Abstract
Objectives
To develop and validate a proof-of-concept convolutional neural network (CNN)–based deep learning system (DLS) that classifies common hepatic lesions on multi-phasic MRI.
Methods
A custom CNN was engineered by iteratively optimizing the network architecture and training cases, finally consisting of three convolutional layers with associated rectified linear units, two maximum pooling layers, and two fully connected layers. Four hundred ninety-four hepatic lesions with typical imaging features from six categories were utilized, divided into training (n = 434) and test (n = 60) sets. Established augmentation techniques were used to generate 43,400 training samples. An Adam optimizer was used for training. Monte Carlo cross-validation was performed. After model engineering was finalized, classification accuracy for the final CNN was compared with two board-certified radiologists on an identical unseen test set.
Results
The DLS demonstrated a 92% accuracy, a 92% sensitivity (Sn), and a 98% specificity (Sp). Test set performance in a single run of random unseen cases showed an average 90% Sn and 98% Sp. The average Sn/Sp on these same cases for radiologists was 82.5%/96.5%. Results showed a 90% Sn for classifying hepatocellular carcinoma (HCC) compared to 60%/70% for radiologists. For HCC classification, the true positive and false positive rates were 93.5% and 1.6%, respectively, with a receiver operating characteristic area under the curve of 0.992. Computation time per lesion was 5.6 ms.
Conclusion
This preliminary deep learning study demonstrated feasibility for classifying lesions with typical imaging features from six common hepatic lesion types, motivating future studies with larger multi-institutional datasets and more complex imaging appearances.
Key Points
• Deep learning demonstrates high performance in the classification of liver lesions on volumetric multi-phasic MRI, showing potential as an eventual decision-support tool for radiologists.
• Demonstrating a classification runtime of a few milliseconds per lesion, a deep learning system could be incorporated into the clinical workflow in a time-efficient manner.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Liver cancer is the second leading cause of cancer-related deaths worldwide and hepatocellular carcinoma (HCC) represents the most common primary liver cancer [1, 2]. Contrary to many other cancer types, HCC incidence rates continue to rise [3]. Rapid and reliable detection and diagnosis of HCC may allow for earlier treatment onset and better outcomes for these patients. As the availability and quality of cross-sectional imaging have improved, the need for invasive diagnostic biopsies has decreased, propelling imaging-based diagnosis to a more central role, with a unique status especially for primary liver cancer. However, the radiological diagnosis of potentially malignant hepatic lesions remains a challenging task. In this setting, standardized image analysis and reporting frameworks such as the Liver Imaging Reporting and Data System (LI-RADS) can improve radiological diagnosis by reducing imaging interpretation variability, improving communication with referring physicians, and facilitating quality assurance and research [4]. However, the increasing complexity of LI-RADS has made its implementation less feasible in a high-volume practice, leaving an unmet clinical need for computational decision-support tools to improve workflow efficiency.
Machine learning algorithms have achieved excellent performance in the radiological classification of various diseases and may potentially address this gap [5,6,7]. In particular, a deep learning system (DLS) based on convolutional neural networks (CNNs) can attain such capabilities after being shown imaging examples with and without the disease. Unlike other machine learning methods, CNNs do not require definition of specific radiological features to learn how to interpret images, and they may even discover additional differential features not yet identified in current radiological practice [8]. However, such capabilities have not yet been fully demonstrated in the realm of HCC imaging. Most prior machine learning studies classified liver lesions on 2D CT slices and ultrasound images [9,10,11,12,13,14]. However, higher performance may be achieved with a model that analyzes 3D volumes of multi-phasic contrast-enhanced MRI, which is the reference standard for image-based diagnosis.
Therefore, this study aimed to develop a preliminary CNN-based DLS that demonstrates proof-of-concept for classifying six common types of hepatic lesions with typical imaging appearances on contrast-enhanced MRI, and to validate performance with comparison to experienced board-certified radiologists.
Materials and methods
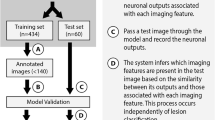
This was a single-center engineering development and validation study compliant with the Health Insurance Portability and Accountability Act and the Standards for Reporting of Diagnostic Accuracy guidelines. The study was approved by the institutional review board and informed consent was waived. The two components of the study involved (1) engineering a CNN-based liver tumor classifier, followed by (2) proof-of-concept validation of the final optimized CNN by comparison with board-certified radiologists on an identical unseen dataset. An overview of the model training and validation portions is illustrated in Fig. 1.
Establishment of “ground truth” cases
A medical student (CH) searched the picture archiving and communication system (PACS) for abdominal MRI examinations between 2010 and 2017 depicting one of the following hepatic lesions: simple cyst, cavernous hemangioma, focal nodular hyperplasia (FNH), HCC, intrahepatic cholangiocarcinoma (ICC), and colorectal cancer (CRC) metastasis. Due to the nature of a single-institution investigation with limited availability of pathological proof, lesions were restricted to those displaying typical imaging features, incorporating clinical criteria to maximize the certainty of definite diagnosis. Table S1 contains the selected criteria for the “ground truth” utilized for each lesion type. Diagnosed lesions formally described by radiology faculty on official reports were double-checked post hoc according to these criteria with another radiological reader (BL), and lesions were excluded if they contained discrepancies or displayed poor image quality. Up to three imaging studies per patient were included as long as studies were more than 3 months apart. Up to nine different lesions were used in each study. The majority of included lesions were untreated; treated lesions were only included if the selected lesion showed progression, or the patient underwent loco-regional therapy more than 1 year ago and now presented with residual tumor. Patients younger than 18 years were excluded.
MRI acquisition protocol
This study involved MRI examinations performed from 2010 to 2017 available throughout the institutional PACS, designed to include a heterogeneous collection of MRI scanners and imaging studies. This incorporated both 1.5-T and 3-T MR scanners, including Siemens Aera, Espree, Verio, Avanto, Skyra, and Trio Tim and GE Discovery and Signa Excite scanners. Multi-phasic contrast-enhanced T1-weighted breath-hold sequences from standard institutional liver MR imaging protocols were used with acquisition times of 12–18 s. Several different gadolinium-based contrast agents were used (dosed at 0.1 mmol/kg), including Dotarem (Guerbet), Gadavist (Bayer), Magnevist (Bayer), ProHance (Bracco Diagnostics), and Optimark (Covidien). Post-contrast images were analyzed, including late arterial phase (~ 20 s post-injection), portal venous phase (~ 70 s post-injection), and delayed venous phase (~ 3 min post-injection). Imaging parameters varied across different scanners and time frames; however, the majority were in the range of TR 3–5 ms, TE 1–2 ms, flip angle 9–13°, bandwidth 300–500 Hz, slice thickness 3–4 mm, image matrix 256 × 132 to 320 × 216, and field-of-view 300 × 200 mm to 500 × 400 mm.
Image processing
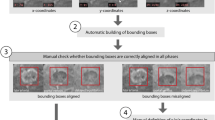
Eligible MRI studies were downloaded from the PACS and stored as DICOM files. The location and size of a 3D bounding box around the target lesion were manually recorded on the x-, y-, and z-axis. The images were processed and automatically cropped to show only the lesion of interest using code written in the programming language Python 3.5 (Python Software Foundation). The cropped image was then resampled to a resolution of 24 × 24 × 12 voxels (Fig. 2). To minimize bias field effects, cropped images were normalized to intensity levels from − 1 to 1. Affine registration with a mutual information metric was used to register portal venous and delayed phase MRI studies to the arterial phase. Ten lesions from each class were randomly selected to comprise the test set (12% of the entire dataset) using Monte Carlo cross-validation and the remaining lesions comprised the training set. Each image in the training set was augmented by a factor of 100 using established techniques [15] to increase the number of training samples, which allows the model to learn imaging features that are invariant to rotation or translation. During augmentation, images randomly underwent rotation, translation, scaling, flipping, interphase translation, intensity scaling, and intensity shifting.
Deep learning model development
The CNN model was trained on a GeForce GTX 1060 (NVIDIA) graphics processing unit. The model was built using Python 3.5 and Keras 2.2 (https://keras.io/) [16] running on a Tensorflow backend (Google, https://www.tensorflow.org/). Model engineering consisted of iteratively adjusting the network architecture (number of convolutional layers, pooling layers, fully connected layers, and filters for each layer, along with parameter optimization) and training cases (removing cases with poor imaging quality or ambiguous imaging features and increasing the number of training samples for lesion classes demonstrating lower performance). The final CNN consisted of three convolutional layers, where the first layer had 64 convolutional filters for each of the three phases in the original image, and the other two had 128 filters across all phases. Each filter generated filtered images by convolving voxels in 3 × 3 × 2 blocks. The model also contained two maximum pooling layers (size 2 × 2 × 2 and 2 × 2 × 1 respectively), which reduce the resolution of filtered images to provide spatial invariance (i.e., a feature that is shifted by a voxel can still be represented by the same neuron, which facilitates learning). The final CNN contained two fully connected layers, one with 100 neurons and the second with a softmax output to six categories that corresponded to the lesion types (Fig. 3). The selected imaging studies spanned 296 patients (155 male/141 female) (Table 1). A total of 334 imaging studies were selected, with a combined total of 494 lesions (74 cysts, 82 cavernous hemangiomas, 84 FNHs, 109 HCCs, 58 ICCs, 87 CRC metastases). The average diameter of all lesions used was 27.5 ± 15.9 mm, ranging from 21.7 ± 15.5 mm for simple cysts to 45 ± 16.8 mm for ICCs (Table 2). The CNN used rectified linear units after each convolutional layer and the first fully connected layer, which helps the model to learn non-linear features [15]. These are used in conjunction with batch normalization and dropout, which are regularization techniques that help the model to generalize beyond the training data [17]. Each CNN was trained with an Adam optimizer using minibatches of five samples from each lesion class. Hyperparameters were chosen via an exhaustive search through a manually specified portion of the search, an approach known in the literature as a grid search [18]. Samples were chosen randomly from the augmented dataset. The model was then tested on its ability to correctly classify 60 lesions in the test dataset (10 from each lesion class) and performance was averaged over 20 independent training iterations with different groupings of training and test datasets to gain a more accurate assessment.
Reader study validation
After development of the CNN model was complete, the classification accuracy of the final CNN was compared with two board-certified radiologists, using an identical set of randomly selected lesions that were unseen by either the CNN model or the radiologists. The two radiologists (39 and 7 years of experience) did not take part in the model training process and were blinded to the lesion selection. The reader study was conducted on an OsiriX MD (v.9.0.1, Pixmeo SARL) workstation. To provide even comparison of input data available to the CNN model, the simulated ready study contained several differences compared to actual clinical practice. The imaging studies were anonymized, and the radiologists were fully blinded to clinical data as well as MRI sequences not utilized for the CNN training. The test set for the reader study consisted of 10 randomly selected lesions of each class, 60 lesions in total, while the remaining lesions were assigned to the training set. The randomization was based on Monte Carlo cross-validation and the results of the reader study were compared after a single iteration to mimic their “first exposure” to the images. Each radiologist independently classified the 60 lesions characterized by the model in the test set based on the original three contrast-enhanced MRI phases (late arterial, portal venous, and delayed/equilibrium). Their performance was evaluated in distinguishing the six lesion entities as well as three broader categories that simulate the application of a deep learning model to an HCC diagnostic imaging framework such as LI-RADS. The three broader derived categories were HCCs (corresponding to LR-5), benign lesions (grouping cysts, hemangiomas, and FNHs, corresponding to LR-1), and malignant non-HCC lesions (grouping ICCs and CRC metastases, corresponding to LR-M). The radiologists did not scroll any further than the superior and inferior margins of the lesion in order to avoid revealing possible other lesions within the liver and thereby biasing the read. The time from opening the MRI phases until classification of the lesion was recorded.
Statistics
The performance of the model was evaluated by averaging the sensitivity, specificity, and overall accuracy over 20 iterations, as described above. For validation of the CNN with radiological readings, the performances of both the model and the radiologists were computed by evaluating sensitivity, specificity, and overall accuracy on the same single randomly selected test set of unseen cases. Prevalence-based parameters such as positive predictive value and negative predictive value were not applicable for this study. A receiver operating characteristic curve was plotted to compare the model and radiologist performance in identifying HCC masses.
Results
Deep learning model
The final CNN demonstrated a training accuracy of 98.7% ± 1.0 (8567/8680 volumetric samples) across six lesion types and 99.1% ± 0.7 (8602/8680) according to the three general derived LI-RADS categories (Table 3). The average test accuracy was 91.9% ± 2.9 (1103/1200) among individual lesions and 94.3% ± 2.9 (1131/1200) across the three broader categories. The time to initially train the DLS was 29 ± 4 min. Once the model was trained, the actual runtime to classify each lesion in the test dataset was 5.6 ± 4.6 ms.
For the 20 iterations, the average model sensitivity across the six lesion types was 92%, with an average specificity of 98% (Table 4). The model sensitivity for individual lesion types ranged from 89% (177/200) for CRC metastases to 99% (197/200) for simple cysts (Table 4). The corresponding model specificity for individual lesions ranged from 97% (965/1000) for ICC to 100% (1000/1000) for simple cysts. HCC lesions demonstrated a sensitivity of 94% (187/200) and specificity of 98% (984/1000). For the case of the three broader categories, the sensitivity ranged from 94% (187/200 for HCC, 563/600 for benign lesions) to 95% (381/400 for malignant non-HCC lesions). The corresponding specificity ranged from 96% (770/800 for malignant non-HCC lesions, and 577/600 for benign lesions) to 98% (984/1000 for HCC). The study was conducted using the same number of lesions from each class, and thus does not reflect the actual prevalence of each lesion type.
Reader study
Classification of unseen randomly selected lesions included in the reader study demonstrated an average model accuracy of 90% (55/60 lesions). Radiologist accuracy was 80% (48/60) and 85% (51/60) on these same lesions, respectively (Table 3). The model accuracy for the three broader categories was 92% (58/60), compared with 88% (53/60) for both radiologists. The total elapsed time analyzing each lesion was 0.8 ms for the classification model versus 14 ± 10 s and 17 ± 24 s for the radiologists.
Lesions included in the reader study showed an average CNN model sensitivity of 90% ± 14 (9/10) and specificity of 98% ± 2 (49/50) across the six lesion types. This compared to an average sensitivity of 80% ± 16 (8/10) and 85% ± 15 (8.5/10) and specificity of 96% ± 3 (48/50) 97% ± 3 (48.5/50) for the two radiologists respectively (Table 4). The model sensitivity ranged from 70% (7/10 for FNH) to 100% (10/10 for simple cysts and hemangiomas) with a specificity ranging from 92% (46/50 for HCC) to 100% (50/50 for simple cysts, hemangiomas, and ICC). Radiologist sensitivity ranged from 50% (5/10 for CRC metastases) to 100% (10/10 for simple cysts, hemangiomas), with specificity ranging from 92% (46/50 for CRC metastases) to 100% (50/50 for HCC and ICC). The average model sensitivity for three broader categories was 92% with a specificity of 97%. This compared to the radiologists’ sensitivity of 88% and specificity of 89% and 91%, respectively. The model demonstrated highest sensitivity for malignant non-HCC lesions at 95% (19/20) compared to 85% (17/20) for both radiologists, whereas radiologists attained highest sensitivity for benign lesions at 97% (29/30) and 100% (30/30), compared to 90% (27/30) for the CNN.
A receiver operating characteristic curve was constructed by varying the probability threshold at which the CNN would classify a lesion as HCC, with an area under the curve of 0.992 (Fig. 4). This included a true positive rate of 93.5% (187/200) averaged over 20 iterations and a false positive rate of 1.6% (16/1000). When including only lesions within the reader study, the model true positive rate was 90% (9/10), and the false positive rate was 2% (1/50). Radiologists had a true positive rate of 60% and 70% (6/10 and 7/10, respectively) and a false positive rate of 0% (0/50).
Discussion
This study demonstrates a deep learning–based prototype for classification of liver lesions with typical imaging features from multi-phasic MRI, demonstrating high performance and time efficiency. While the study did not simulate clinical practice conditions, comparison with equivalent data input showed the potential of DL systems to eventually aid in improving radiological diagnosis of six classes of hepatic lesions (model accuracy of 92%, radiologist accuracy of 80% and 85%), as well as three broader categories of benign, HCC, and malignant non-HCC lesions (model accuracy of 94%, radiologist accuracy of 88%), with a classification time of 5.6 ms per lesion.
Building upon prior 2D CT and ultrasound models, the inherent improved soft tissue contrast resolution of MRI can enable this CNN to capture a wider variety of imaging features [14]. Additionally, the 3D volumetric approach may improve detection of inhomogeneous growth or enhancement patterns that may be relevant to lesion classification, while removing the model’s variability and dependence on manual slice selection [19, 20]. Furthermore, the use of heterogeneous imaging sources demonstrated the robustness of DLS in the setting of different MRI scanners and acquisition protocols.
Previous studies have paved the way for computational classification of diverse lesion types by grouping hepatic lesion entities into three to five classes [11, 13, 14]. Moving towards clinical implementation, classification becomes increasingly challenging when lesions are ungrouped and single entities are differentiated. In this case, a higher number of differential features must be learned with a lower chance of guessing correctly. The present study included six ungrouped lesion classes, demonstrating a high accuracy level of 91.9%. As expected, the overall accuracy was higher with three grouped classes (94.3%).
Since single-center developmental efforts often suffer from limited datasets, selection of idealized cases is often necessary, making the interpretation of classification results ambiguous. The direct comparison between the DLS and two radiologists allows for better interpretation of performance and potential clinical value. High sensitivity for HCC and CRC metastases was demonstrated relative to radiologists. The radiologists tended to misclassify HCCs with faint enhancement as CRC metastases and HCCs with unclear washout as FNHs, whereas the DLS could more reliably make use of other features to correctly identify the HCCs. Similarly, radiologists misclassified CRC metastases without clear progressive enhancement with cysts, and those with heterogeneous, nodular appearances were misclassified for ICCs, whereas the computational predictions were likely more robust to the absence of these features. Still, the radiologists’ diagnostic accuracy may have matched or exceeded the DLS’s accuracy if given access to clinical information or additional imaging sequences. As a proof-of-concept study with limited sequences, this simulated environment provided unbiased comparison between the DLS and radiologists with the same available input data.
These performance metrics suggest that a DLS could serve as a quick and reliable “second opinion” for radiologists in the diagnosis of hepatic lesions, helping to reduce interpretation difficulty and inter-reader variability when imaging features are more ambiguous. In HCC diagnosis, most inter-reader studies demonstrated a moderate level of reliability in determining LI-RADS classes [21,22,23,24,25,26], and the rigor and complexity of LI-RADS constitutes a major barrier for broad adoption [25, 27]. The DLS classified lesions into benign, HCC, and malignant non-HCC lesions (roughly corresponding to LR-1, LR-5, and LR-M respectively) with an accuracy of 94.3%. While this is a preliminary feasibility study with many limitations, it suggests that a DLS could potentially interface with LI-RADS, for example, by averaging the model and radiologist predictions to score lesions that are suspicious for HCC but lack a definite benign/malignant appearance (i.e., LR-2/3/4). Such an implementation could reduce rote manual tasks, helping to simplify LI-RADS for clinical workflow integration [27].
While these results are promising, there are several limitations that make this a preliminary feasibility study. As a single-center investigation, only a limited number of imaging studies were available for each class. Thus, only lesions with typical imaging features on MRI were used, excluding lesions with more ambiguous features or poor image quality as well as more complex lesion types such as infiltrative HCC or complicated cysts. Additionally, LI-RADS is only applicable to patients at high risk for HCC. However, because non-HCC lesions are much less common in cirrhotic livers, this study also included lesions in livers without cirrhotic background or hepatitis-B/C, and thus the input does not identically conform to current consensus. Additionally, due to limited data from a single institution, pathological proof was not available for all lesions. Thus, “ground truth” criteria were carefully selected and defined for each lesion type as thoroughly outlined in Table S1. Notably, for lesions without pathological diagnosis, “ground truth” was established by analyzing all available clinical and imaging data, including T1 pre-contrast, T2, and other sequences. However, these sequences were not used in the model training and subsequent reader study, and thus their potential additive value for the CNN performance needs to be evaluated in further studies. Additionally, the simulated reader comparison did not reflect conditions in clinical practice, as the test set contained equal numbers of each lesion type and participants did not have access to ancillary information such as clinical data. However, this allowed for initial validation of the CNN with radiologists using the same conditions and input data for a more equivalent comparison. Within these limitations, this approach met the study’s purpose to demonstrate initial feasibility of a liver MRI lesion classification prototype from available data at one large academic medical center, providing motivation for the establishment of larger multi-institutional databases.
In summary, this preliminary study provides proof of principle for a DLS that classifies six hepatic lesion types on multi-phasic MRI, demonstrating high performance when validated by comparison with board-certified radiologists. As the demands of radiological practice continue to increase, a synergistic workflow that combines the experience and intuition of radiologists with the computational power of DL decision-support tools may offer higher-quality patient care in a time-efficient manner.
Abbreviations
- CNN:
-
Convolutional neural network
- CRC:
-
Colorectal carcinoma
- DL:
-
Deep learning
- DLS:
-
Deep learning system
- FNH:
-
Focal nodular hyperplasia
- HCC:
-
Hepatocellular carcinoma
- ICC:
-
Intrahepatic cholangiocarcinoma
- LI-RADS:
-
Liver Imaging Reporting and Data System
- PACS:
-
Picture archiving and communication system
- Sn:
-
Sensitivity
- Sp:
-
Specificity
References
El–Serag HB, Rudolph KL (2007) Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132:2557–2576
Wang H, Naghavi M, Allen C et al (2016) Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388:1459–1544
Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66:7–30
Mitchell DG, Bruix J, Sherman M, Sirlin CB (2015) LI-RADS (Liver Imaging Reporting and Data System): summary, discussion, and consensus of the LI-RADS Management Working Group and future directions. Hepatology 61:1056–1065
Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286:887–896. https://doi.org/10.1148/radiol.2017170706
Grewal M, Srivastava MM, Kumar P, Varadarajan S (2018) RADnet: radiologist level accuracy using deep learning for hemorrhage detection in CT scans2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp 281–284
Klöppel S, Stonnington CM, Barnes J et al (2008) Accuracy of dementia diagnosis—a direct comparison between radiologists and a computerized method. Brain 131:2969–2974
Greenspan H, Van Ginneken B, Summers RM (2016) Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35:1153–1159
Shiraishi J, Sugimoto K, Moriyasu F, Kamiyama N (2008) Computer-aided diagnosis for the classification of focal liver lesions by use of contrast-enhanced ultrasonography. Med Phys 35:1734–1746
Sugimoto K, Shiraishi J, Moriyasu F, Doi K (2010) Computer-aided diagnosis for contrast-enhanced ultrasound in the liver. World J Radiol 2:215
Hwang YN, Lee JH, Kim GY, Jiang YY, Kim SM (2015) Classification of focal liver lesions on ultrasound images by extracting hybrid textural features and using an artificial neural network. Biomed Mater Eng 26:S1599–S1611
Virmani J, Kumar V, Kalra N, Khandelwa N (2013) PCA-SVM based CAD system for focal liver lesions using B-mode ultrasound images. Def Sci J 63:478
Acharya UR, Koh JEW, Hagiwara Y et al (2018) Automated diagnosis of focal liver lesions using bidirectional empirical mode decomposition features. Comput Biol Med 94:11–18
Rajpurkar P, Irvin J, Ball RL et al (2018) Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 15:e1002686. https://doi.org/10.1371/journal.pmed.1002686
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. pp 1097–1105
Chollet F (2015) Keras. https://keras.io/. Accessed 15 Oct 2018
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:150203167
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint arXiv:14126980
Chapiro J, Lin M, Duran R, Schernthaner RE, Geschwind J-F (2015) Assessing tumor response after loco-regional liver cancer therapies: the role of 3D MRI. Expert Rev Anticancer Ther 15:199
Chapiro J, Wood LD, Lin M et al (2014) Radiologic-pathologic analysis of contrast-enhanced and diffusion-weighted MR imaging in patients with HCC after TACE: diagnostic accuracy of 3D quantitative image analysis. Radiology 273:746–758
Barth B, Donati O, Fischer M et al (2016) Reliability, validity, and reader acceptance of LI-RADS-an in-depth analysis. Acad Radiol 23:1145
Bashir M, Huang R, Mayes N et al (2015) Concordance of hypervascular liver nodule characterization between the organ procurement and transplant network and liver imaging reporting and data system classifications. J Magn Reson Imaging 42:305
Davenport MS, Khalatbari S, Liu PS et al (2014) Repeatability of diagnostic features and scoring systems for hepatocellular carcinoma by using MR imaging. Radiology 272:132
Ehman EC, Behr SC, Umetsu SE et al (2016) Rate of observation and inter-observer agreement for LI-RADS major features at CT and MRI in 184 pathology proven hepatocellular carcinomas. Abdom Radiol (NY) 41:963–969
Fowler KJ, Tang A, Santillan C et al (2018) Interreader reliability of LI-RADS version 2014 algorithm and imaging features for diagnosis of hepatocellular carcinoma: a large international multireader study. Radiology 286:173–185
Liu W, Qin J, Guo R et al (2017) Accuracy of the diagnostic evaluation of hepatocellular carcinoma with LI-RADS. Acta Radiol. https://doi.org/10.1177/0284185117716700:284185117716700
Sirlin CB, Kielar AZ, Tang A, Bashir MR (2018) LI-RADS: a glimpse into the future. Abdom Radiol (NY) 43:231–236
Funding
BL and CW received funding from the Radiological Society of North America (RSNA Research Resident Grant no. RR1731). JD, JC, ML, and CW received funding from the National Institutes of Health (NIH/NCI R01 CA206180).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Brian Letzen.
Conflict of interest
The authors of this manuscript declare relationships with the following companies: JW: Bracco Diagnostics, Siemens AG; ML: Pro Medicus Limited; JC Koninklijke Philips, Guerbet SA, Eisai Co.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• experimental
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 21.4 kb)
Rights and permissions
About this article
Cite this article
Hamm, C.A., Wang, C.J., Savic, L.J. et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol 29, 3338–3347 (2019). https://doi.org/10.1007/s00330-019-06205-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06205-9