the Korean population. Clin Orthop Relat Res 466(5):1041–1046. https://doi.org/10.1007/s11999-008-0147-1
Gagala J, Buraczynska M, Mazurkiewicz T, Ksiazek A (2013) Endothelial nitric oxide synthase gene intron 4 polymorphism in non-traumatic osteonecrosis of the femoral head. Int Orthop 37(7):1381–1385. https://doi.org/10.1007/s00264-013-1892-7
Lee YJ, Lee JS, Kang EH, Lee YK, Kim SY, Song YW, Koo KH (2012) Vascular endothelial growth factor polymorphisms in patients with steroid-induced femoral head osteonecrosis. J Orthop Res 30(1):21–27. https://doi.org/10.1002/jor.21492
Pandey S, Ranjan R, Pandey S, Mishra RM, Seth T, Saxena R (2012) Effect of ANXA2 gene single nucleotide polymorphism (SNP) on the development of osteonecrosis in Indian sickle cell patient: a PCR-RFLP approach. Indian J Exp Biol 50(7):455–458
Karol SE, Yang W, Van Driest SL, Chang TY, Kaste S, Bowton E, Basford M, Bastarache L, Roden DM, Denny JC, Larsen E, Winick N, Carroll WL, Cheng C, Pei D, Fernandez CA, Liu C, Smith C, Loh ML, Raetz EA, Hunger SP, Scheet P, Jeha S, Pui CH, Evans WE, Devidas M, Mattano LA Jr, Relling MV (2015) Genetics of glucocorticoid-associated osteonecrosis in children with acute lymphoblastic leukemia. Blood. https://doi.org/10.1182/blood-2015-05-643601
Chen J, Liu W, Cao Y, Zhang X, Guo Y, Zhu Y, Li J, Du J, Jin T, Wang G, Wang J (2017) MMP-3 and MMP-8 single-nucleotide polymorphisms are related to alcohol-induced osteonecrosis of the femoral head in Chinese males. Oncotarget 8(15):25177–25188. https://doi.org/10.18632/oncotarget.15587
Du J, Liu W, Jin T, Zhao Z, Bai R, Xue H, Chen J, Sun M, Zhang X, Wang G, Wang J (2016) A single-nucleotide polymorphism in MMP9 is associated with decreased risk of steroid-induced osteonecrosis of the femoral head. Oncotarget 7(42):68434–68441. https://doi.org/10.18632/oncotarget.12034
Li Y, Wang Y, Guo Y, Wang Q, Ouyang Y, Cao Y, Jin T, Wang J (2016) OPG and RANKL polymorphisms are associated with alcohol-induced osteonecrosis of the femoral head in the north area of China population in men. Medicine (Baltimore) 95(25):e3981. https://doi.org/10.1097/MD.0000000000003981
Kim TH, Hong JM, Oh B, Cho YS, Lee JY, Kim HL, Lee JE, Ha MH, Park EK, Kim SY (2008) Association of polymorphisms in the Interleukin 23 receptor gene with osteonecrosis of femoral head in Korean population. Exp Mol Med 40(4):418–426. https://doi.org/10.3858/emm.2008.40.4.418
Kuribayashi M, Fujioka M, Takahashi KA, Arai Y, Hirata T, Nakajima S, Yoshimura N, Satomi Y, Nishino H, Kondo K, Fukushima W, Hirota Y, Kubo T (2008) Combination analysis of three polymorphisms for predicting the risk for steroid-induced osteonecrosis of the femoral head. J Orthop Sci 13(4):297–303. https://doi.org/10.1007/s00776-008-1244-4
Kim TH, Hong JM, Oh B, Cho YS, Lee JY, Kim HL, Shin ES, Lee JE, Park EK, Kim SY (2008) Genetic association study of polymorphisms in the catalase gene with the risk of osteonecrosis of the femoral head in the Korean population. Osteoarthr Cartil 16(9):1060–1066. https://doi.org/10.1016/j.joca.2008.02.004
Liu Y, Zhang Z, Liu S, Su X, Zhou S (2015) Association between VEGF -634G/C polymorphism and osteonecrosis of the femoral head susceptibility: a meta analysis. Int J Clin Exp Med 8(7):10979–10985
Hernigou P, Daltro G (2014) Osteonecrosis in Sickle-Cell Disease. In: Koo K-H, Mont MA, Jones LC (eds) Osteonecrosis. Springer Berlin Heidelberg, Berlin, pp 125–131. https://doi.org/10.1007/978-3-642-35767-1_16
Daltro G, Franco BA, Faleiro TB, Rosario DAV, Daltro PB, Fortuna V (2018) Osteonecrosis in sickle cell disease patients from Bahia, Brazil: a cross-sectional study. Int Orthop 42(7):1527–1534. https://doi.org/10.1007/s00264-018-3905-z
Gurzu S, Turdean SG, Pop ST, Zazgyva A, Roman CO, Opris M, Jung I (2017) Different synovial vasculogenic profiles of primary, rapidly destructive and osteonecrosis-induced hip osteoarthritis. An immunohistochemistry study. Int Orthop 41(6):1107–1112. https://doi.org/10.1007/s00264-016-3302-4
Kabata T, Matsumoto T, Yagishita S, Wakayama T, Iseki S, Tomita K (2008) Vascular endothelial growth factor in rabbits during development of corticosteroid-induced osteonecrosis: a controlled experiment. J Rheumatol 35(12):2383–2390. https://doi.org/10.3899/jrheum.070838
Varoga D, Drescher W, Pufe M, Groth G, Pufe T (2009) Differential expression of vascular endothelial growth factor in glucocorticoid-related osteonecrosis of the femoral head. Clin Orthop Relat Res 467(12):3273–3282. https://doi.org/10.1007/s11999-009-1076-3
Asano T, Takahashi KA, Fujioka M, Inoue S, Satomi Y, Nishino H, Tanaka T, Hirota Y, Takaoka K, Nakajima S, Kubo T (2003) Genetic analysis of steroid-induced osteonecrosis of the femoral head. J Orthop Sci 8(3):329–333. https://doi.org/10.1007/s10776-003-0646-7
He N, Li S, Liu H (2009) High-throughput SNP detection based on PCR amplification on magnetic nanoparticles using dual-color hybridization. Methods Mol Biol 578:393–402. https://doi.org/10.1007/978-1-60327-411-1_24
Zhang Y, Kong X, Wang R, Li S, Niu Y, Zhu L, Chen W, Lin N (2014) Genetic association of the P-glycoprotein gene ABCB1 polymorphisms with the risk for steroid-induced osteonecrosis of the femoral head in Chinese population. Mol Biol Rep 41(5):3135–3146. https://doi.org/10.1007/s11033-014-3173-y
Han N, Yan Z, Guo CA, Shen F, Liu J, Shi Y, Zhang Z (2010) Effects of p-glycoprotein on steroid-induced osteonecrosis of the femoral head. Calcif Tissue Int 87(3):246–253. https://doi.org/10.1007/s00223-010-9385-9
Krupoves A, Mack D, Seidman E, Deslandres C, Amre D (2011) Associations between variants in the ABCB1 (MDR1) gene and corticosteroid dependence in children with Crohn's disease. Inflamm Bowel Dis 17(11):2308–2317. https://doi.org/10.1002/ibd.21608
Zhou Z, Hua Y, Liu J, Zuo D, Wang H, Chen Q, Zheng L, Cai Z (2015) Association of ABCB1/MDR1 polymorphisms in patients with glucocorticoid-induced osteonecrosis of the femoral head: evidence for a meta-analysis. Gene 569(1):34–40. https://doi.org/10.1016/j.gene.2015.03.023
Zhang Y, Xie H, Zhao D, Wang B, Yang L, Meng Q (2017) Association of ABCB1 C3435T polymorphism with the susceptibility to osteonecrosis of the femoral head: a meta-analysis. Medicine (Baltimore) 96(20):e6049. https://doi.org/10.1097/MD.0000000000006049
Samara S, Kollia P, Dailiana Z, Chassanidis C, Papatheodorou L, Koromila T, Malizos KN (2012) Predictive role of cytokine gene polymorphisms for the development of femoral head osteonecrosis. Dis Markers 33(4):215–221. https://doi.org/10.3233/DMA-2012-0928
Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K, Zonin F, Vaisberg E, Churakova T, Liu M, Gorman D, Wagner J, Zurawski S, Liu Y, Abrams JS, Moore KW, Rennick D, de Waal-Malefyt R, Hannum C, Bazan JF, Kastelein RA (2000) Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 13(5):715–725
Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, Pflanz S, Zhang R, Singh KP, Vega F, To W, Wagner J, O'Farrell AM, McClanahan T, Zurawski S, Hannum C, Gorman D, Rennick DM, Kastelein RA, de Waal Malefyt R, Moore KW (2002) A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol 168(11):5699–5708
Wang Y, Yang X, Shi J, Zhao Y, Pan L, Zhou J, Wang G, Wang J (2017) Combination analysis of NOS3, ABCB1 and IL23R polymorphisms with alcohol-induced osteonecrosis of the femoral head risk in Chinese males. Oncotarget 8(20):33770–33778. https://doi.org/10.18632/oncotarget.16809
Zheng L, Wang W, Ni J, Li Z, Xiao T, Zhang Q, Mao X, He A (2014) Plasma interleukin 33 level in patients with osteonecrosis of femoral head: an alarmin for osteonecrosis of the femoral head? J Investig Med 62(3):635–637. https://doi.org/10.2310/JIM.0000000000000050
Ma J, Guo W, Li Z, Wang B, Li S, Wang P (2017) Hip osteonecrosis is associated with increased plasma IL-33 level. Mediat Inflamm 2017:1732638. https://doi.org/10.1155/2017/1732638
Dai JC, He P, Chen X, Greenfield EM (2006) TNFalpha and PTH utilize distinct mechanisms to induce IL-6 and RANKL expression with markedly different kinetics. Bone 38(4):509–520. https://doi.org/10.1016/j.bone.2005.10.007
El-Tahan RR, Ghoneim AM, El-Mashad N (2016) TNF-alpha gene polymorphisms and expression. Springerplus 5(1):1508. https://doi.org/10.1186/s40064-016-3197-y
Wei BF, Feng Z, Wei W, Chen X (2017) Associations of TNF-alpha -238 A/G and IL-10 -1082 G/A genetic polymorphisms with the risk of NONFH in the Chinese population. J Cell Biochem. https://doi.org/10.1002/jcb.26167
Peng Y, Liu Y, Huang D, Huang W, Shao Z (2018) Association of TNF-alpha-308(G/A) and −238(G/A) polymorphisms with non-traumatic osteonecrosis of the femoral head risks: a meta-analysis. Int Orthop. https://doi.org/10.1007/s00264-018-3859-1
Ulug P, Vasavda N, Awogbade M, Cunningham J, Menzel S, Thein SL (2009) Association of sickle avascular necrosis with bone morphogenic protein 6. Ann Hematol 88(8):803–805. https://doi.org/10.1007/s00277-008-0659-5
Dar AA, Majid S, Nosrati M, de Semir D, Federman S, Kashani-Sabet M (2010) Functional modulation of IGF-binding protein-3 expression in melanoma. J Invest Dermatol 130(8):2071–2079. https://doi.org/10.1038/jid.2010.70
Nadeau M, Seguin C, Theodoropoulos JS, Harvey EJ (2007) Short term clinical outcome of a porous tantalum implant for the treatment of advanced osteonecrosis of the femoral head. Mcgill J Med 10(1):4–10
Kuroda Y, Matsuda S, Akiyama H (2016) Joint-preserving regenerative therapy for patients with early-stage osteonecrosis of the femoral head. Inflamm Regen 36:4. https://doi.org/10.1186/s41232-016-0002-9
Kuroda Y, Asada R, So K, Yonezawa A, Nankaku M, Mukai K, Ito-Ihara T, Tada H, Yamamoto M, Murayama T, Morita S, Tabata Y, Yokode M, Shimizu A, Matsuda S, Akiyama H (2016) A pilot study of regenerative therapy using controlled release of recombinant human fibroblast growth factor for patients with pre-collapse osteonecrosis of the femoral head. Int Orthop 40(8):1747–1754. https://doi.org/10.1007/s00264-015-3083-1
Andriolo L, Merli G, Tobar C, Altamura SA, Kon E, Filardo G (2018) Regenerative therapies increase survivorship of avascular necrosis of the femoral head: a systematic review and meta-analysis. Int Orthop 42(7):1689–1704. https://doi.org/10.1007/s00264-018-3787-0
Brakspear KS, Mason DJ (2012) Glutamate signaling in bone. Front Endocrinol (Lausanne) 3:97. https://doi.org/10.3389/fendo.2012.00097
Acknowledgements
The authors gratefully acknowledge Dr. Terry Chow for reviewing the manuscript. This work was supported by awards to C. Séguin from the Fonds de la Recherche en Santé du Québec (FRQ-S) and from the Montreal General Hospital Foundation. C. Séguin is also supported by awards from the Canadian and US Leukemia & Lymphoma Societies.
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript. TW performed the literature search and participated in writing the first draft, BA finalized the literature search and participated in writing of the manuscript, finalizing the last version. TW and BA participated equally to this manuscript. EJH and DR helped to draft the manuscript and made an extensive revision. CS participated in the design of the review, its coordination and helped to draft the manuscript and finalizing the last version.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Wang, T., Azeddine, B., Mah, W. et al. Osteonecrosis of the femoral head: genetic basis. International Orthopaedics (SICOT) 43, 519–530 (2019). https://doi.org/10.1007/s00264-018-4172-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00264-018-4172-8