Abstract
Osteonecrosis of the femoral head (ONFH) is a complex, polygenic, or multifactorial disease which is caused by a number of genetic factors of relatively smaller effects and environmental factors. Diverse conditions have been implicated in the development of ONFH. There are several well-accepted common associations: corticosteroids use [1], alcohol abuse [2], systemic lupus erythematosus [3], Legg-Calve-Perthes disease [4], sickle cell disease [5], radiation [6], cytotoxic agents [7], Gaucher disease [8], dysbarism [9], HIV [10], hyperlipidemia [11], pancreatitis [12], and gout [13]. Idiopathic ONFH refers only when there are no identifiable factors identified.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Vascular Endothelial Growth Factor
- Sickle Cell Disease
- Gauche Disease
- Korean Population
- Tissue Factor Pathway Inhibitor
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Osteonecrosis of the femoral head (ONFH) is a complex, polygenic, or multifactorial disease which is caused by a number of genetic factors of relatively smaller effects and environmental factors. Diverse conditions have been implicated in the development of ONFH. There are several well-accepted common associations: corticosteroids use [1], alcohol abuse [2], systemic lupus erythematosus [3], Legg-Calvé-Perthes disease [4], sickle cell disease [5], radiation [6], cytotoxic agents [7], Gaucher disease [8], dysbarism [9], HIV [10], hyperlipidemia [11], pancreatitis [12], and gout [13]. Idiopathic ONFH defines only when there are no identifiable factors identified.
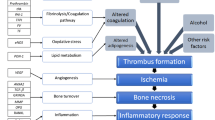
The etiology of ONFH involves (1) vascular insult by trauma such as femoral neck fracture or hip dislocation, (2) intravascular coagulation, and (3) fat emboli by alcohol or corticosteroids. The pathogenesis involves vascular interruption by trauma, thrombotic occlusion by intravascular coagulation, and extravascular compression by fat emboli. Steroids enhance adipogenesis and hinder osteogenesis as well as angiogenesis by marrow stem cells [14]. Alcohol stimulates adipogenesis and inhibits osteogenesis by marrow stromal cells [15]. The pathophysiology involves decreased blood flow and ischemia by the above pathogenesis. Ischemia leading to death of osteocytes followed by repairing process, subsequent structural changes, and progressive collapse of the femoral head followed by degenerative arthritis of the hip joint (Fig. 9.1). In this chapter, we are going to review the genetic studies performed by a few technology in literatures and introduce future genetic studies by recent developed techniques to identify more gene variants associated with ONFH. These findings will help the early diagnosis and appropriate therapeutics of ONFH also.
2 Methods of Genetic Study
Various genetic studies had been developed and changed rapidly to identify disease-associated loci or to identify causal variation for specific disease. Single nucleotide polymorphisms (SNPs) are the most common form of DNA sequence variations. They can be very useful markers to map genes that modify the susceptibility of specific disease or those related drug responsiveness or side effect. Discovery of large numbers of SNPs is necessary to narrow down the genetic loci of specific disease susceptibility to small region of one gene by linkage disequilibrium mapping. Over the last few decades, genetic studies performing linkage analysis by positional cloning have identified about 3,000 genes as being inherited causing diseases. Linkage analysis which requires family pedigree is good for initial detection and, especially, is powerful for rare variants. To the contrary, candidate gene approach for association analysis was done to identify less than 1,000 SNPs using genotyping technology between a few hundred disease cases and controls under the concept of common variants implicated as common diseases. It is good for fine mapping, but poor for initial detection, and powerful for common variants.
Human genome project (HGP) identified 3,000,000,000 base pair DNA sequence. The reference sequence of genome constructed by the HGP is very informative about the vast majority of bases that are invariant across individuals. Subsequently, HapMap focusing on DNA sequence differences among individuals could inform substantial correlations of SNPs with many of their neighbors by recombination of linkage disequilibrium and low haplotype diversity. After international HapMap projects (Phase 1), genome-wide association study (GWAS) was used extensively through the world. GWAS enabled high-throughput genotyping more than 500,000 SNPs to identify disease-associated loci and rarely to identify causal variation of disease. It consisted of phenotyping (recruitment of thousands of cases and controls), genotyping of SNPs of individuals, and mapping to determine linkage disequilibrium block by statistical analysis. GWAS mapped a huge number of susceptibility genes for common diseases. However, because the proportion of heritability explained by GWAS was relatively low for common diseases, it had many false positives, and genetic markers applied in GWAS are common variants with minor allele frequency (MAF) greater than 5 % and odds ratio of phenotype less than 1.5. Genome-wide association meta-analysis (GWA MA) imputed 1.5–3 million genotyped SNPs. Next-generation sequencing (NGS) such as targeted resequencing for disease loci, whole-exome sequencing (WES) selective capturing and sequencing of all annotated protein-coding genes of whole exons, and whole-genome sequencing are expected to find to identify low-frequency (0.5 % < MAF <5 %) or rare (MAF <0.5 %) variants and high odds ratio for phenotype of disease in the near future(Table 9.1).
3 Genetic Studies in Osteonecrosis of the Femoral Head
The cause of ONFH is multifactorial. Both genetic predisposition and exposure to certain environmental factors are associated. The incidence or prevalence of idiopathic ONFH reflects ethnic differences [16]. Some patients who have taken high dose of corticosteroids or excessive alcohol intake for long period develop ONFH, but rare cases of ONFH occur after short corticosteroid treatment indicating the presence of differences in susceptibility to risk factors as well as genetic predisposition to ONFH between individuals [17, 18]. Particularly, idiopathic ONFH in identical twins or a clustering of cases in families implicates involvement of genetic factors [19, 20].
3.1 Hypercoagulopathy and Genetic Alteration
Increased intravascular coagulation, hypercoagulability, seems to be associated with the development of ON [11]. The role of sickle cell disease and other hemoglobinopathies in the development of ONFH had been documented as undergoing the pathway of intravascular coagulation.
There were two studies using pedigrees linkage analysis followed by positional cloning on familial autosomal-dominant ONFH in Taiwanese. Chen et al. mapped the presence of mutations in the protein C, protein S, and PAI-1 protein to the 2q14-q14, 3q11.1-q11.2, and 7q21.3-q22 chromosomal segments, respectively. Liu et al. showed that the mutation in COL2A2 (mapped on chromosome 12q13) is associated with the genetic cause of ONFH. COL2A2 mutation, a major structural protein in the extracellular matrix of cartilage, was identified in three families with idiopathic ONFH showing autosomal-dominant inheritance [21].
Thrombophilia due to protein C and protein S deficiency was reported to have an association with ONFH. Pierre-Jacques et al. reported a familial heterozygous protein S deficiency in a patient with multifocal ON [22].
Factor V Leiden (G1691A, Arg506Gln) through generation of coagulation factor V results in a hypercogulable state. Homozygosity in the mutation is associated with much higher increase in the lifetime risk of venous thrombosis than heterozygosity in the mutation(100-fold versus 7-8 fold) [23].
Mutations in the factor V Leiden (FVL; G1691A) were significantly more common in patients with idiopathic ON than in a healthy population. Glueck et al. compared the prevalence of heterozygote for the FVL mutation between 244 ON patients (161 idiopathic and 83 secondary ON) and 104 controls [24]. Twenty-three of 244 patients with ON (9.4 %) had heterozygote for FVL, whereas 2 of 104 (1.9 %) controls had heterozygote for it. Fifteen of 161 patients with idiopathic ON (9.3 %) and 8 of 83 (9.6 %) patients with secondary ON had heterozygote for FVL. In contrast with this study, our study comparing the incidence of heterozygote in Korean population (423 patients with ON versus 348 controls), however, could not find significant difference (unpublished data). Therefore, the prevalence of the FVL mutation seems to be different in ethnic groups.
The substitution of G for A at nucleotide position 20210 in prothrombin gene (PTG) which results in increased plasma prothrombin levels has been reported to be associated with increased risk of thrombosis [25]. Almost all of studies reported negative association between G20210A and primary ON [26–30], only one reported the presence of G20210A associated with ON of knee with an OR of 3.6 compared with control subjects [31].
Plasminogen-activating inhibitor-1 (PAI-1) is a critical factor that regulates coagulation and fibrinolytic systems. Homozygosity or heterozygosity for 4G allele (4G/4G) has been reported to increase the levels of PAI-I levels, and this leads to reduced plasma fibrinolytic activity [29, 32]. PAI-1 gene is reported to be polymorphic, especially in rs1799889 (−675 4G/5G SNP) of the promoter region. Homozygosity for the 4G allele (4G/4G) has been reported to significantly increase the plasma PAI-1. Recently, Kim et al. reported prevalence of rs1799889 (−675; 4G/5G) and rs2227631 (−844G/A) in the promoter, and +107009 C/T in the 3′UTR (rs11178) SNP of PAI-1was significantly high in 206 patients with ONFH than that of 251 controls [33]. PAI-1 4G/5G polymorphism, particularly 4G allele, may be a risk factor for ON [29, 30, 33].
The 5,10-methylenetetrahydrofolate reductase (MTHFR) is the metabolic enzyme involved in the conversion of homocysteine to methionine via the remethylation pathway. Reduced levels of MTHFR activity lead to elevated plasma concentrations of homocysteine, a condition referred to as hyperhomocysteinemia. Hyperhomocysteinemia has also been identified as an independent risk factor for thrombotic events and ON [30]. Several case-control studies have examined the association between the C677T polymorphism in MTHFR and ONFH. However, reports concerning the role of this polymorphism in the pathogenesis ONFH have been inconsistent [29, 34, 35]. The association between the MTHFR C677T gene polymorphism and the development of ON was reported in a small number of Caucasian people [29, 34, 35]; we could not find any association between the MTHFR C677T gene polymorphism and the development of ON in 443 Korean patients with ONFH and 273 controls [35]. Recent meta-analysis with eight study samples reported that there was an association between MTHFR C677T polymorphism and ONFH in non-Asian population, but not for Asian population, although study samples were not considered etiology [36].
Tissue factor pathway inhibitor (TFPI) is an important regulator of blood coagulation pathway, and mutations of TFPI genes can increase the risk of thrombin generation and venous thrombosis. The haplotypes of AAAT and GAAT of TFPI were associated with the risk of ONFH in 474 Korean population with ONFH compared with 349 normal controls [37].
3.2 Genetic Risk Factors for Glucocorticoid-Induced Osteonecrosis
The high-dose corticosteroid use (~2 g of prednisone within 2–3 months) is the most common risk factor accounting for almost 10–30 % of ON cases [1, 38, 39]. However, only 8–10 % of patients exposed to corticosteroid therapy may develop ON [1]. Since not all patients who are treated with steroids develop ONFH, the presence of additional risk factors or individual variation of sensitivity for glucocorticoids has been suggested. However, no definite risk factor has been confirmed to date [40]. Genetic studies in subgroups of patients with steroid-induced ON first focused on coagulation, fibrinolytic factors, and homocysteine metabolism. Some authors reported a positive association of PAI and MTHFR [30] or factor V Leiden [31] polymorphisms with steroid-induced ON, but these studies included a relatively small number of patients. Moreover, results in these studies were not replicated in other studies [27, 34].
The 3435TT genotype and homozygosity in the mutant 2677T/A variants in the multidrug resistance gene 1(ABCB1; MDR1) had a protective effect against the development of ONFH after renal transplantation. These two polymorphisms were found to be in linkage dis-equilibrium [41]. The ABCB1 polymorphism was associated with ONFH in patients with SLE in Chinese people [42]. The 3435TT genotype individuals had increased pump activity of P-gp which might prevent the accumulation of steroids in bone cells. These results suggested genes related to steroid metabolism might influence sensitivity for steroid in patients with ONFH. However, polymorphisms in the CYP3A4, CYP2D6, and CYP2C19 isoforms and the CYP3A4 promoter region of cytochrome P450, participating in the drug metabolism of steroid, had not been associated with the increased risk of ON [43].
The development of ONFH increases in renal transplanted patients treated with glucocorticoids [43, 44]. Ferrari et al. reported a positive association of PAI 4G/4G genotype with ON [32]. However, other studies have not replicated this result [44, 45].
Recent meta-analysis showed there was no evidence for significant association between MTFHR C677T and ABCB1 G2677/A polymorphisms and risk of steroid-induced ON. However, the PAI-1 4G/5G and ABCB1 C3435T may be risk factors for steroid-induced ON.
3.3 Genetic Risk Factors for Alcohol-Induced Osteonecrosis
Liver alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) converts ethanol to acetic acid. Acetaldehyde quickly forms free radicals which are highly toxic to the kidney and liver. The liver ADH, ALDH, and cytochrome P4502E1 are polymorphic at the ADH2, ADH3, ALDH2 loci, and the 5′-flank region of the P4502E1. Individuals with ADH2 2/2 genotypes were reported to have faster rates of alcohol metabolism. The ADH2*1 allele was lower in the ONFH patients than in the cirrhosis subgroup [46].
3.4 Other Genetic Abnormalities
In recent years, we have focused on new candidate genes participating in hypoxia, angiogenesis, adipogenesis, and osteogenesis for genetic association with ONFH.
A vascular hypothesis is considered to be the most persuasive in explaining pathogenesis of ONFH. It assumes that if thrombosis occurs, it is followed by a sequential process of blood flow obstruction, increased venous pressure, impaired arterial flow, osseous hypoxia, and bone death [27, 31], which appear to be important in ONFH development; hypoxia-inducible factor 1 (HIF1) [47], vascular endothelial growth factor (VEGF) [40, 48], annexin 6 [49], and catalase (CAT) [50] showed significant and meaningful association with risk of ONFH.
Hypoxia can induce both apoptosis as well as necrosis of cells and is associated with vascular disease. HIF1 is a master transcription regulator induced by hypoxia and critically acts in a wide range of cellular regulation processes including glycolysis, apoptosis, erythropoiesis, and angiogenesis [51]. We firstly examined association between polymorphisms of HIF1-α and ONFH using 384 ONFH patients and 237 normal controls. Our study suggested that genetic variations in HIF1 alpha (−89A/T, −20T > C, +3033C/T, +14539A > T, +22348C > T, +244413T > C) were associated with the risk of ONFH in 443 Korean population with ONFH compared with 273 normal controls [47].
Oxidative stress is one of the factors inducing vascular injury and apoptosis. Alcohol promotes the generation of ROS resulting in oxidative stress. Steroid-induced ON in the rabbit demonstrated that administration of steroids promotes to the development of oxidative stress and oxidative injury in bone tissue soon thereafter [52]. A number of polymorphisms in the CAT have been described as being associated with several diseases, such as osteoporosis, hypertension, diabetes mellitus, Alzheimer’s disease, and vitiligo. Six polymorphisms (−89A/T, −20T > C, +3033C/T, +14539A > T, +22348C > T) of CAT gene were associated with the risk of ONFH in 443 Korean population with ONFH compared with 273 normal controls [50]. These results suggest that oxidative stress may play an important role in the pathogenesis of ONFH.
Nitric oxide synthesized by eNOS has vasodilatory effects on vascular tone, inhibits platelet aggregation, and modulates smooth muscle proliferation [53]. Many studies have been carried out to determine the associations between genetic polymorphisms in the eNOS gene and vascular diseases, including coronary artery disease or myocardial infarction, hypertension, stroke, and renal diseases. A few studies have showed an association between eNOS polymorphisms and ONFH. Allele 4a of a VNTR polymorphism in intron 4 and T-786C polymorphism in the eNOS gene were associated with idiopathic ONFH [54, 55]. Furthermore, we also found that Asp258Asp (rs1549758) and Glu298Asp (rs1799983) polymorphisms of eNOS gene were significantly associated with the risk of ONFH in Korean SLE patients [56].
VEGF, a major inducer of angiogenesis, is important for bone formation processes, such as normal growth plate morphogenesis, including blood vessel invasion and cartilage remodeling [57], and it has also been implicated in bone repair [58]. During hypoxia, HIF binds to the hypoxia-responsive elements and the expression of VEGF can be induced. This leads to the stimulation of angiogenesis [59]. The VEGF gene was reported to be polymorphic, especially in the promoter region (−2578, −1154, etc.), the 5′-UTR (−634, −7), and the 3′-UTR (+936). Several studies have shown that polymorphisms within the 5′-UTR have led to differences in VEGF expression and that they could influence the etiology of a variety of pathological conditions such as diabetic retinopathy, prostate cancer, and breast cancer. Our study showed that the −634G > C polymorphism in the VEGF promoter was significantly associated with an increased susceptibility of ONFH in 317 Korean patients with ONFH compared with 497 normal controls. Lee et al. also found that −1154A/G in promoter was associated with 74 steroid-induced ONFH compared with 160 age- and gender-matched normal controls [40]. Additionally, Liu et al. reported the VEGF −634G/C polymorphism was associated with ONFH in 220 Chinese patients with ONFH compared with 220 normal controls [60]. Also, rs1485766 and rs3775203 SNPs of VEGF-C gene were associated with the risk of ONFH in 460 Korean patients with ONFH compared with 300 normal controls, especially in idiopathic and steroid-induced ON [61].
Annexins have been implicated in inhibition of coagulation and have been reported as major components of matrix vesicles. SNPs and haplotypes of annexin A2 were associated with sickle cell necrosis [5]. rs9324679, rs9324677, rs10037814, and rs11960458 SNPs of annexin A6 gene were associated with the risk of ONFH in 443 Korean population with ONFH compared with 273 normal controls [48].
Recently, Hirata et al. examined the differences of gene polymorphism frequencies of apolipoprotein B (ApoB) and apolipoprotein A1 (ApoA1), which are important proteins for lipid transport, as well as of lipid parameters, between ONFH cases and referent patients among those who were subjected to renal transplantation [62]. The results showed that a higher frequency of 7623TT or CT of the ApoB gene was observed in ONFH cases than in referent patients. Regarding lipid parameters, a higher value of ApoB/ApoA1 ratio was observed in cases. Miyanishi et al. also reported the relationship between ONFH and serum ApoB/ApoA1 ratio in a study of Japanese subjects [63].
Peroxisome proliferator-activated receptor-gamma (PPAR-γ) is predominantly expressed in adipose tissue. It regulates the lipid homeostasis and angiogenesis. −769 > G, +34C > G, and +82466c > T SNPs of PPAR-γ gene were associated with the risk of ONFH in 448 Korean population with ONFH compared with 336 normal controls [64].
SREBP-2 plays central role in the maintenance of lipid homeostasis through stimulating expression of genes associated with cholesterol biosynthetic pathway. rs2267439, rs2269567, rs1052717, and rs2267443 SNPs of SREBP-2 were significantly associated with the risk of ONFH in 443 Korean population with ONFH compared with 273 normal controls [65].
SREBP-1, sterol regulatory element-binding protein, activates genes regulating lipid biosynthesis. IVS7 + 117A > G SNPs of SREBP-1 genes were significantly associated with the risk of ONFH in 423 Korean population with ONFH compared with 348 normal controls, especially in male [15].
We investigate SNPs of other genes relating angiogenesis and hypoxia. rs1880669, rs2692695, and rs2718806 SNPs of transferring (TF) gene were significantly associated with the risk of ONFH in 460 Korean population with ONFH compared with 300 normal controls, especially in idiopathic ON. rs4309, rs4344, and rs4461142 SNPs of angiotensin-1-converting enzyme gene were associated with the risk of ONFH in 460 Korean population with ONFH compared with 300 normal controls, especially in steroid-induced ON. The most significant association with risk of ONFH was an SNP (rs2453839) of the insulin-like growth factor binding protein-3, especially in idiopathic and alcohol-induced ON. Two SNPs (rs6837735 and rs1870377) of kinase insert domain receptor and three SNPs (rs12573218, rs12358370, rs2269091) of neutrophilin 1 were associated with the reduction of ONFH risk [61].
Immunologic factors involving interleukins and cytokines might influence the development of ONFH [66]. Also autoimmune disorders including SLE, polymyalgia rheumatica, rheumatoid arthritis, and ulcerative colitis are related to the development of osteonecrosis. Kim et al. reported that three SNPs (rs4655686, rs1569922, and rs7539625) of IL-23 receptor gene were significantly associated with the risk of ONFH in 443 Korean population with ONFH compared with 273 normal controls, especially idiopathic ON [67].
Samara et al. reported the CT and GA genotypes of the IL-1A (−889) and TNF-a genes were higher and the G to C polymorphism in the homozygous state in TGF-b codon 25 and GC genotype of IL-10 (−1082) gene were lower in 112 ON patients than 438 healthy donors [68].
In addition to mentioned above, many genetic association studies have been reported about ON in sickle cell disease patients, Legg-Calvé-Perthes disease, and malignancy such as acute leukemia or acute lymphoblastic lymphoma [69].
4 Future Genetic Study of ONFH
We are living in era of genome, and the recent advent of next-generation sequencing technologies has dramatically changed the nature of biomedical research. ONFH is believed to be a multifactorial disease associated in some cases with both genetic predilection and exposure to certain risk factors (environmental factors). The genetic studies about ONFH had been performed using linkage analysis on familial ONFH and candidate genes approach identifying the difference of SNPs between patients with ONFH and controls in limited numbers. The genetic susceptibility to ONFH probably involves many genes, most of which have very small effects. This fact represents the importance of the identification of large numbers of SNPs throughout the genome. Traditionally, the use of linkage analysis of families with extreme disease phenotypes has been successful in gene mutation discovery in ONFH. GWAS provides an important method for undertaking an evaluation of the association between common genetic variants and risk of disease. However, GWAS explained only a small fraction of the disease risk and showed problems of false-positive or false-negative [70]. As a result, a greater attention has been drawn to profiling rare variants. Advent of next-generation sequencing (NGS) such as targeted resequencing for disease loci, whole-exome sequencing (WES) selective capturing and sequencing of all annotated protein-coding genes of whole exons, and whole-genome sequencing are expected to find to identify low-frequency (0.5 % < MAF < 5 %) or rare (MAF < 0.5 %) variants and high odds ratio for phenotype of disease in the near future (Table 9.1). This approach would not be limited by the choice of candidate genes and cover the complete spectrum of coding and noncoding variants. Unlike GWASs, it would be able to test both rare and common variants for roles in ONFH.
Next, to study the genetic basis of disease, it is essential to determine the appropriate sample size and sampling design for well-designed study. Genetic associations can be real but nonetheless not reproducible if the underlying genetic effect is weak. As summarized in reported genetic associations with ONFH have become numerous but are mostly of small sample sizes. Even some association studies about same gene showed conflicting results. Increasing the size of the patient cohort will strengthen the power to discover variants. The lack of consistency across these studies may be the result of the geographic and ethnic variability of populations or the probability of a type II error resulting from small sample sizes.
Lastly, population stratification, which occurs when the cases and controls are unintentionally drawn from two or more ethnic groups or subgroups, also linked to false appearance of association [71]. Possible approach is to reevaluate the clinical diagnosis in the cohort by correlating subtle differences of clinical measurements and genetic variants in order to focus the analysis on a more clinically homogeneous set of patients as a separate cohort.
Furthermore, replication of findings in independent data sets is now widely regarded as a prerequisite for convincing evidence of association. Systematic reviews and meta-analyses have become a common approach to summarize gene-disease associations [72].
Therefore, genetic studies in the future should be performed in a cohort with more clinically homogeneous set of patients and normal controls and should be well designed. Global collaboration is essential to replicate the results across the ethnics. Next generation sequencing will provide tremendous genetic information about ONFH which can be applied to early diagnosis, prediction of the disease, reinvention of treatment practice, and the development of personalized medicine (Fig. 9.2).
References
Koo KH, Kim R, Kim YS, Ahn IO, Cho SH, Song HR, Park YS, Kim H, Wang GJ. Risk period for developing osteonecrosis of the femoral head in patients on steroid treatment. Clin Rheumatol. 2002;21(4):299–303.
Jones Jr JP. Alcoholism, hypercortisonism, fat embolism and osseous avascular necrosis. 1971. Clin Orthop Relat Res. 2001;393:4–12.
Mont MA, Glueck CJ, Pacheco IH, Wang P, Hungerford DS, Petri M. Risk factors for osteonecrosis in systemic lupus erythematosus. J Rheumatol. 1997;24(4):654–62.
Koo KH, Song HR, Ha YC, Kim JR, Kim SJ, Kim KI, Chang KC, Ahn IO, Cho SH. Role of thrombotic and fibrinolytic disorders in the etiology of Perthes’ disease. Clin Orthop Relat Res. 2002;399:162–7.
Baldwin C, Nolan VG, Wyszynski DF, Ma QL, Sebastiani P, Embury SH, Bisbee A, Farrell J, Farrer L, Steinberg MH. Association of klotho, bone morphogenic protein 6, and annexin A2 polymorphisms with sickle cell osteonecrosis. Blood. 2005;106(1):372–5.
Morrish Jr RB, Chan E, Silverman Jr S, Meyer J, Fu KK, Greenspan D. Osteonecrosis in patients irradiated for head and neck carcinoma. Cancer. 1981;47(8):1980–3.
Hancock BW, Huck P, Ross B. Avascular necrosis of the femoral head in patients receiving intermittent cytotoxic and corticosteroid therapy for Hodgkin’s disease. Postgraduate medical journal. 1978;54(634):545–6.
Amstutz HC. The hip in Gaucher’s disease. Clin Orthop Relat Res. 1973;90:83–9.
Slichter SJ, Stegall P, Smith K, Huang TW, Harker LA. Dysbaric osteonecrosis: a consequence of intravascular bubble formation, endothelial damage, and platelet thrombosis. J Lab Clin Med. 1981;98(4):568–90.
Glesby MJ, Hoover DR, Vaamonde CM. Osteonecrosis in patients infected with human immunodeficiency virus: a case-control study. J Infect Dis. 2001;184(4):519–23.
Jones Jr JP. Fat embolism and osteonecrosis. Orthop Clin North Am. 1985;16(4):595–633.
Koseki H, Tsurumoto T, Osaki M, Shindo H. Multifocal osteonecrosis caused by traumatic pancreatitis in a child. A case report. J Bone Joint Surg Am. 2009;91(9):2229–31.
Hungerford DS, Lennox DW. The importance of increased intraosseous pressure in the development of osteonecrosis of the femoral head: implications for treatment. Orthop Clin North Am. 1985;16(4):635–54.
Kawai K, Tamaki A, Hirohata K. Steroid-induced accumulation of lipid in the osteocytes of the rabbit femoral head. A histochemical and electron microscopic study. J Bone Joint Surg Am. 1985;67(5):755–63.
Lee HJ, Choi SJ, Hong JM, Lee WK, Baek JI, Kim SY, Park EK, Kim SY, Kim TH, Kim UK. Association of a polymorphism in the intron 7 of the SREBF1 gene with osteonecrosis of the femoral head in Koreans. Ann Hum Genet. 2009;73(1):34–41.
Kim S-Y, Rubash HE. Avascular necrosis of the femoral head: the Korean experience. In: The adult hip. 2nd ed. Philadelphia: Lippincott Williams & Wilkins; 2006. p. 1078–86.
Orlic D, Jovanovic S, Anticevic D, Zecevic J. Frequency of idiopathic aseptic necrosis in medically treated alcoholics. Int Orthop. 1990;14(4):383–6.
Jones LC, Hungerford DS. The pathogenesis of osteonecrosis. Instr Course Lect. 2007;56:179–96.
Glueck CJ, Glueck HI, Welch M, Freiberg R, Tracy T, Hamer T, Stroop D. Familial idiopathic osteonecrosis mediated by familial hypofibrinolysis with high levels of plasminogen activator inhibitor. Thromb Haemost. 1994;71(2):195–8.
Nobillot R, Le Parc JM, Benoit J, Paolaggi JB. Idiopathic osteonecrosis of the hip in twins. Ann Rheum Dis. 1994;53(10):702.
Liu YF, Chen WM, Lin YF, Yang RC, Lin MW, Li LH, Chang YH, Jou YS, Lin PY, Su JS, Huang SF, Hsiao KJ, Fann CS, Hwang HW, Chen YT, Tsai SF. Type II collagen gene variants and inherited osteonecrosis of the femoral head. N Engl J Med. 2005;352(22):2294–301.
Pierre-Jacques H, Glueck CJ, Mont MA, Hungerford DS. Familial heterozygous protein-S deficiency in a patient who had multifocal osteonecrosis. A case report. J Bone Joint Surg Am. 1997;79(7):1079–84.
Koster T, Rosendaal FR, de Ronde H, Briet E, Vandenbroucke JP, Bertina RM. Venous thrombosis due to poor anticoagulant response to activated protein C: Leiden Thrombophilia Study. Lancet. 1993;342(8886–8887):1503–6.
Glueck CJ, Freiberg RA, Boriel G, Khan Z, Brar A, Padda J, Wang P. The role of the factor V Leiden mutation in osteonecrosis of the hip. Clin Appl Thromb Hemost. 2013;19(5):499–503.
Poort SR, Rosendaal FR, Reitsma PH, Bertina RM. A common genetic variation in the 3′-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood. 1996;88(10):3698–703.
Bjorkman A, Burtscher IM, Svensson PJ, Hillarp A, Besjakov J, Benoni G. Factor V Leiden and the prothrombin 20210A gene mutation and osteonecrosis of the knee. Arch Orthop Trauma Surg. 2005;125(1):51–5.
Bjorkman A, Svensson PJ, Hillarp A, Burtscher IM, Runow A, Benoni G. Factor V leiden and prothrombin gene mutation: risk factors for osteonecrosis of the femoral head in adults. Clin Orthop Relat Res. 2004;425:168–72.
Chang JD, Hur M, Lee SS, Yoo JH, Lee KM. Genetic background of nontraumatic osteonecrosis of the femoral head in the Korean population. Clin Orthop Relat Res. 2008;466(5):1041–6.
Glueck CJ, Fontaine RN, Gruppo R, Stroop D, Sieve-Smith L, Tracy T, Wang P. The plasminogen activator inhibitor-1 gene, hypofibrinolysis, and osteonecrosis. Clin Orthop Relat Res. 1999;366:133–46.
Glueck CJ, Freiberg RA, Fontaine RN, Tracy T, Wang P. Hypofibrinolysis, thrombophilia, osteonecrosis. Clin Orthop Relat Res. 2001;386:19–33.
Zalavras CG, Vartholomatos G, Dokou E, Malizos KN. Genetic background of osteonecrosis: associated with thrombophilic mutations? Clin Orthop Relat Res. 2004;422:251–5.
Ferrari P, Schroeder V, Anderson S, Kocovic L, Vogt B, Schiesser D, Marti HP, Ganz R, Frey FJ, Kohler HP. Association of plasminogen activator inhibitor-1 genotype with avascular osteonecrosis in steroid-treated renal allograft recipients. Transplantation. 2002;74(8):1147–52.
Kim H, Cho C, Cho Y, Cho S, Yoon K, Kim K. Significant associations of PAI-1 genetic polymorphisms with osteonecrosis of the femoral head. BMC Musculoskelet Disord. 2011;12:160.
Zalavras CG, Malizos KN, Dokou E, Vartholomatos G. The 677C- > T mutation of the methylene-tetrahydrofolate reductase gene in the pathogenesis of osteonecrosis of the femoral head. Haematologica. 2002;87(1):111–2.
Kim TH, Hong JM, Kim HJ, Park EK, Kim SY. Lack of association of MTHFR gene polymorphisms with the risk of osteonecrosis of the femoral head in a Korean population. Molecules and cells. 2010;29(4):343–8.
Shang X-f, Su H, Chang W-w, Wang C-c, Han Q, Xu Z-w. Association between MTHFR C677T polymorphism and osteonecrosis of the femoral head: a meta-analysis. Mol Biol Rep. 2012;39(6):7089–94.
Dai XL, Hong JM, Oh B, Cho YS, Lee JY, Park EK, Kim CY, Kim SY, Kim TH. Association analysis of tissue factor pathway inhibitor polymorphisms and haplotypes with osteonecrosis of the femoral head in the Korean population. Mol Cells. 2008;26(5):490–5.
Griffith JF, Antonio GE, Kumta SM, Hui DS, Wong JK, Joynt GM, Wu AK, Cheung AY, Chiu KH, Chan KM, Leung PC, Ahuja AT. Osteonecrosis of hip and knee in patients with severe acute respiratory syndrome treated with steroids. Radiology. 2005;235(1):168–75.
Wang GJ, Cui Q, Balian G. The Nicolas Andry award. The pathogenesis and prevention of steroid-induced osteonecrosis. Clin Orthop Relat Res. 2000;370:295–310.
Lee YJ, Lee JS, Kang EH, Lee YK, Kim SY, Song YW, Koo KH. Vascular endothelial growth factor polymorphisms in patients with steroid-induced femoral head osteonecrosis. J Orthop Res. 2012;30(1):21–7.
Asano T, Takahashi KA, Fujioka M, Inoue S, Okamoto M, Sugioka N, Nishino H, Tanaka T, Hirota Y, Kubo T. ABCB1 C3435T and G2677T/A polymorphism decreased the risk for steroid-induced osteonecrosis of the femoral head after kidney transplantation. Pharmacogenetics. 2003;13(11):675–82.
Yang XY, Xu DH. MDR1(ABCB1) gene polymorphisms associated with steroid-induced osteonecrosis of femoral head in systemic lupus erythematosus. Pharmazie. 2007;62(12):930–2.
Asano T, Takahashi KA, Fujioka M, Inoue S, Satomi Y, Nishino H, Tanaka T, Hirota Y, Takaoka K, Nakajima S, Kubo T. Genetic analysis of steroid-induced osteonecrosis of the femoral head. J Orthop Sci. 2003;8(3):329–33.
Celik A, Tekis D, Saglam F, Tunali S, Kabakci N, Ozaksoy D, Manisali M, Ozcan MA, Meral M, Gulay H, Camsari T. Association of corticosteroids and factor V, prothrombin, and MTHFR gene mutations with avascular osteonecrosis in renal allograft recipients. Transplant Proc. 2006;38(2):512–6.
Asano T, Takahashi KA, Fujioka M, Inoue S, Ueshima K, Hirata T, Okamoto M, Satomi Y, Nishino H, Tanaka T, Hirota Y, Kubo T. Relationship between postrenal transplant osteonecrosis of the femoral head and gene polymorphisms related to the coagulation and fibrinolytic systems in Japanese subjects. Transplantation. 2004;77(2):220–5.
Chao YC, Wang SJ, Chu HC, Chang WK, Hsieh TY. Investigation of alcohol metabolizing enzyme genes in Chinese alcoholics with avascular necrosis of hip joint, pancreatitis and cirrhosis of the liver. Alcohol Alcohol. 2003;38(5):431–6.
Hong JM, Kim TH, Chae SC, Koo KH, Lee YJ, Park EK, Choi JY, Ryoo HM, Kim SY. Association study of hypoxia inducible factor 1alpha (HIF1alpha) with osteonecrosis of femoral head in a Korean population. Osteoarthritis Cartilage. 2007;15(6):688–94.
Kim TH, Hong JM, Lee JY, Oh B, Park EK, Lee CK, Bae SC, Kim SY. Promoter polymorphisms of the vascular endothelial growth factor gene is associated with an osteonecrosis of the femoral head in the Korean population. Osteoarthritis Cartilage. 2008;16(3):287–91.
Kim TH, Hong JM, Shin ES, Kim HJ, Cho YS, Lee JY, Lee SH, Park EK, Kim SY. Polymorphisms in the Annexin gene family and the risk of osteonecrosis of the femoral head in the Korean population. Bone. 2009;45(1):125–31.
Kim TH, Hong JM, Oh B, Cho YS, Lee JY, Kim HL, Shin ES, Lee JE, Park EK, Kim SY. Genetic association study of polymorphisms in the catalase gene with the risk of osteonecrosis of the femoral head in the Korean population. Osteoarthritis Cartilage. 2008;16(9):1060–6.
Semenza GL. HIF-1 and human disease: one highly involved factor. Genes Dev. 2000;14(16):1983–91.
Ichiseki T, Kaneuji A, Katsuda S, Ueda Y, Sugimori T, Matsumoto T. DNA oxidation injury in bone early after steroid administration is involved in the pathogenesis of steroid-induced osteonecrosis. Rheumatology (Oxford). 2005;44(4):456–60.
Cooke JP, Dzau VJ. Nitric oxide synthase: role in the genesis of vascular disease. Annu Rev Med. 1997;48:489–509.
Koo KH, Lee JS, Lee YJ, Kim KJ, Yoo JJ, Kim HJ. Endothelial nitric oxide synthase gene polymorphisms in patients with nontraumatic femoral head osteonecrosis. J Orthop Res. 2006;24(8):1722–8.
Glueck CJ, Freiberg RA, Oghene J, Fontaine RN, Wang P. Association between the T-786C eNOS polymorphism and idiopathic osteonecrosis of the head of the femur. J Bone Joint Surg Am. 2007;89(11):2460–8.
Kim HS, Bae SC, Kim TH, Kim SY. Endothelial nitric oxide synthase gene polymorphisms and the risk of osteonecrosis of the femoral head in systemic lupus erthematosus. Int Orthop. 2013;37(11):2289–96.
Schipani E, Ryan HE, Didrickson S, Kobayashi T, Knight M, Johnson RS. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 2001;15(21):2865–76.
Komatsu DE, Hadjiargyrou M. Activation of the transcription factor HIF-1 and its target genes, VEGF, HO-1, iNOS, during fracture repair. Bone. 2004;34(4):680–8.
Treins C, Giorgetti-Peraldi S, Murdaca J, Van Obberghen E. Regulation of vascular endothelial growth factor expression by advanced glycation end products. J Biol Chem. 2001;276(47):43836–41.
Liu B, Cao Y, Wang D, Yao G, Bi Z. Vascular endothelial growth factor -634G/C polymorphism associated with osteonecrosis of the femoral head in a Chinese population. Genet Test Mol Biomarkers. 2012;16(7):739–43.
Hong JM, Kim TH, Kim HJ, Park EK, Yang EK, Kim SY. Genetic association of angiogenesis- and hypoxia-related gene polymorphisms with osteonecrosis of the femoral head. Exp Mol Med. 2010;42(5):376–85.
Hirata T, Fujioka M, Takahashi KA, Arai Y, Asano T, Ishida M, Kuribayashi M, Akioka K, Okamoto M, Yoshimura N, Satomi Y, Nishino H, Fukushima W, Hirota Y, Nakajima S, Kato S, Kubo T. ApoB C7623T polymorphism predicts risk for steroid-induced osteonecrosis of the femoral head after renal transplantation. J Orthop Sci. 2007;12(3):199–206.
Miyanishi K, Yamamoto T, Irisa T, Noguchi Y, Sugioka Y, Iwamoto Y. Increased level of apolipoprotein B/apolipoprotein A1 ratio as a potential risk for osteonecrosis. Ann Rheum Dis. 1999;58(8):514–6.
Kim TH, Hong JM, Park EK, Kim SY. Peroxisome proliferator-activated receptor-gamma gene polymorphisms are not associated with osteonecrosis of the femoral head in the Korean population. Mol Cells. 2007;24(3):388–93.
Kim TH, Baek JI, Hong JM, Choi SJ, Lee HJ, Cho HJ, Park EK, Kim UK, Kim SY. Significant association of SREBP-2 genetic polymorphisms with avascular necrosis in the Korean population. BMC Med Genet. 2008;9:94.
Boss JH. Experimental models of osteonecrosis of the femoral head. J Orthop Sci. 2004;9(5):533–4.
Kim TH, Hong JM, Oh B, Cho YS, Lee JY, Kim HL, Lee JE, Ha MH, Park EK, Kim SY. Association of polymorphisms in the Interleukin 23 receptor gene with osteonecrosis of femoral head in Korean population. Exp Mol Med. 2008;40(4):418–26.
Samara S, Kollia P, Dailiana Z, Chassanidis C, Papatheodorou L, Koromila T, Malizos KN. Predictive role of cytokine gene polymorphisms for the development of femoral head osteonecrosis. Dis Markers. 2012;33(4):215–21.
Hadjigeorgiou G, Dardiotis E, Dardioti M, Karantanas A, Dimitroulias A, Malizos K. Genetic association studies in osteonecrosis of the femoral head: mini review of the literature. Skeletal Radiol. 2008;37(1):1–7.
Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13(2):135–45.
Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4(2):45–61.
Ioannidis JP, Gwinn M, Little J, Higgins JP, Bernstein JL, Boffetta P, Bondy M, Bray MS, Brenchley PE, Buffler PA, Casas JP, Chokkalingam A, Danesh J, Smith GD, Dolan S, Duncan R, Gruis NA, Hartge P, Hashibe M, Hunter DJ, Jarvelin MR, Malmer B, Maraganore DM, Newton-Bishop JA, O’Brien TR, Petersen G, Riboli E, Salanti G, Seminara D, Smeeth L, Taioli E, Timpson N, Uitterlinden AG, Vineis P, Wareham N, Winn DM, Zimmern R, Khoury MJ. A road map for efficient and reliable human genome epidemiology. Nat Genet. 2006;38(1):3–5.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Kim, SY., Kim, TH. (2014). Genetic Studies in Osteonecrosis of the Femoral Head. In: Koo, KH., Mont, M., Jones, L. (eds) Osteonecrosis. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-35767-1_9
Download citation
DOI: https://doi.org/10.1007/978-3-642-35767-1_9
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-35766-4
Online ISBN: 978-3-642-35767-1
eBook Packages: MedicineMedicine (R0)