Abstract
Purpose
The ability to reliably distinguish benign from malignant solid liver lesions on ultrasonography can increase access, decrease costs, and help to better triage patients for biopsy. In this study, we used deep learning to differentiate benign from malignant focal solid liver lesions based on their ultrasound appearance.
Methods
Among the 596 patients who met the inclusion criteria, there were 911 images of individual liver lesions, of which 535 were malignant and 376 were benign. Our training set contained 660 lesions augmented dynamically during training for a total of 330,000 images; our test set contained 79 images. A neural network with ResNet50 architecture was fine-tuned using pre-trained weights on ImageNet. Non-cystic liver lesions with definite diagnosis by histopathology or MRI were included. Accuracy of the final model was compared with expert interpretation. Two separate datasets were used in training and evaluation, one with all lesions and one with lesions deemed to be of uncertain diagnosis based on the Code Abdomen rating system.
Results
Our model trained on the complete set of all lesions achieved a test accuracy of 0.84 (95% CI 0.74–0.90) compared to expert 1 with a test accuracy of 0.80 (95% CI 0.70–0.87) and expert 2 with a test accuracy of 0.73 (95% CI 0.63–0.82). Our model trained on the uncertain set of lesions achieved a test accuracy of 0.79 (95% CI 0.69–0.87) compared to expert 1 with a test accuracy of 0.70 (95% CI 0.59–0.78) and expert 2 with a test accuracy of 0.66 (95% CI 0.55–0.75). On the uncertain dataset, compared to all experts averaged, the model had higher test accuracy (0.79 vs. 0.68, p = 0.025).
Conclusion
Deep learning algorithms proposed in the current study improve differentiation of benign from malignant ultrasound-captured solid liver lesions and perform comparably to expert radiologists. Deep learning tools can potentially be used to improve the accuracy and efficiency of clinical workflows.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Benign and malignant focal solid liver lesions have very different prognosis and management [1]. Benign lesions such as hemangiomas [2] are often observed, while malignant lesions such as hepatocellular carcinoma (HCC) [3] have a variety of treatment options depending on stage at diagnosis. Similarly, metastatic involvement of the liver by cancer elsewhere in the body portends a worse prognosis and dictates different treatment strategies [4]. There are currently three main ways to diagnose liver lesions: CT, MRI, and biopsy [5, 6]. CT exposes patients to radiation and can be nondiagnostic [7], while MRI is expensive and may not be available in resource limited areas [8]. Furthermore, contrast used in CT and MRI can be contraindicated in patients with poor renal function [9, 10]. Ultrasound-guided percutaneous liver biopsy is considered the gold standard for diagnosing solid liver lesions. However, the procedure is invasive and carries the risk of complications such as bleeding [11].
Ultrasound is often the first line imaging method to screen the abdomen. As an imaging modality, abdominal ultrasound is cheap, widely available, does not expose patients to ionizing radiation, and is non-invasive [6]. Patients with a history of viral hepatitis or liver cirrhosis are recommended to have semi-annual ultrasound screenings for early diagnosis of liver lesions [12]. Patients with elevated liver function tests or abdominal pain may also be imaged. Additionally, ultrasound studies often reveal liver lesions incidentally. A major drawback of ultrasound for the evaluation of focal liver lesions is that it is sometimes difficult to make a definitive diagnosis, and additional workup is frequently required in the form of CT, MRI, or in some cases, subsequent liver biopsy for definitive diagnosis.
Deep learning is an increasingly popular and powerful technique for image pattern recognition, with modern approaches at the level of or exceeding expert physician interpretation [13,14,15,16]. One commonly used neural network architecture is the Residual Network (ResNet), which has been shown to be effective and stable during training [17]. This model introduces the concept of residual connections between convolutional layers which allows models to be trained to much deeper depths while still maintaining a low complexity. A recent study has applied deep learning for the diagnosis of liver tumors on CT [18]. To our knowledge, no study in the literature has investigated the use of deep learning in diagnosing focal solid liver lesion on routine abdominal ultrasound. In the current study, we trained a ResNet model to differentiate benign from malignant focal solid liver lesions based on their appearance on ultrasound and compared our model accuracies with those of experts.
Methods
Code abdomen
The Code Abdomen diagnostic system was developed in 2014 at our institution and helps radiologists communicate malignancy risk of lesions found in four abdominal organs including the liver, adrenal glands, pancreas, and kidneys to ordering physicians [19]. The scale ranges from category 0 to category 7 with 99 being nondiagnostic. Table 1 lists different categories and descriptions associated with each category.
Patient cohort
Patients who had abdominal ultrasound from 2014 to 2018 with Code Abdomen liver categories 2, 3, 4, and 5 (C2–C5) were included in this current study. US units in this study included Philips Medical Systems model iU22 and model Epiq (Philips Ultrasound, Bothell, WA). Patients who did not undergo further work up by MRI or histopathology were excluded [3]. Category 0, 1, and 7 were excluded as presence of a lesion was required for training of our model and changes due to treatment may confound our model. Category 6, known cancer, were included in our training set.
Among the 596 patients who met the inclusion criteria, there were 911 images of individual lesions. Of the 596 patients, 300 had benign lesions while 296 had malignant lesions. Of the 911 lesions, 535 were malignant and 376 were benign based on MRI or histopathology. The diagnosis of benign versus malignant was established by histopathology in 265 patients and MRI in 331 patients [5, 20,21,22,23,24,25]. MRI was performed on 1.5 or 3.0 T scanners, with standard T2-weighted sequences, diffusion-weighted imaging and T1-weighted sequences including gradient-echo in-phase and out-of-phase sequences, gadolinium-enhanced three-dimensional fat-suppressed multiphasic sequences. Every patient with benign or malignant lesion definitely diagnosed on MRI had typical imaging features of a benign or malignant solid liver lesion, as interpreted in the original radiology report and subsequently reviewed and confirmed by a radiologist (JW). Detailed make and model of MRI scanners are in Supplementary Table S1. Malignant lesions were diagnosed on MRI based on clearly defined criteria such as enhancement and washout time in HCC [26]. Benign lesions had imaging follow-up lasting at least 24 months to ensure that they were benign.
There were 159 images in Code Abdomen liver category 2, 238 in category 3, 217 in category 4, 256 in category 5. C2 and C3 lesions were more likely to have been confirmed by MRI, while C4 and C5 lesions were more likely to have been confirmed by biopsy (Table 1). The complete set was divided into a training set of 660 lesions with 3,30,000 augmented images, validation set of 172 lesions, and a test set of 79 lesions. The detailed clinical characteristics of the patient cohort is shown in Supplementary Table S2. The uncertain diagnosis set was divided by patient into a training set of 314 lesions with 157,000 augmented images, a validation set of 80 lesions, and a test set of 82 lesions.
Image segmentation
All images were downloaded in JPEG format at their original dimensions and resolution. A novel website application was developed using Python, the web framework Flask, javascript, and the javascript framework React. With this tool, a radiologist specialized in abdominal imaging (JW) manually cropped downloaded ultrasound images to select the region of interest. Two segmentation schemes were used for all images: the first was a free crop of the lesion itself where a lesion was isolated in a square crop bounded tight to visualized lesion margins; the second was a fixed crop that was normalized to three real world physical centimeters across the x and y dimensions, centered on the lesion. Fixed crop images were normalized to three centimeters using ultrasound tick marks found in the images.
Model building
The imaging data were split into training, validation, and testing groups at a 7:2:1 ratio. Subgroup analysis with C3 and C4 lesions (uncertain diagnosis set) was split into 3:1:1 given smaller sample sizes. When multiple lesions originated from the same patient, these lesions were kept together during the randomized validation/training/testing split; this ensures that the model was never evaluated during validation or testing on a patient that it saw when training. Model building was performed on the segmented images using the two methods described above. During training, images were rescaled to 200 by 200 pixel squares, then augmented in real-time with random horizontal/vertical flips, shearing, and zooming to augment the size of the training set [27]. Models were trained with a batch size of 16, and training was stopped after 50 epochs with no improvement in the validation accuracy. Training was capped at a maximum of 500 epochs. After 100 training trials, the model with the best validation accuracy was selected.
Model architecture
The model was based on the ResNet50 architecture [17] with the following modifications: the 1000-class softmax fully-connected layer was replaced with a multi-layer perceptron, five fully-connected layers of decreasing width (256, 128, 64, 32, 16) with ReLU activations, and a single sigmoid output neuron for probability output and binary classification (benign or malignant); in the subgroup analysis of uncertain diagnosis set lesions, the learning rate was weighted by the reciprocal of the class frequency in the dataset. Pre-trained weights from ImageNet were used [28]. Hyperparameters, including batch size (8, 16, 32), base learning rate (0.001, 0.0001, 0.00001), percent of pre-trained weights and biases frozen (100%, 75%, 25%, 0%), and presence of the top multi-layer perceptron were all trialed, with the combination that performed best on the validation dataset selected (batch size = 16, learning rate = 0.0001, 0% pre-trained weights frozen, using top multi-layer perceptron).
Expert evaluation
Two expert radiologists (QP and DC), with 21 and 22 years of experience reading abdominal ultrasound, respectively, blindly evaluated unsegmented images for malignancy through our web application. The experts were given the age, gender and the clinical indication for the scan. The model’s results were compared to these expert evaluations to assess model performance.
Model assessment
Each trained model was assessed for its performance based on its accuracy, sensitivity, specificity, and area under the curve (AUC) of its receiver operating characteristic curve (ROC). In addition, the activations from the last convolutional layer of the best performing models were visualized by t-distributed Stochastic Neighbor Embedding (t-SNE) [29]. Representative images from the free crop uncertain diagnosis set were visualized with Grad-CAM overlay [30, 31].
Code availability
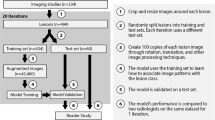
Our implementation was based on the Keras package [32] with the Tensorflow library as our backend [33]. Models were trained on a computer with an NVidia GTX 1080Ti GPU. To allow other researchers to develop their models, the code is publicly available on Github at https://github.com/intrepidlemon/deep-ultrasound. Figure 1 summarizes our data collection, annotation, and model training methodology in a graphical format.
Data collection and model training pipeline. Images were collected in JPEG format. Images were processed through a custom web application accessed by radiologists to provide segmentation and expert evaluation. Four experiments were set up comparing fixed segmentation vs. free segmentation as well as uncertain images vs. all images. Images and malignancy labels were used to train a convolutional neural network
Results
Performance
Performance characteristics of our model trained on the complete set (C2–C5) and on the uncertain diagnosis set (C3–C4) using both free crop and fixed crop segmentation methods are summarized in Table 2.
The model trained on all free segmentation images achieved a test accuracy of 0.84 (95% CI 0.74–0.90), F1 score of 0.86, precision recall AUC of 0.86, sensitivity of 0.87 (95% CI 0.74–0.94), and specificity of 0.78 (95% CI 0.61–0.89). The model trained on all fixed segmentation images achieved a test accuracy of 0.80 (95% CI 0.70–0.87), F1 score of 0.84, precision recall AUC of 0.87, sensitivity of 0.91 (95% CI 0.80–0.97), and specificity of 0.62 (95% CI 0.45–0.77).
The model trained on uncertain free segmentation images achieved a test accuracy of 0.79 (95% CI 0.69–0.87), F1 score of 0.80, precision recall AUC of 0.75, sensitivity of 0.80 (95% CI 0.66–0.90), and specificity of 0.78 (95% CI 0.63–0.88). The model trained on uncertain fixed segmentation images achieved a test accuracy of 0.71 (95% CI 0.60–0.80), F1 score of 0.73, precision recall AUC of 0.77, sensitivity of 0.78 (95% CI 0.63–0.88), and specificity of 0.63 (95% CI 0.48–0.76).
In comparison, on the complete set of all images, expert 1 achieved a test accuracy of 0.80 (95% CI 0.70–0.87), F1 score of 0.84, and sensitivity of 0.87 (95% CI 0.74–0.94), and specificity of 0.69 (95% CI: 0.51–0.82); expert 2 had a test accuracy of 0.73 (95% CI 0.63–0.82), F1 score of 0.78, and sensitivity of 0.81 (95% CI 0.67–0.90), and specificity of 0.62 (95% CI 0.45–0.77). On the uncertain set of images, expert 1 achieved a test accuracy of 0.70 (95% CI 0.59–0.78), F1 score of 0.71, and sensitivity of 0.76 (95% CI 0.60–0.86), and specificity of 0.63 (95% CI 0.48–0.76) and expert 2 achieved a test accuracy of 0.66 (95% CI 0.55–0.75), F1 score of 0.66, and sensitivity of 0.66 (95% CI 0.50–0.79), and specificity of 0.66 (95% CI 0.50–0.79).
Compared to a baseline zero rule algorithm, the free segmentation deep learning model had higher test accuracy (0.84 vs. 0.59, p < 0.0001). On the complete dataset, compared to all experts averaged, the free segmentation deep learning model had similar test accuracy (0.84 vs. 0.77, p = 0.18), similar test sensitivity (0.87 vs. 0.84, p = 0.69) and similar test specificity (0.78 vs. 0.66, p = 0.19) and the fixed segmentation model had similar test accuracy (0.80 vs. 0.77, p = 0.60), similar test sensitivity (0.91 vs. 0.84, p = 0.23) and similar test specificity (0.62 vs. 0.66, p = 0.71). On the uncertain dataset, compared to all experts averaged, the free segmentation deep learning model had higher test accuracy (0.79 vs. 0.68, p = 0.025), similar test sensitivity (0.80 vs. 0.71, p = 0.23) and similar test specificity (0.78 vs. 0.65, p = 0.074) and the fixed segmentation model had similar test accuracy (0.71 vs. 0.68, p = 0.64), similar test sensitivity (0.78 vs. 0.71, p = 0.39) and similar test specificity (0.63 vs. 0.65, p = 0.87). Figure 2 shows the ROC curves of all models overlaid with expert performance.
Figure 3 breaks down accuracy performance of models and experts by Code Abdomen category. t-SNE representation of the final dense layer of ResNet demonstrates good separation of malignant and benign lesions by the model when compared to histopathological diagnosis (Fig. 4). Confusion matrices for all models and experts is shown in Supplementary Fig. S1.
TSNE representation of neural network. Figure 3 shows a TSNE transformed representation of the final layer of the neural network before the classification node for every image in the validation dataset color coded by a model prediction and b biopsy or MRI diagnosis
Discussion
In the current study, ResNet models were trained to distinguish benign from malignant solid liver lesions on routine abdominal ultrasound images. Overall, these models achieved high test accuracy on the complete set along with high sensitivity, which is important for not missing a malignant diagnosis at a time of presentation where intervention may have been possible. At the same time, on the uncertain diagnosis set containing C3 (indeterminate) and C4 (suspicious for malignancy) lesions where usually a subsequent MRI and/or biopsy is recommended for further evaluation, the free crop model performed significantly better than experts in terms of accuracy and was trending toward statistical significance in specificity. High specificity is crucial in a screening setting where appropriate triage to subsequent MRI or biopsy can decrease cost and spare patients from unnecessary invasive procedures for patients with truly benign lesions.
Comparing segmentation methods, fixed crop methods trended toward performing worse. One possible explanation is fixed crop images contained varying amounts of surrounding tissue. Free crop images maintained an approximately consistent ratio of surrounding tissue to lesion tissue. In contrast, fixed crop images can range from including no surrounding tissue (when the lesion is wider and taller than 3 cm), to consisting of mostly surrounding tissue (when the lesion is much smaller than 3 cm). Nevertheless, it seems that the model architecture is robust to the difference in availability of surrounding tissue to some degree, i.e., the model trained on images using the fixed cropped method still achieved a high accuracy within the 95% confidence interval of the free crop model in both complete and uncertain diagnosis sets. This is likely due to the use of zoom augmentation during training.
Although models built on the complete set and the uncertain diagnosis set using the free crop segmentation method both performed well, achieving similar test accuracies, differences appear when broken down by code abdomen category. The free crop model seemed to perform better than the fixed crop model on C2 and C3 lesions while the two methods were similar in C4 and C5 lesions. This suggests that the fixed crop model’s incorporation of more surrounding tissue may have made a benign, simpler lesion seem more complex and malignant. Compared to experts, the free crop model excelled at identifying benign C2 lesions, and performed similar to experts in all other code categories. As for the uncertain diagnosis set, the free crop and fixed crop models both demonstrated a substantial increase in accuracy in C3 lesions when trained only on uncertain diagnosis lesions (C3 and C4), which suggests that training focused on uncertain lesions may lead to more clinically useful models.
Figure 3 shows that both experts and models generally performed better on C2 and C5 lesions and worse on C3 and C4 lesions. This is expected as C2 and C5 are respectively defined as “benign” and “highly suspicious” lesions, while C3 and C4 are defined as “indeterminate” and “suspicious”. There were also differences in the performance of the two experts: expert 1 had significantly higher accuracy than expert 2.
The t-SNE visualization (Fig. 4) of the final neural network layer weights demonstrates clear clustering between malignant and benign lesions. This representation offers a glimpse into the hyperspace of features for each lesion at the final neural network layer. The borders along which the final classifier is categorizing lesions is visible. Most importantly, the t-SNE demonstrates that the gold standard labels also cluster and correspond well with the features derived in the neural network.
The free crop segmentation method matched expert radiologist performance by every metric, although none was statistically significant. The fixed crop segmentation method outperformed experts in most metrics. In the uncertain diagnosis set, every malignant lesion that the model predicted incorrectly as benign was also predicted incorrectly as benign by expert 1. This demonstrates that our model allows for interpretation of liver lesions that matches and trend toward exceeding a radiologist’s expertise.
Machine learning models have been trained with ultrasound images in a number of organ systems including the thyroid [34], breast [35], and liver [36]. These previous studies most often extracted features from ultrasound images that were then fed into a support vector machine (SVM) or other traditional machine learning classification model [35,36,37]. Chi et al. used deep learning to remove image artifacts and to extract features from thyroid ultrasound images; these features were then fed into a random forest model for classification [34]. Shan et al. identifies features most important in classifying BI-RADS categories from breast ultrasound images in a number of different model architectures including neural network, decision tree and random forest [35]. Xian et al. reported high accuracy in distinguishing benign from malignant liver lesions from ultrasound data using image features fed into a fuzzy SVM [37]. However, the authors failed to assess their model using a separate validation set so its generalizability is unknown.
Previous studies achieved good results in using deep learning to categorize liver lesions on other imaging modalities. Yasaka et al. used deep learning to differentiate among liver lesions on CT [18]. Wu et al. trained a deep learning model on contrast enhancement time series data extracted from ultrasound videos to classify focal liver lesions in a small patient sample set [38]. Our study improves upon previous studies in a number of ways. By using only ultrasound images with no contrast, our method of image capture is the least invasive, most accessible, and safest. CT scans expose patients to radiation, while MRI is expensive and may not be available in resource limited areas. Our method eliminates the need for contrast injection which can be contraindicated in some people. In addition, our model is based solely on a single captured image from routine abdominal ultrasound using a well-validated deep neural network architecture, and thus can be easily integrated into routine clinical workflow.
Our study has several limitations. First, our cohort size is limited since the Code Abdomen system was only implemented at our institution since 2014, limiting the pool of available annotated ultrasound images. Second, while our models performed well without clinical data, the addition of clinical data such as history of viral hepatitis infection or cirrhosis may further improve accuracy. Third, the quality of an ultrasound image depends to a certain degree on body habitus, background liver disease, machine functionality, and operator skill. Thus, our deep learning model may not perform as well on images of lower quality as result of machine type or low operator skill. Fourth, our model currently depends on human segmentation of lesions, whereas the most ideal pipeline would accept raw ultrasound images as input. Given the wide variation in inclusion of surrounding tissue by operators, using whole images without a manual segmentation step is not likely to result in optimal performance. It may be possible to use deep learning to develop an automated segmentation algorithm that may be used in tandem with our proposed neural network model, and such a study would be the next step for this work. Lastly, only 271 of 609 patients were diagnosed via histopathology. However, only lesions with typical malignant imaging features on MRI as defined in the guidelines were included in the analysis, while benign lesions had a reasonable period of follow-up to ensure that they were benign.
Conclusion
Through this study, we have shown that a deep learning model can be trained to distinguish benign from malignant solid liver lesion visualized using ultrasound to a skill level that matches that of our expert radiologists. Given that this model has shown potential for this clinical application, it may be integrated into clinical workflow of ultrasound practitioners to increase access, decrease cost and facilitate triage.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- ResNet:
-
Residual Network
- AUC:
-
Area under the curve
- ROC:
-
Receiver operating characteristic curve
- t-SNE:
-
T-Distributed stochastic neighbor embedding
- SVM:
-
Support vector machine
References
Bonder A, Afdhal N (2012) Evaluation of liver lesions. Clinics in liver disease 16:271–283
Toro A, Mahfouz A-E, Ardiri A, et al (2014) What is changing in indications and treatment of hepatic hemangiomas. A review. Annals of hepatology 13:327–339
Marrero JA, Ahn J, Reddy KR (2014) ACG clinical guideline: The diagnosis and management of focal liver lesions. The American journal of gastroenterology 109:1328
Dietrich CF, Kratzer W, Strobel D, et al (2006) Assessment of metastatic liver disease in patients with primary extrahepatic tumors by contrast-enhanced sonography versus ct and mri. World Journal of Gastroenterology: WJG 12:1699
Semelka RC, Martin DR, Balci C, Lance T (2001) Focal liver lesions: Comparison of dual-phase ct and multisequence multiplanar mr imaging including dynamic gadolinium enhancement. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine 13:397–401
Harvey CJ, Albrecht T (2001) Ultrasound of focal liver lesions. European radiology 11:1578–1593
Sodickson A, Baeyens PF, Andriole KP, et al (2009) Recurrent ct, cumulative radiation exposure, and associated radiation-induced cancer risks from ct of adults. Radiology 251:175–184
Ginde AA, Foianini A, Renner DM, et al (2008) Availability and quality of computed tomography and magnetic resonance imaging equipment in us emergency departments. Academic emergency medicine 15:780–783
Stevens MA, McCullough PA, Tobin KJ, et al (1999) A prospective randomized trial of prevention measures in patients at high risk for contrast nephropathy: Results of the prince study. Journal of the American College of Cardiology 33:403–411
Perez-Rodriguez J, Lai S, Ehst BD, et al (2009) Nephrogenic systemic fibrosis: Incidence, associations, and effect of risk factor assessment—report of 33 cases. Radiology 250:371–377
Thampanitchawong P, Piratvisuth T (1999) Liver biopsy: Complications and risk factors. World journal of gastroenterology 5:301
Sherman M, Peltekian KM, Lee C (1995) Screening for hepatocellular carcinoma in chronic carriers of hepatitis b virus: Incidence and prevalence of hepatocellular carcinoma in a north american urban population. Hepatology 22:432–438
Esteva A, Kuprel B, Novoa RA, et al (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542:115
Lakhani P, Sundaram B (2017) Deep learning at chest radiography: Automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284:574–582
Brown JM, Campbell JP, Beers A, et al (2018) Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA ophthalmology
Gulshan V, Peng L, Coram M, et al (2016) Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama 316:2402–2410
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the ieee conference on computer vision and pattern recognition. pp 770–778
Yasaka K, Akai H, Abe O, Kiryu S (2017) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced ct: A preliminary study. Radiology 286:887–896
Zafar HM, Chadalavada SC, Kahn Jr CE, et al (2015) Code abdomen: An assessment coding scheme for abdominal imaging findings possibly representing cancer. Journal of the American College of Radiology: JACR 12:947
Strauss E, de Ferreira A SP, França AVC, et al (2015) Diagnosis and treatment of benign liver nodules: Brazilian society of hepatology (sbh) recommendations. Arquivos de gastroenterologia 52:47–54
Anderson SW, Kruskal JB, Kane RA (2009) Benign hepatic tumors and iatrogenic pseudotumors. Radiographics 29:211–229
Qian H, Li S, Ji M, Lin G (2016) MRI characteristics for the differential diagnosis of benign and malignant small solitary hypovascular hepatic nodules. European journal of gastroenterology & hepatology 28:749
Albiin N (2012) MRI of focal liver lesions. Current medical imaging reviews 8:107–116
Fowler KJ, Brown JJ, Narra VR (2011) Magnetic resonance imaging of focal liver lesions: Approach to imaging diagnosis. Hepatology 54:2227–2237
Itai Y, Ohtomo K, Furui S, et al (1985) Noninvasive diagnosis of small cavernous hemangioma of the liver: Advantage of mri. American journal of roentgenology 145:1195–1199
Willatt JM, Hussain HK, Adusumilli S, Marrero JA (2008) MR imaging of hepatocellular carcinoma in the cirrhotic liver: Challenges and controversies. Radiology 247:311–330
Chang K, Bai HX, Zhou H, et al (2018) Residual convolutional neural network for the determination of idh status in low-and high-grade gliomas from mr imaging. Clinical Cancer Research 24:1073–1081
Deng J, Dong W, Socher R, et al (2009) Imagenet: A large-scale hierarchical image database. In: Computer vision and pattern recognition, 2009. CVPR 2009. IEEE conference on. Ieee, pp 248–255
Maaten L van der, Hinton G (2008) Visualizing data using t-sne. Journal of machine learning research 9:2579–2605
Selvaraju RR, Cogswell M, Das A, et al (2017) Grad-cam: Visual explanations from deep networks via gradient-based localization. In: ICCV. pp 618–626
Lee H, Yune S, Mansouri M, et al (2018) An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nature Biomedical Engineering 1
Chollet F, others (2015) Keras
Abadi M, Barham P, Chen J, et al (2016) Tensorflow: A system for large-scale machine learning. In: OSDI. pp 265–283
Chi J, Walia E, Babyn P, et al (2017) Thyroid nodule classification in ultrasound images by fine-tuning deep convolutional neural network. Journal of digital imaging 30:477–486
Shan J, Alam SK, Garra B, et al (2016) Computer-aided diagnosis for breast ultrasound using computerized bi-rads features and machine learning methods. Ultrasound in medicine & biology 42:980–988
Yeh W-C, Huang S-W, Li P-C (2003) Liver fibrosis grade classification with b-mode ultrasound. Ultrasound in medicine & biology 29:1229–1235
Xian G-m (2010) An identification method of malignant and benign liver tumors from ultrasonography based on glcm texture features and fuzzy svm. Expert Systems with Applications 37:6737–6741
Wu K, Chen X, Ding M (2014) Deep learning based classification of focal liver lesions with contrast-enhanced ultrasound. Optik-International Journal for Light and Electron Optics 125:4057–4063
Acknowledgements
We are sincerely grateful to Dr. Qinghai Peng and Dr. Danming Cao from the Second Xiangya Hospital for their work in expert evaluation.
Funding
This project was funded by RSNA Research Fellow Grant (ID: RF1802) and SIR Foundation Radiology Resident Research Grant to HXB.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study. ILX, HXB and ZZ contributed to conception and design. ILX, JW, JG and HXB contributed to acquisition of data. ILX and HXB contributed to analysis and interpretation of data. ILX, JW, HXB, JG, PJZ, SCH, MCS and ZZ contributed to drafting the article or revising it for important intellectual content. All authors approved the final version to be published.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
Institutional Review Boards of Hospital of University of Pennsylvania was obtained for the study cohort with waiver of consent.
Ethics approval
The study was approved by the Institutional Review Boards of Hospital of University of Pennsylvania.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
261_2020_2564_MOESM1_ESM.doc
Supplementary file1 Supplementary Fig S1: Confusion matrices for all models and experts. Legend: Confusion matrices of the test datasets for both models are shown. Confusion matricies for both experts are shown as well (DOC 88 kb)
261_2020_2564_MOESM3_ESM.doc
Supplementary file3 Supplementary Table S2: Clinical characteristics of the overall cohort by Code Abdomen categories (DOC 69 kb)
Rights and permissions
About this article
Cite this article
Xi, I.L., Wu, J., Guan, J. et al. Deep learning for differentiation of benign and malignant solid liver lesions on ultrasonography. Abdom Radiol 46, 534–543 (2021). https://doi.org/10.1007/s00261-020-02564-w
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-020-02564-w