Abstract
We previously developed an efficient deletion system for streptomycetes based on the positive selection of double-crossover events using bpsA, a gene for producing the blue pigment indigoidine. Using this system, we removed interfering secondary metabolite clusters from Streptomyces lividans TK24, resulting in RedStrep strains with dramatically increased heterologous production of mithramycin A (up to 3-g/l culture). This system, however, required a time-consuming step to remove the resistance marker genes. In order to simplify markerless deletions, we prepared a new system based on the plasmid pAMR18A. This plasmid contains a large polylinker with many unique restriction sites flanked by apramycin and kanamycin resistance genes and the bpsA gene for selecting a double-crossover event. The utility of this new markerless deletion system was demonstrated by its deletion of a 21-kb actinorhodin gene cluster from Streptomyces lividans TK24 with 30% efficiency. We used this system to efficiently remove the matA and matB genes in selected RedStrep strains, resulting in biotechnologically improved strains with a highly dispersed growth phenotype involving non-pelleting small and open mycelia. No further increase in mithramycin A production was observed in these new RedStrep strains, however. We also used this system for the markerless insertion of a heterologous mCherry gene, an improved variant of the monomeric red fluorescent protein, under the control of the strong secretory signal sequence of the subtilisin inhibitor protein, into the chromosome of S. lividans TK24. The resulting recombinant strains efficiently secreted mCherry into the growth medium in a yield of 30 mg/l.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gram-positive bacteria of the genus Streptomyces are one of the most important producers of biologically active secondary metabolites, including many antibiotics important in human medicine and agriculture. Antibiotic biosynthesis genes are generally clustered and regulated by specific transcriptional regulators that are located within these gene clusters (Hopwood 2007). Robust genomic sequencing of streptomycetes revealed a wealth of biosynthetic gene clusters for unknown secondary metabolites; however, many of them are silent under laboratory conditions. In order to identify new biologically active secondary metabolites, genetic manipulations are applied to activate these silent clusters or to improve their productivity (Baltz 2016). In addition to secondary metabolite production, some Streptomyces strains, like Streptomyces lividans, the most efficient one, have been used for the secretion of heterologous proteins. In general, the heterologous genes are fused to signal peptide sequences from highly secreted Streptomyces proteins to target them to the Sec pathway. These heterologous proteins are then efficiently secreted directly into the growth medium (Anne et al. 2012).
To increase production of secondary metabolites and enzymes, efficient techniques for deleting specific genes or gene clusters or for inserting heterologous genes into Streptomyces chromosomes are desired. Many methods based on the highly conserved process of homologous recombination (HR) have already been developed (Kieser et al. 2000). One of the most powerful is the PCR-targeting system, which has been widely used in Streptomyces. However, this system requires an ordered cosmid library for gene deletion, which may be an obstacle for Streptomyces strains that have not been well-characterised genetically (Gust et al. 2003). In general, these techniques are based on non-replicative or suicide vectors carrying the targeted DNA fragments flanking the gene(s) to be deleted together with an inserted antibiotic resistance marker gene. In addition, they also carry a second antibiotic resistance gene for selecting the integration of the vector into the chromosome by HR (a single-crossover event). Replacement of the desired gene(s) in the Streptomyces chromosome is subsequently accomplished by a double-crossover event, with selection for loss of the selectable marker (the second antibiotic resistance marker gene). These techniques are rather time consuming, however, and require investigating hundreds of clones. Inserting a suitable chromogenic selection marker gene into the suicide vector to distinguish between single-crossover and double-crossover mutants by producing colonies of different colours could simplify the procedure and significantly reduce the time needed for selection. The gusA chromogenic selection marker gene, encoding a β-glucuronidase, has already been used for a counterselection gene inactivation system in Streptomyces. However, it requires an external supply of the rather expensive substrate X-Gluc (Myronovskyi et al. 2014). Moreover, these genetic deletion strategies result in the replacement of the target gene(s) with a selectable resistance marker gene, which may have a polar effect on the expression of flanking genes. Several methods have already been established to remove the antibiotic marker gene, but they leave a scar sequence of about 50 bp in the Streptomyces chromosome (Fedoryshyn et al. 2008; Myronovskyi et al. 2014; Siegl and Luzhetskyy 2012). Like the HR-based methods, these techniques are also time consuming, requiring the investigation of hundreds of clones. Moreover, the scar sequence contains the integration FRT site for Flp recombinase, which contains a longer inverted repeat, which may affect neighbouring genes.
We previously established and optimised an easy gene deletion method based on selecting blue or white colonies for single- and double-crossover events using the bpsA gene, which encodes a protein for synthesising the blue pigment indigoidine (Knirschova et al. 2015). This method allowed double-crossover clones, which manifested white colonies, to be very easily selected from amongst hundreds of blue colonies (single-crossover clones), thereby enabling the fast and reliable deletion of several interfering secondary metabolite gene clusters in S. lividans TK24 to give several RedStrep-deleted strains. These were then used for the efficient, heterologous production of the anticancer compound mithramycin A in yields of up to 3 g/l. Unfortunately, this method required a laborious and time-consuming removal of the antibiotic resistance marker genes using the phiC31 recombinase (Novakova et al. 2018). In the present study, we prepared and optimised a new vector, pAMR18A (Fig. 1), for the easy and efficient screening of markerless deletions based on the selection of blue or white colonies. In addition, we also used it for the markerless insertion of a heterologous gene (mCherry) under the control of strong secretory regulatory elements into the chromosome of S. lividans TK24.
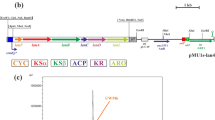
Restriction map of the plasmid pAMR18A. This plasmid contains the promoterless bpsA gene, the Tn5 (neo) kanamycin resistance gene, the aac3(IV) apramycin resistance gene with oriT origin of transfer and FRT regions from the plasmid pIJ773 (Gust et al. 2003), the E. coli ColE1 replication origin and the ampicillin resistance gene (bla) from pBluescript II SK+. The single restriction sites in the polylinker which can be used for cloning are in bold, red characters
Materials and methods
Bacterial strains, plasmids and culture conditions
S. lividans TK24 (Kieser et al. 2000) was used as wild-type strain for these studies. Its genomic sequence (GenBank Acc. No. CP009124) has been previously published (Rückert et al. 2015). Bennet solid medium (Horinouchi et al. 1983), MM and SFM solid media (Kieser et al. 2000) were used to propagate the strains and for selection of apramycin- and kanamycin-resistant clones after conjugation. Liquid inoculation medium (Knirschova et al. 2007), Bennet medium (Horinouchi et al. 1983), NMP and TSB media (Kieser et al. 2000) were used to check the growth and heterologous production of mCherry. SM17 production medium was used for the heterologous production of mithramycin A as recently described (Novakova et al. 2018). The growth of cultures was evaluated by measurement of dry mass of mycelium. Five-millilitre aliquots were taken from the culture at particular time points and filtrated through GF/C glass filters (Whatman). The filters were air-dried for 24 h and weighted. All cloning procedures were performed in E. coli DH5ɑ (ThermoFisher Scientific, Waltham, MA, USA), while E. coli ET12567/pUZ8002 was used for the conjugative transfer of plasmid DNA into S. lividans TK24 (Kieser et al. 2000). E. coli plasmid pBluescript II SK+ (Stratagene, La Jolla, CA, USA) was used for the E. coli cloning experiments and pmCherry (Clontech, Mountain view, CA, USA) was used as a template for PCR amplification of the mCherry gene. E. coli BL21(DE3) pLysS and the plasmid pET28a (Novagen, Madison, WI, USA) were used for mCherry overexpression. The plasmid pIJ773 containing the apramycin resistance gene aac3(IV) with oriT and FRT regions (Gust et al. 2003) was kindly provided by Dr. Bertold Gust, John Innes Centre, Norwich, UK. The conditions for E. coli growth and transformation were as described in Ausubel et al. (1995). When required, media were supplemented with 100 μg/ml ampicillin (Sigma-Aldrich), 50 μg/ml kanamycin (Sigma-Aldrich), 50 μg/ml apramycin (Sigma-Aldrich), 25 μg/ml chloramphenicol (Sigma-Aldrich) and 20 μg/ml nalidixic acid (Sigma-Aldrich).
DNA manipulations
DNA manipulations in E. coli were done as described in Ausubel et al. (1995). Nucleotide sequencing was performed with the ABI PRISM™ Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) and analysed on an Applied Biosystems model 373 DNA sequencer. Chromosomal DNA from all S. lividans TK24 strains was isolated according to Kieser et al. (2000). For Southern blot hybridization analysis, 1 μg of the isolated DNA was digested with the appropriate restriction endonuclease, separated by electrophoresis in a 0.8% (w/v) agarose gel in TAE and transferred to Hybond N (Amersham) as described in Ausubel et al. (1995). The membrane was hybridised with random-primed, DIG-labelled DNA probes (Roche, Mannheim, Germany) according to the manufacturer’s instructions. The probes were prepared by PCR amplification from the S. lividans TK24 chromosomal DNA using the primers indicated and PCR DIG Probe Synthesis Kit (Roche, Mannheim, Germany) at the conditions suggested by manufacturer.
Construction of pAMR18A
The plasmid pAMR4 (Knirshova et al. 2015), containing the promoterless bpsA gene, the kanamycin resistance gene from Tn5 (neo), the apramycin resistance gene aac3(IV) with oriT flanked by the FRT regions and polylinkers in the backbone of the E. coli plasmid pBluescript II SK+, was used as a source for the preparation of pAMR18A. First, the apramycin resistance cassette was removed as a 1.4-kb XbaI DNA fragment from pAMR4. Next, the same 1.4-kb XbaI fragment was inserted in the border XbaI site of the resulting plasmid, but in the opposite orientation from the kanamycin resistance gene, resulting in pAMR18A (Fig. 1). All cloning regions, together with a long polylinker containing many restriction sites for the rare restriction enzymes in Streptomyces, were confirmed by nucleotide sequencing.
Deletion of the act cluster in S. lividans TK24
To prepare a S. lividans TK24 markerless act cluster deletion mutant, two DNA fragments flanking the act cluster were PCR amplified from the S. lividans TK24 genomic DNA. High-fidelity Pfu DNA polymerase (Thermo Scientific) was used for each PCR cloning amplification at the conditions recommended by supplier. The first one, a 3-kb DNA fragment flanking the 3′ end of the SLIV_12925 gene (Fig. 2a), was amplified with the primers ActMfe and ActClaF (Table 1). This DNA fragment was digested using MfeI and ClaI. The second one, a 3-kb DNA fragment flanking the 3′ end of the SLIV_13030 gene (Fig. 2a), was amplified with the primers ActHind and ActClaR (Table 1). This DNA fragment was digested using HindIII and ClaI. Both DNA fragments were successively cloned into a pAMR18A digested with the same enzymes, resulting in pAMR18A-act (Fig. 2a). Both DNA fragments were verified by nucleotide sequencing. The plasmid was transformed into the non-methylating E. coli ET12567/pUZ8002 strain and introduced into S. lividans TK24 by conjugation (Kieser et al. 2000) in solid SFM medium. After 24 h, the plates were overlied with 1 ml sterile water with 1.25 mg/ml apramycin, 1.25 mg/ml kanamycin and 0.5 mg/ml nalidixic acid and grown for 7 days at 28 °C. A single blue colony (representing a single-crossover event) was picked and sporulated twice on solid Bennet medium. Spores were diluted and spread to Bennet plates to examine the blue and white colonies. The spores from 10 white colonies were dispersed in 100 μl of TE buffer, boiled 5 min and chilled on ice. A 1-μl aliquot was used as a template for a 20 μl PCR with 5 pmol of the primers Act1 and Act2 (Table 1) (Fig. 2b). The correct deletion of the act cluster in the positive colonies was further confirmed by Southern blot hybridization (Fig. 2c).
a Scheme of the markerless deletion of the actinorhodin act cluster in S. lividans TK24 using the plasmid pAMR18A-Act, which contains 3-kb DNA regions upstream and downstream of the act cluster cloned in pAMR18A (details in the materials and methods section). Thick arrows denote the direction and size of genes; red arrows represent the act genes. Gene labelling is based on the genomic sequence of S. lividans TK24 (GenBank Acc. No. CP009124). The thin red arrows below the map represent the position of the Act1 and Act2 primers used for PCR verification of the markerless deletion. The black bars below the maps represent the probe used for Southern blot hybridization analysis. Relevant restriction sites are indicated. A solid Bennet medium plate with colonies following two non-selective sporulations is shown to distinguish between double-crossover clones or wild-type revertant clones (white colonies) and single-crossover clones (blue colonies). b PCR analysis of ten randomly selected white clones for the markerless deletion of the act cluster using the primers Act1 and Act2 (Table 1). c Southern blot hybridization analysis of chromosomal DNA from these ten white clones. One microgram of DNA from the corresponding clone was digested with the restriction enzymes indicated, separated by electrophoresis in 0.8% (w/v) agarose gel and transferred on Hybond N (Amersham) as described in Ausubel et al. (1995). Hybridization followed the standard DIG protocol (Roche, Mannheim, Germany) using DIG-labelled probes. A Lambda DNA-BstEII digest was used as the size standard
Deletion of the matAB genes in S. lividans RedStrep deletion strains
To prepare a markerless matAB deletion mutant, a similar procedure was followed as for the act cluster deletion. Two, 2-kb DNA fragments flanking the matAB (SLIV_22885, SLIV_22890) genes were PCR amplified from the genomic DNA of S. lividans TK24. The first one, flanking the 5′ end of the SLIV_22885 gene (Fig. 3a), was amplified with the primers MatAMfe and MatABam (Table 1). This DNA fragment was digested using MfeI and BamHI. The second one, flanking the 3′ end of the SLIV_22890 gene (Fig. 3a), was amplified with the primers MatBHind and MatBAfl (Table 1). This DNA fragment was digested using HindIII and AflII. Both DNA fragments were successively cloned into a pAMR18A digested with the same enzymes, resulting in pAMR18A-matAB (Fig. 3a). Both DNA fragments were verified by nucleotide sequencing. The plasmid was introduced into S. lividans RedStrep 1.6 and 1.7 deletion strains (Novakova et al. 2018) in a similar way, by conjugation with selection for apramycin and kanamycin resistance. A similar procedure as above was used to identify a correct markerless deletion of matAB with the primer combinations 22880 + 22995r and 22880 + 22895r2. The correct matAB deletion was also confirmed by nucleotide sequencing of the PCR amplified DNA fragment (Fig. 3c).
a Scheme of the markerless deletion of the matAB operon (SLIV_22885, SLIV_22890) in S. lividans RedStrep 1.6 and 1.7 (Novakova et al. 2018) using the plasmid pAMR18A-matAB, which contains 2-kb DNA regions upstream and downstream of the matAB operon cloned in pAMR18A (details in the materials and methods section). Thick arrows denote the direction and size of genes; red arrows represent the matA and matB genes. Gene labelling is based on the genomic sequence of S. lividans TK24 (GenBank Acc. No. CP009124). The thin red arrows below the map represent the positions of the 22880, 22809r and 22895r2 primers used for PCR verification of the markerless deletion. b PCR analysis of ten randomly selected white clones from each of the strains used for conjugation of pAMR18A-matAB using the primer pairs 22880 + 22895r and 22880 + 22895r2 (Table 1). A Lambda DNA-BstEII digest was used as the size standard
Integration of the secretion ssi-mCherry fusion in S. lividans TK24
Likewise, to insert the ssi-mCherry fusion into the S. lividans TK24 chromosome, a procedure similar to that used for act cluster deletion was performed. Two, 2-kb DNA fragments flanking the subtilisin inhibitor ssi gene (SLIV_34120) were PCR amplified from the S. lividans TK24 genomic DNA. The first one, flanking the 5′ end of the SLIV_34120 gene with signal sequence and an inserted NheI fusion site (Fig. 5a), was amplified with the primers 34120Spe and 34120Nhe (Table 1). This DNA fragment was digested using SpeI and NheI. The second one, flanking the 3′ end of the SLIV_34120 gene (Fig. 5a), was amplified with the primers 34120Hind and 34120Afl (Table 1). This DNA fragment was digested using HindIII and AflII. The third 720-bp DNA fragment, containing the mCherry gene, was amplified from the pmCherry template plasmid with the primers mCherryNhe and mCherryNde (Table 1), to remove the mCherry start ATG codon and replace it with an in-frame NheI site. All three DNA fragments were successively cloned into a pAMR18A digested with the same enzymes, resulting in pAMR18A-secA2AM (Fig. 5a). All DNA fragments and fusions were verified by nucleotide sequencing. As before, the plasmid was introduced into S. lividans TK24 by conjugation with selection for apramycin and kanamycin resistance. A similar procedure as above was used to identify the correct markerless insertion of an ssi-mCherry fusion with primers 34120up and 34120down. The correct fusions were additionally confirmed by Southern blot hybridization (Fig. 5c).
Fluorescence measurement of intracellular and extracellular mCherry
Spores from the S. lividans ssi-mCherry and S. lividans TK24 strains were inoculated into 2-ml inoculation medium in 100-ml Erlenmayer flasks and grown for 24 h at 270 rpm and 28 °C. TSB medium of 20 ml was subsequently added and growth continued under the same conditions. After 24 and 42 h, 1-ml culture aliquots were taken and centrifuged for 2 min at 12,000×g. The supernatant was removed and designated the mCherry export fraction. The cell pellet was washed with ice-cold 0.9% NaCl to remove residual medium and thoroughly suspended in 1-ml buffer I (50 mM Na2HPO4/NaH2PO4 pH 7, 5 mM DTT, 0.1% Triton X100) to homogeneity, giving the intracellular mCherry fraction. One hundred-microlitre aliquots of both the export and intracellular fractions were added to a black 96-well plate, and dual fluorescence was measured with a Synergy microplate reader (BioTek, Winooski, VT, USA) at the optimal conditions for mCherry observation (Duellman et al. 2015): excitation wavelength/bandwith of 590/20 nm and emission wavelength/bandwith of 645/40 nm. The measured fluorescence units (FU) were normalised to 20 mg of mycelium dry mass. To measure the mycelium dry mass, 5-ml samples were taken from 24- and 42-h cultures, filtrated through GF/C glass filters, washed with water, dried at room temperature for 24 h and weighted. Twenty-microlitre aliquots of the exported fractions were resolved by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli 1970). Following electrophoresis, the proteins were visualised by Coomassie brilliant blue staining.
Overexpression of the mCherry gene in E. coli and protein purification
The mCherry gene was amplified by PCR using the plasmid pmCherry as a template and the primers mCherryDir and mCherryRev (Table 1), which introduced an NdeI site overlapping the translation initiation codon and a NotI site downstream of the stop codon. A 720-bp mCherry-containing DNA fragment was digested with NdeI and NotI and cloned into a pET28a cut with the same enzymes, resulting in pET-mCherry, which was verified by nucleotide sequencing. The host strain for pET series expression plasmids (E. coli BL21 (DE3) pLysS) transformed with the plasmid pET-mCherry was grown in LB medium containing 30 μg/ml chloramphenicol and 40 μg/ml kanamycin at 30 °C to an OD600 of 0.5. Expression was induced with 1 mM IPTG, and the cells were harvested after 3 h by centrifugation at 12,000×g for 10 min and washed with ice-cold 0.9% (w/v) NaCl. Cell lysis and native purification of His-tagged mCherry on His-Tag Bind resin (Novagen) were carried out as directed by the manufacturer. The eluted protein was dialysed overnight at 4 °C against the storage buffer (12.5 mM Tris-HCl pH 7.9, 60 mM KCl, 1 mM EDTA, 1 mM DTT, 50% (v/v) glycerol), cleared by centrifugation at 30,000×g for 10 min and stored at − 20 °C. The overproduction and purity of the eluted protein were confirmed by SDS-PAGE. A prominent band (Mr = 30,000) was identified after IPTG induction and isolated using His-Tag Bind resin (data not shown). This value corresponded to the calculated Mr of the His-tagged mCherry protein (28,903 Da). Protein concentration was determined according to Bradford (1976) with BSA as a standard. The His-tagged mCherry protein, after serial dilution in the storage buffer without glycerol, was used to determine a fluorescence calibration curve under the same conditions described above. Measurements were done in triplicate. In addition, 20-μl aliquots of serially diluted His-tagged mCherry protein were resolved by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualised by Coomassie brilliant blue staining.
Results
Preparation of the pAMR18A plasmid for markerless deletion of genes in streptomycetes
We previously developed an efficient deletion system for streptomycetes based on the positive selection of double-crossover events using bpsA, a gene for producing the blue pigment indigoidine. In this system, blue colonies indicate single-crossover recombination by the homologous recombination of the DNA fragments flanking the region to be deleted which is cloned in the plasmid pAMR4; uncoloured colonies indicate double-crossover events, resulting in the replacement of the deleted region by an antibiotic resistance marker gene (Knirschova et al. 2015). Using this system, we recently removed interfering secondary metabolite clusters from Streptomyces lividans TK24, thereby dramatically increasing the heterologous production of mithramycin A to as high as 3-g/l culture. However, this system required an additional step to remove the resistance marker gene using the phiC31 recombinase, which was both time consuming and left a short RR region (Novakova et al. 2018). In order to reduce the time needed for a particular deletion without leaving behind a resistance marker gene, we prepared a new system based on the pAMR18A plasmid. This system contains a large polylinker with many unique restriction sites flanked by apramycin and kanamycin resistance genes (used for single-crossover selection) and the bpsA gene for double-crossover selection (Fig. 1). To delete a gene of interest, two PCR-amplified DNA fragments from the flanking regions to the deleted gene(s) are cloned into the large polylinker of pAMR18A, leaving just a single restriction site between them. Because pAMR18A lacks a Streptomyces replication origin, it will be integrated into streptomycetes by the HR of one of the cloned Streptomyces DNA fragments. Following two rounds of non-selective sporulation, a double-crossover mutant or reverted wild-type colony can be easily selected based on the colony colour (blue or white), and subsequently examined by PCR to distinguish between these two events.
Deletion of the actinorhodin cluster in S. lividans TK24
We tested this new markerless deletion system by deleting the 22 genes of the act cluster, which is responsible for the biosynthesis of the blue-pigmented polyketide antibiotic actinorhodin in S. lividans TK24. DNA regions of 3 kb upstream and downstream of the act cluster were PCR-amplified and cloned using the unique restriction sites of the pAMR18A plasmid (Fig. 2a). The resulting construct, pAMR18A-Act, was conjugated into S. lividans TK24. About 1000 apramycin- and kanamycin-resistant blue colonies producing indigoidine were identified, indicating the successful integration of the recombinant plasmid into S. lividans TK24 through the homologous recombination of one region adjacent to the act cluster. A single blue colony was taken and sporulated twice on solid Bennet medium to ensure that either the double-crossover event or reversion to wild-type occurred. Ten randomly selected white and apramycin- and kanamycin-sensitive colonies, representing either act deletion mutants or wild-type revertants, were analysed by PCR using primers from the act flanking regions. Of the analysed clones, three were positive, containing the correct 500-bp amplified DNA fragment, thus indicating the correct double-crossover and markerless deletion of the act cluster (Fig. 2b). To additionally confirm the deletion of act, Southern blot hybridization analysis was performed with chromosomal DNA from these ten colonies using an act-flanking probe. All three positive clones contained the correctly deleted act cluster (Fig. 2c), indicating about 30% efficiency in double-crossover recombination in the selected white colonies. Actinorhodin is produced by S. lividans strains grown on minimal medium with glycerol as the sole carbon source (Kim et al. 2001). While it is consistently produced under these conditions by wild-type S. lividans TK24, no actinorhodin was detected in all three act-deleted S. lividans strains (data not shown).
Deletion of the matAB genes in S. lividans RedStrep deletion strains
Recently, using the deletion system based on the bpsA gene, we prepared a collection of S. lividans strains with several interfering secondary metabolite gene clusters sequentially deleted (S. lividans RedStrep 1.1 to 1.8), which dramatically increased their heterologous production of mithramycin A (Novakova et al. 2018). To further increase mithramycin production and improve the growth characteristics of these RedStrep strains, we used the new markerless pAMR18A-based system described here to remove the matA and matB genes in selected RedStrep strains. Deletion of matA or matB, which encode putative polysaccharide synthases, in S. lividans 66 was previously shown to affect mycelial aggregation, resulting in a highly dispersed growth phenotype with non-pelleting small and open mycelia. This biotechnologically improved Streptomyces strain had an increased overproduction of heterologously expressed genes (van Dissel et al. 2015). As in the case of act deletion, 2 kb DNA regions upstream and downstream of the matAB operon (SLIV_22885, SLIV_22890) were PCR-amplified and cloned using the unique restriction sites of the pAMR18A plasmid, resulting in pAMR18A-matAB (Fig. 3a), which was conjugated into S. lividans RedStrep 1.6 and 1.7 strains. We selected these strains as main representatives of RedStrep strains (Table 2), which also differ in the sporulation phenotype. The deletion of the cpk cluster in RedStrep 1.7 partially affected sporulation (Novakova et al. 2018). Likewise, ten randomly selected white and apramycin- and kanamycin-sensitive colonies were analysed by PCR using two primer combinations (22880 + 22895r and 22880 + 22895r2) from the matAB flanking regions, which will produce different length of amplicons. Four positive clones were found in both S. lividans RedStrep 1.6 and 1.7 strains, containing the correct 470 bp DNA fragment amplified with primers 22880 + 22895r and a 740 bp DNA fragment amplified with primers 22880 + 22895r2 (Fig. 3b). This indicated the correct double crossover leading to the markerless deletion of the matAB operon. In both strains, therefore, the efficiency of double crossover leading to deletion of the matAB operon was 40%. The correct deletion was confirmed by sequencing the PCR-amplified DNA fragment (data not shown).
When grown in liquid media, both new S. lividans strains RedStrep 1.9 and 1.10 (Table 2) showed dispersed growth with a non-pelleting phenotype, as previously described for the S. lividans 66 matA and matB mutant strains (van Dissel et al. 2015). Light microscopy clearly confirmed this phenotype. While the parent S. lividans RedStrep 1.6 strain grew as large pellets with an average diameter of about 150 μm, the pellets of both RedStrep 1.9 and 1.10 were much smaller and less compact, with a diameter of only about 30 μm (Fig. 4). Biomass measurements, however, showed that they grew comparably as their corresponding progenitor strains, S. lividans RedStrep 1.6 and 1.7 (data not shown). The heterologous production of mithramycin A was estimated in both matAB mutant strains after conjugation with the pMTMF plasmid in SM17 production medium, as described previously (Novakova et al. 2018); average mithramycin A production in eight pMTMF exconjugants of each strain was similar to both progenitor RedStrep 1.6 and 1.7 strains conjugated with the same plasmid (data not shown).
Liquid-culture morphology of the progenitor strain S. lividans RedStrep 1.6, and the matAB mutant strains S. lividans RedStrep 1.9 and 1.10. Strains were grown for 48 h at 30 °C and 250 rpm in 50-ml SM17 medium in 250-ml Erlenmayer flasks and analysed under a Leica DMi phase-contrast microscope. The scale bar represents 200 μm
Integration of the secretion fusion ssi-mCherry into S. lividans TK24
In addition to the deletion of genes in Streptomyces spp., the pAMR18A-based system can also be used for the efficient and stable markerless integration of foreign genes into any neutral position of the Streptomyces chromosome. Since S. lividans TK24 is a widely used host for heterologous protein secretion (Anne et al. 2012), we investigated the ability of this system to integrate a gene encoding an improved variant of the monomeric red fluorescent protein mCherry (Shaner et al. 2004) under the control of the promoter and secretory signal sequence of the subtilisin inhibitor protein (SSI) into the S. lividans TK24 chromosome. The mCherry protein has been shown to be secreted in an active form from E. coli and eukaryotic cells (Chen et al. 2005; Duellman et al. 2015). The SSI secretion signal from several Streptomyces strains was previously used for the efficient secretion of several heterologous proteins (Lammertyn et al. 1997; Sianidis et al. 2006; Taguchi et al. 1992), and determining the N-terminal sequence of the exported S. lividans SSI (SLYAPSALVLT) revealed the cleavage site of the SSI signal sequence (Lammertyn et al. 1997; Strickler et al. 1992).
In order to insert the ssi-mCherry fusion into the chromosome of S. lividans TK24, 2-kb DNA regions upstream and downstream of the S. lividans TK24 ssi gene (SLIV_34120) were PCR-amplified, fused with the mCherry gene through an NheI site inserted downstream of the ssi cleavage site and cloned into pAMR18A. The resulting plasmid, pAMR18A-secA2AM (Fig. 5a), was conjugated into S. lividans TK24. As above, a single blue apramycin- and kanamycin-resistant colony was selected for sporulation twice on solid Bennet medium, and ten randomly selected white, apramycin- and kanamycin-sensitive colonies were subsequently analysed by PCR using the primer combination 34120up + 34120down from the SLIV_34120 flanking region. Seven of the analysed clones contained a 575-bp amplified DNA fragment corresponding to the wild-type ssi allele and three clones lacked this DNA fragment (Fig. 5b). In order to confirm correct integration, a Southern blot hybridization analysis was performed with chromosomal DNA from these three colonies using the SLIV_34120-flanking probe. All three positive clones contained the correctly inserted ssi-mCherry fusion (Fig. 5c), indicating about 30% efficiency for the correct double crossover.
a Scheme of the markerless ssi-mCherry secretory fusion insertion in S. lividans TK24 using the pAMR18A-secA2AM plasmid, which contains 2-kb DNA regions upstream and downstream of the SLIV_34120 (ssi) gene fused with the mCherry gene through an inserted NheI site downstream of the ssi cleavage site and cloned into pAMR18A (details in the materials and methods section). The nucleotide sequence of the SLIV_34120 (ssi) signal sequence region and the ssi-mCherry fusion through the inserted NheI site are shown above the map. The inferred protein products are given in the single-letter amino acid code above the second position of each codon. The mCherry gene is inserted from the second codon (VSKGEE). The vertical arrow indicates the cleavage site of the secreted protein (Lammertyn et al. 1997; Strickler et al. 1992). Thick arrows denote the direction and size of genes. Gene labelling is based on the genomic sequence of S. lividans TK24 (GenBank Acc. No. CP009124). The red arrow represents the mCherry gene and the green arrow the position of the ssi gene. The thin bent green arrow denotes the position of the ssi promoter. The thin red arrows below the map represent the positions of the primers 34120up and 34120down used for the PCR verification of the markerless insertion of the ssi-mCherry fusion. The black bars below the maps represent the probe used for Southern blot hybridization analysis. The relevant restriction sites are indicated. b PCR analysis of ten selected white clones for the markerless ssi-mCherry secretory fusion insertion using the primers 34120up and 34120down (Table 1). c Southern blot hybridization analysis of chromosomal DNA from three positive clones. One microgram of DNA from the indicated clone was digested with the restriction enzymes indicated, separated by electrophoresis in 0.8% (w/v) agarose gel and transferred on Hybond N (Amersham) as described in Ausubel et al. (1995). Hybridization followed the standard DIG protocol (Roche, Mannheim, Germany) using DIG-labelled probes. A Lambda DNA-BstEII digest was used as the size standard
All three positive S. lividans ssi-mCherry strains (clones 3, 5 and 10) were phenotypically similar in growth and sporulation to S. lividans TK24 (data not shown). The intracellular and extracellular (exported) levels of mCherry from these three strains as well as the wild-type S. lividans TK24 strain were fluorescently measured. The control S. lividans TK24 strain had only low background fluorescence in both intracellular and extracellular fractions, but in all three S. lividans ssi-mCherry strains, the main part of the mCherry activity was exported (Fig. 6a). SDS-PAGE analysis of the medium (Fig. 6b) revealed a band of about 28 kDa, which is similar to the theoretical molecular weight of mCherry from the second amino acid (26,591). A previous report showed that the fluorescence of mCherry measured at the optimal wavelengths (590 nm/646 nm) is directly proportional to the amount of protein present, even after fusion to other proteins (Duellman et al. 2015). We therefore prepared a similar calibration curve using our purified mCherry protein under our measurement conditions and found that the relation remained linear up to 22.4 mg of mCherry (Fig. 6c, d). Based on this calibration curve, the estimated amount of secreted mCherry was 30 mg/l of growth medium.
a Intracellular and extracellular (export) activity of mCherry in the wild-type S. lividans TK24 and three S. lividans ssi-mCherry strains. The activity was estimated in three independent clones and expressed as fluorescence units (FU) in 100 μl of TSB medium and corresponding cell suspension per 20 mg dry weight. Values are means of three independent determinations; error bars show the standard deviation. b 12.5% SDS-PAGE gel from 20 μl of the TSB medium (extracellular secreted fraction) from selected strains grown in TSB medium. c Fluorescence intensity of a serially diluted, purified His-tagged mCherry protein. Data are means of three independent experiments. They were fit to a linear equation by regression giving the equations shown with the indicated R2 values. d 12.5% SDS-PAGE gel with 20 μl of serially diluted purified His-tagged mCherry protein
Discussion
In this paper, we demonstrate the utility of the bpsA gene, encoding the blue pigment indigoidine biosynthetic enzyme from S. aureofaciens CCM3239 (Novakova et al. 2010), for creating an efficient markerless system for the easy deletion from or insertion of genes into Streptomyces chromosomes, based on an easy distinction between blue single-crossover colonies and white double-crossover (or wild-type revertant) colonies. The utility of this system, based on the Streptomyces non-replicative plasmid pAMR18A, was demonstrated by the easy and efficient deletion of the 22 genes of the actinorhodin biosynthetic gene cluster act in S. lividans TK24. In this case, 30% of the white colonies possessed the correct markerless deletion of the act gene cluster.
Using the bpsA gene, we had previously developed an efficient deletion system for streptomycetes, where deleted genes were replaced by an antibiotic marker resistance gene, but a time-consuming procedure was required to remove this marker gene afterward and left a scar sequence of about 50 bp (Knirschova et al. 2015; Novakova et al. 2018). At the same time and independently, a similar blue-white deletion system, based on the homologous indigoidine biosynthetic gene idgS from S. lavendulae CGMCC 4.1386, was published by another group (Li et al. 2015). Comparing the abilities of both previous systems based on pAMR4 and the new system based on pAMR18A to delete the act cluster, we find that the newer system takes a month less and leaves no scar in the deleted region. This new pAMR18A-based system could therefore be easily and efficiently used for the markerless deletion of genes and even larger regions in Streptomyces.
Several counterselection gene inactivation systems for streptomycetes have already been developed (Dubeau et al. 2009; Gust et al. 2003; Kieser et al. 2000; Myronovskyi et al. 2014), but all have some restrictions, e.g., a requirement for an ordered cosmid library, specific mutated strains, or an external substrate supply. Furthermore, removal of the antibiotic marker gene in these systems requires the conjugation of an additional replicative plasmid containing a recombinase gene essential for removing the marker gene (Fedoryshyn et al. 2008; Myronovskyi et al. 2014; Siegl and Luzhetskyy 2012). Besides this additional, time-consuming step, it is possible that some Streptomyces strains may fail to accept these replicative plasmids. Similar problems may also arise when using the yeast I-SceI meganuclease recombination system for markeless deletion, which also requires the additional conjugation of a replicative plasmid with a Streptomyces codon-optimised I-sceI gene (Fernandez-Martinez and Bibb 2014; Siegl et al. 2010). Likewise, several of the methods reported previously for efficient Streptomyces deletions using an optimised CRISPR/Cas gene editing system (Cobb et al. 2015; Huang et al. 2015; Zheng et al. 2015) use additional replicative plasmids, which, again, may cause similar problems. In contrast, our new, easy and efficient pAMR18A-based markerless system does not have any additional specific requirements. The only restrictions might be its use in actinomycetes strains which possess homologues of bpsA. However, homologues of this gene are rather rare among streptomycetes (Li et al. 2015). In addition to our bpsA gene, only four other homologous genes have been reported: the idgS gene in S. lavendulae CGMCC 4.1386 (Li et al. 2015), the Sc-indC gene in S. chromofuscus ATCC 49982 (Yu et al. 2013), the SSHG_00313 gene in S. albus J1074 (Olano et al. 2014) and the bpsA gene in S. lavendulae ATCC 11924 (Takahashi et al. 2007). In all these cases, the gene was silent under laboratory conditions, meaning it is unlikely to interfere with the blue-white screening.
Using this new pAMR18A-based markerless deletion system, we were also able to delete the matAB operon (SLIV_22885, SLIV_22890), encoding two putative polysaccharide synthases, with similar efficiency (40%) in two selected S. lividans RedStrep strains which had previously been prepared by the deletion of interfering secondary metabolite gene clusters (S. lividans RedStrep 1.6 and 1.7), which had dramatically increased their heterologous production of mithramycin A (Novakova et al. 2018). This deletion should improve the biotechnological properties of the RedStrep strains, since matAB deletion in S. lividans 66 affected mycelial aggregation, giving the resulting mutant a highly dispersed growth phenotype with non-pelleting small and open mycelia together with an increased overproduction of heterologously expressed genes (van Dissel et al. 2015). Our new S. lividans RedStrep 1.9 and 1.10 strains with markerless deletions of matAB had likewise had a dispersed growth phenotype, but this deletion did not further increase mithramycin A production. These new mutated RedStrep strains may nevertheless have biotechnological potential as hosts for the heterologous overproduction of biotechnologically relevant proteins without interference from the coloured antibiotics actinorhodin and undecylprodigiosin.
Besides deletions, this pAMR18A-based system can also be used for the stable markerless insertion of heterologous genes into Streptomyces chromosomes. We tested this possibility with mCherry, a gene for an improved variant of the monomeric red fluorescent protein (Shaner et al. 2004). Since S. lividans TK24 is widely used as a host for heterologous protein secretion (Anne et al. 2012), we tested the insertion of mCherry under the control of the promoter and efficient secretory signal sequence of the subtilisin inhibitor protein (encoded by the ssi gene, SLIV_34120) into the chromosome of S. lividans TK24. The secretion signal for homologous ssi genes from several Streptomyces strains was previously used for the efficient secretion of several heterologous proteins (Lammertyn et al. 1997; Sianidis et al. 2006; Taguchi et al. 1992). In addition, the signal peptidase cleavage site in SSI has been determined in S. lividans TK24 (Lammertyn et al. 1997; Strickler et al. 1992). The correct integration of the ssi-mCherry fusion by double-crossover HR into S. lividans TK24 occurred with 30% efficiency, a level similar to that for the deletions. All resulting recombinant strains efficiently secreted active mCherry into the growth medium. Moreover, SDS-PAGE analysis of the medium revealed a band of about 28 kDa, which is similar to the theoretical molecular weight of mCherry from the second amino acid (26,591). This small difference might be caused by the partially aberrant mobility of mCherry. Therefore, these results confirm that mCherry is efficiently secreted from the S. lividans ssi-mCherry strain. This markerless system can therefore also be used to efficiently insert foreign genes into Streptomyces chromosomes. This pAMR18A-based system may also have a biotechnological impact, since the most common integration systems, based on Streptomyces phage and plasmid integrative elements (Kieser et al. 2000), integrate the whole integrative plasmid together with an E. coli replicon, integrase gene and a resistance marker gene. It is a significant biotechnological obstacle, as biotechnological production strains should avoid antibiotic resistance genes, and expression of the integrase gene and the E. coli plasmid replication gene may affect stability of constructs. Although these integrative systems are quite stable, some loss was still reported (Baltz 2012; Myronovskyi and Luzhetskyy 2013; Siegl et al. 2010; Siegl and Luzhetskyy 2012). We previously also found that E. coli origin of replication caused dramatic instability after integration in some Streptomyces chromosomes (Kormanec et al. 1993). In comparison with the integrative plasmids, our markerless system based on HR has several advantages:it is more stable and can integrate foreign DNA into any neutral chromosome position. This system could also be used to integrate biotechnologically relevant genes under the control of heterologous strong regulatory elements (promoters, RBS, secretory signal sequences). Since the bpsA homologous gene idgS has previously been found to be active in other actinomecetes (Li et al. 2015), this pAMR18A-based markerless system can reasonably be expected to also work in other actinomycetes.
In conclusion, we prepared a new efficient blue-white screening system for markerless deletions and stable integrations in Streptomyces chromosomes based on the plasmid pAMR18A containing blue pigment indigoidine biosynthetic gene bpsA. The system was verified by deletion of the actinorhodin gene cluster from S. lividans TK24 and matA and matB genes in selected S. lividans RedStrep strains. In addition, we used this system for the markerless insertion of the mCherry gene under the control of the subtilisin inhibitor protein secretory signal sequence into the chromosome of S. lividans TK24. The resulting recombinant strains efficiently secreted mCherry into the growth medium.
References
Anne J, Maldonado B, van Impe J, Mellart v, Bernaerts K (2012) Recombinant protein production and streptomycetes. J Biotechnol 158:159–167
Ausubel FM, Brent R, Kingston RE, Moore DO, Seidman JS, Smith JA, Struhl K (1995) Current protocols in molecular biology. Wiley, New York
Baltz RH (2012) Streptomyces temperate bacteriophage systems for stable genetic engineering of actinomycetes (and other organisms). J Ind Microbiol Biotechnol 39:661–672
Baltz RH (2016) Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Microbiol Biotechnol 43:343–370
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chen JC, Viollier PH, Shapiro L (2005) A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol 55:1085–1103
Cobb RE, Wang Y, Zhao H (2015) High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol 4:723–728
Dubeau M-P, Ghinet MG, Jacques P-E, Clermont N, Beaulieu C, Brzezinski R (2009) Cytosine deaminase as a negative selection marker for gene disruption and replacement in the genus Streptomyces and other actinobacteria. Appl Environ Microbiol 75:1211–1214
Duellman T, Burnett J, Yang J (2015) Quantitation of secreted proteins using mCherry fusion construct and a fluorescent microplate reader. Anal Biochem 473:34–40
Fedoryshyn M, Petzke L, Welle E, Bechthold A, Luzhetskyy A (2008) Marker removal from actinomycetes genome using Flp recpombinase. Gene 419:43–47
Fernandez-Martinez LT, Bibb MJ (2014) Use of the meganuclease I-SceI of Saccharomyces cerevisiae to select for gene deletions in actinomycetes. Sci Rep 4:7100
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the esquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 18:1541–1548
Hopwood DA (2007) Streptomyces in nature and Mecicine. In: The antibiotic makers. Oxford University Press, New York
Horinouchi S, Hara O, Beppu T (1983) Cloning of a pleiotropic gene that positively controls biosynthesis of A-factor, actinorhodin, and prodigiosin in Streptomyces coelicolor A3(2) and Streptomyces lividans. J Bacteriol 155:1238–1248
Huang H, Zheng G, Jiang W, Hu H, Lu Y (2015) One-step high-efficiency CRISPR/Cas9- mediated genome editing in Streptomyces. Acta Biochim Biophys Sin 47:231–243
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. The John Innes Foundation, Norwich
Kim E-S, Hong H-J, Choi C-Y, Cohen SN (2001) Modulation of actinorhodin biosynthesis in Streptomyces lividans by glucose repression of afsR2 gene transcription. J Bacteriol 183:2197–2203
Knirschova R, Novakova R, Feckova L, Timko J, Turna J, Bistakova J, Kormanec J (2007) Multiple regulatory genes in the salinomycin biosynthetic gene cluster of Streptomyces albus CCM 4719. Folia Microbiol 52:359–365
Knirschova R, Novakova R, Mingyar E, Bekeova C, Homerova D, Kormanec J (2015) Utilization of a reporter system based on the blue pigment indigoidine biosynthetic gene bpsA for detection of promoter activity and deletion of genes in Streptomyces. J Microbiol Methods 113:1–3
Kormanec J, Rezuchova B, Farkasovsky M (1993) Optimization of Streptomyces aureofaciens transformation and disruption of the hrdA gene encoding a homologue of the principal sigma factor. J Gen Microbiol 139:2525–2529
Laemmli UK (1970) Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680–685
Lammertyn E, Van Mellaert L, Schacht S, Dillen C, Sablon E, Van Broekhoven A, Anne J (1997) Evaluation of a novel subtilisin inhibitor gene and mutant derivatives for the expression and secretion of mouse tumor necrosis factor alpha by Streptomyces lividans. Appl Environ Microbiol 63:1808–1813
Li P, Li J, Guo Z, Tang W, han J, meng X, Hao T, Zhu Y, Zhang L, Chen Y (2015) An efficient blue-white screening based gene inactivation system for Streptomyces. Appl Microbiol Biotechol 99:1923–1933
Myronovskyi M, Luzhetskyy A (2013) Genome engineering in actinomycetes using site-specific recombinases. Appl Microbiol Biotechnol 97:4701–4712
Myronovskyi M, Rosenkranzer B, Luzhetskyy A (2014) Iterative marker excision system. Appl Microbiol Biotechnol 98:4557–4570
Novakova R, Núñez LE, Homerova D, Knirschova R, Feckova L, Rezuchova B, Sevcikova B, Menéndez N, Morís F, Cortés J, Kormanec J (2018) Increased heterologous production of the Antitumoral polyketide Mithramycin a by engineered Streptomyces lividans TK24 strains. Appl Microbiol Biotechnol 102:857–869
Novakova R, Odnogova Z, Kutas P, Feckova L, Kormanec J (2010) Identification and characterization of an indigoidine-like gene for a blue pigment biosynthesis in Streptomyces aureofaciens CCM3239. Folia Microbiol 55:119–125
Olano C, Garcia I, Gonzales A, Rodriguez M, Rozas D, Rubio J, Sanchez-Hidalgo M, Brana AF, Mendez C, Salas JA (2014) Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol 7:242–256
Rückert C, Albersmeier A, Busche T, Jaenicke S, Winkler A, Friethjonsson OH, Hreggviethsson GO, Lambert C, Badcock D, Bernaerts K, Anne J, Economou A, Kalinowski J (2015) Complete genome sequence of Streptomyces lividans TK24. J Biotechnol 199:21–22
Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572
Sianidis G, Pozidis C, Becker F, Vrancken K, Sjoeholm C, Karamanou S, Takamiya-Wik M, van Mellaert L, Schaefer T, Anne J, Economou A (2006) Functional large-scale production of a novel Jonesia sp. xyloglucanase by heterologous secretion from Streptomyces lividans. J Biotechnol 21:498–507
Siegl T, Luzhetskyy A (2012) Actinomycetes genome engineering approaches. Antonie Van Leeuwenhoek 102:503–516
Siegl T, Petzke L, Welle E, Luzhetskyy A (2010) I-SceI endonuclease: a new tool for DNA repair studies and genetic manipulations in streptomycetes. App Microbiol Biotechnol 87:1525–1532
Strickler JE, Berka TR, Gorniak J, Fornwald J, Keys R, Rowland JJ, Rosenberg M, Taylor DP (1992) Two novel Streptomyces protein protease inhibitors. J Biol Chem 267:3236–3241
Taguchi S, Maeno M, Momose H (1992) Extracellular production system of a heterologous peptide driven by a secretory protease inhibitor of Streptomyces. Appl Microbiol Biotechnol 36:749–753
Takahashi H, Kumagai T, Kitani K, Mori M, Matoba Y, Sugiyama M (2007) Cloning and characterization of a Streptomyces single module type non-ribosomal peptide synthetase catalyzing a blue pigment synthesis. J Biol Chem 282:9073–9081
van Dissel D, Claessen D, Roth M, van Wezel GP (2015) A novel locus for mycelial aggregation forms a gateway to improved Streptomyces cell factories. Microb Cell Factories 14:44
Yu D, Xu F, Valiente J, Wang S, Zhang J (2013) An indigoidine biosynthetic gene cluster from Streptomyces chromofuscus ATCC 49982 contains an unusual IndB homologue. J Ind Microbiol Biotechnol 40:159–168
Zheng H, Wen S, Xu W, He Z, Zhai G, Liu Y, Deng Z, Sun Y (2015) Highly efficient editing of the actinorhodin polyketide chain length factor gene in Streptomyces coelicolor M145 using CRISPR/Cas9-CodA(sm) combined system. Appl Microbiol Biotechnol 99:10575–10585
Acknowledgments
We are grateful to Jacob Bauer for critically reading the manusrtipt and correcting the English. We are also grateful to Dr. Bertold Gust (John Innes Centre, Norwich, UK) for the pIJ773 plasmid.
Funding
This work was supported by the Slovak Research and Development Agency under contract No. APVV-15-0410 and by VEGA grant 2/0002/16 from the Slovak Academy of Sciences. The research leading to these results has received funding from the European Commission’s Seventh Framework Programme (FP7/2007-2013) under the grant agreement STREPSYNTH (project No. 613877). This work was co-funded by the Slovak Research and Development Agency under contract No. DO7RP-0037-12.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Rezuchova, B., Homerova, D., Sevcikova, B. et al. An efficient blue-white screening system for markerless deletions and stable integrations in Streptomyces chromosomes based on the blue pigment indigoidine biosynthetic gene bpsA. Appl Microbiol Biotechnol 102, 10231–10244 (2018). https://doi.org/10.1007/s00253-018-9393-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9393-7