Abstract
Unraveling elevational diversity patterns of plants and animals has long been attracting scientific interests. However, whether soil microorganisms exhibit similar elevational patterns remains largely less explored, especially for functional microbial communities, such as ammonia oxidizers. Here, we investigated the diversity and distribution pattern of ammonia-oxidizing archaea (AOA) in meadow soils along an elevation gradient from 4400 m to the grassline at 5100 m on the Tibetan Plateau using terminal restriction fragment length polymorphism (T-RFLP) and sequencing methods by targeting amoA gene. Increasing elevations led to lower soil temperature and pH, but higher nutrients and water content. The results showed that AOA diversity and evenness monotonically increased with elevation, while richness was relatively stable. The increase of diversity and evenness was attributed to the growth inhibition of warm-adapted AOA phylotypes by lower temperature and the growth facilitation of cold-adapted AOA phylotypes by richer nutrients at higher elevations. Low temperature thus played an important role in the AOA growth and niche separation. The AOA community variation was explained by the combined effect of all soil properties (32.6%), and 8.1% of the total variation was individually explained by soil pH. The total AOA abundance decreased, whereas soil potential nitrification rate (PNR) increased with increasing elevations. Soil PNR positively correlated with the abundance of cold-adapted AOA phylotypes. Our findings suggest that low temperature plays an important role in AOA elevational diversity pattern and niche separation, rising the negative effects of warming on AOA diversity and soil nitrification process in the Tibetan region.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Elevation gradients are one of the most powerful natural experiments to test ecological and evolutionary theories (Koerner 2007), because environmental factors including temperature and precipitation experience substantial change over short spatial distance. Elevational patterns of plants and animals have been extensively explored over the last decades and provided very useful insights into their diversity, distribution, and driving environmental factors (Lomolino 2001). The findings showed that macroorganisms generally exhibit mid-elevation peak or monotonic patterns (Bertuzzo et al. 2016; Lomolino 2001; Novillo and Ojeda 2014). Another elevational pattern was empirically observed in birds in east Andean slope, showing that maximum diversity occurs across lower elevations and then a sharp decrease occurs at the transition from the mid-elevation to high elevations (Herzog et al. 2005).
In contrast, microorganism elevational pattern received few attention, although they play a key role in maintaining biogeochemistry of carbon and nitrogen in soils (McGradySteed et al. 1997). Few existing studies on microorganisms revealed two general elevational patterns, including mid-elevation peak (Singh et al. 2012, 2016) and monotonic decrease (Bryant et al. 2008; Heger et al. 2016; Wang et al. 2015). It was observed that microorganisms (prokaryotes and eukaryotes) did not demonstrate clear patterns along elevation gradients, whereas plants or animals significantly decreased (Fierer et al. 2011; Shen et al. 2014). A recent study demonstrated that ectomycorrhizal fungal diversity did not change with elevation, but their community compositions significantly shifted (Jarvis et al. 2015). These studies have clued that the elevational diversity patterns of microorganism were driven by plant vegetation (Matsuoka et al. 2016), space (Liu et al. 2015a, b), geomorghy (Bertuzzo et al. 2016), climate, and a series of soil factors, particularly pH (Shen et al. 2013; Siles and Margesin 2016; Yashiro et al. 2016). Additionally, it was observed that different microbes exhibited diverse or even reverse patterns along elevations, and the elevational pattern is dependent of microbial groups. For instance, bacteria exhibited a mid-elevation peak, whereas fungi did not show a clear elevational diversity pattern (Meng et al. 2013). Even different Acidobacteria taxa showed contrasting diversity patterns along an elevation gradient (Yashiro et al. 2016). The previous studies focus on general microbial communities, including bacteria, archaea, and fungi; however, the functional microbial diversity pattern along elevations remains poorly studied, such as ammonia-oxidizing microbes (Singh et al. 2016; Yang et al. 2014).
Ammonia oxidation is the rate-limiting step of aerobic nitrification (Kowalchuk and Stephen 2001) and is performed by ammonia-oxidizing archaea (AOA) and bacteria (AOB) (Prosser and Nicol 2008; Treusch et al. 2005; Venter et al. 2004). AOA have a higher affinity to ammonium and are well adapted to low-pH pressure (Martens-Habbena et al. 2009). AOA thus have widely been observed to be the dominant ammonia oxidizers and play a key role in nitrification in acidic (Gubry-Rangin et al. 2010; Zhang et al. 2012; Prosser and Nicol 2012) and Arctic soils (Alves et al. 2013). In contrast, AOB are frequently detected in neutral or alkaline and nitrogen-rich soils (Di et al. 2009; Jia and Conrad 2009). The existing AOA elevational findings showed that both AOA abundance and community structure changed as altitude ascending (Yuan et al. 2015; Zhang et al. 2009), but their elevational diversity patterns have not been articulated. Our previous study showed that Tibetan meadow soils are acidic, with a pH range from 5.1 to 7.1 (Guo et al. 2015); thus, the AOA population was chosen as a model to explore the elevational pattern of functional microbes.
Tibetan Plateau is the highest plateau (>4000 m above sea level, a.s.l.) on Earth and is characterized by harsh conditions including low temperature, high UV, drought, and low oxygen. The harsh conditions result in a unique and fragile ecosystem hosting particular plants, animals, and microbes, which is highly sensitive to global climate change. In this study, we investigated the AOA elevational diversity pattern and their driving factors in meadow soils along an elevation gradient from 4400 m to the grassline at 5100 m on Tibetan Plateau. The elevation gradient exhibited less heterogenity in vegetation community compositions (Wang et al. 2013) and could serve as a perfect model to test microbial elevational pattern by minimizing the effects of vegetation compositions. Our previous findings have demonstrated that soil properties, including pH, water content, temperature, and soil nutrients, significantly changed with the elevation gradient (Guo et al. 2015). We hypothesized that (1) the AOA diversity and abundance would change along the elevation gradient, (2) AOA community structure should shift according to the gradient of environmental factors, and (3) specific AOA phylotypes could inhabit the harsh environments and exhibit different elevational patterns. This study provides fundamental knowledge on the elevational diversity pattern and distribution of soil AOA along an elevation to the grassline where vegetation growth is constrained by harsh environments.
Materials and methods
Study sites and soil
Soil samples were collected at five elevations (4400, 4650, 4800, 4950, and 5100 m, a.s.l.), along the south-facing slope of Nyaiqentanglha Mountains (30° 30′ N, 91° 03′ E). The vegetation and environmental factors in this location have been extensively investigated and showed a gradual increase in vegetation biomass (Wang et al. 2013). Briefly, average precipitation ranged from 227 to 420 mm, and the mean annual temperature dropped from 10.1 to 4.3 °C with increasing elevations. Stipa capillacea and Stipa purpurea dominated the plant communities at the low elevations and Kobresia pygmaea at high elevations. At each elevation, bulk soils were collected in triplicate in August 2012. From each replicate site, seven soil cores (0–10 cm in depth) were collected and mixed. Soil samples were transported to the laboratory in coolers with ice bags. Roots and stones were removed, and soils were passed through a 2.0-mm soil sieve. Soils subsampled for DNA extraction were stored at −80 °C, and the remaining soils were air-dried for soil physicochemical analyses.
Soil DNA extraction
Total DNA was extracted from 0.5 g of soil using the FastDNA® SPIN Kit for Soil (MP Biomedicals, CA, USA) according to the manufacturer’s instruction with few modifications (Kong and Nakatsu 2010). A NanoDrop® ND-2000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE) was used to assess the purity and concentration of the DNA extracted from soil.
qPCR of archaeal amoA gene
The archaeal amoA gene abundance was quantified using the primer set Arch-amoAF/Arch-amoAR, which was designed to specifically target AOA amoA gene (Francis et al. 2005). Quantitative PCR (qPCR) was conducted using SYBR Green Premix (Takara, Japan) in a LightCycler 480II thermocycler (Roche, Switzerland) with a temperature program of denaturation at 94 °C for 1 min and followed by 40 cycles of 30 s at 95 °C, 30 s at 55 °C, and 1 min at 72 °C, and fluorescence intensity was collected at 72 °C. A standard curve was generated using plasmid clones containing the target amoA gene fragments (Kong and Nakatsu 2010). To determine the melting temperature and PCR product specificity, a melting curve was acquired by heating from 50 to 95 °C.
T-RFLP analysis of AOA
The AOA community structure based on amoA gene was screened using the T-RFLP method. The PCR for amoA in AOA was performed in triplicate using Takara Master Mix (Takara, Japan) with a temperature program of 25 cycles of 94 °C for 1 min, 54 °C for 1 min, and 72 °C for 1 min. The primer set was the same as the above, with the exception that the forward primer Arch-amoAF was labeled at the 5′ end with 6-carboxy-fluorescein (FAM). The labeled PCR product with target amoA fragment was analyzed on an agarose gel; the band was cut from the gel and purified (Axygen, CA, USA). An amount of 200 ng of the purified product was digested at 37 °C for 3 h using the restriction enzyme MboI with the digestion site at ^GATC. The digested products were purified using Sigma-Aldrich Spin Post-Reaction Clean-Up columns (Sigma, USA), and a portion was mixed with deionized formamide and the internal standard GeneScan-1000 LIZ (Applied Biosystems, USA). The mixtures were denatured for 3 min at 95 °C, and the DNA fragments were size separated using a 3130xl Genetic Analyzer (Applied Biosystems, USA). T-RFLP profiles of each sample were determined using the GeneScan analysis software (Applied Biosystems, USA). The fragments between 50 and 640 bp and the peak areas of the terminal restriction fragments (T-RFs) >1% in all three replicates were included for community structure analysis.
Cloning, sequencing, and phylogenetic analysis of archaeal amoA gene
To identify the AOA present in the alpine meadow soils, clone libraries for amoA fragments were constructed from two soils at 4400 and 5100 m a.s.l. PCR products were amplified using the same primer set and conditions as for T-RFLP without the FAM label. The purified PCR products were ligated into the pGEM-T Easy Vector System I (Promega, Madison, WI, USA) and transformed into Escherichia coli JM109 competent cells according to the manufacturer’s instructions. A total of 98 positive clones were randomly selected and sequenced using an ABI model 3730xl DNA analyzer (Applied Biosystems, CA). Multiple sequence alignments were performed using CLUSTALW in MEGA6.0 (Tamura et al. 2013), and sequences sharing 97% sequence similarity were classified into operational taxonomic units (OTUs) using Mothur v.1.33.3 (Schloss et al. 2009). BLASTn (www.ncbi.nlm.nih.gov/BLAST/) was employed to search GenBank for the nearest related sequences for each OTU. Phylogenetic tree was constructed using the neighbor-joining method with a bootstrap with 1000 iterations in MEGA6.0. Sequences generated in this study have been deposited in the National Center for Biotechnology Information GenBank under accession numbers from KP984479 to KP984498. These sequences were subjected to virtual digestion and matched to distinct T-RFs by searching the enzyme digestion site (^GATC), which allowed identifying taxonomic affiliations for all the T-RFs (Ives et al. 2004).
Measurement of potential nitrification rate
Potential nitrification rate (PNR) was measured using the chlorate inhibition method as described previously (Chen et al. 2014; Kurola et al. 2005). Briefly, 5.0 g of fresh soil was added to 50-ml centrifuge tube containing 20 ml of phosphate buffer solution (PBS) (NaCl, 8.0 g l−1; KCl, 0.2 g l−1; Na2HPO4, 0.2 g l−1; NaH2PO4, 0.2 g l−1; pH 7.4) with 1 mM (NH4)2SO4. Potassium chlorate at a final concentration of 10 mM was added to the tube to inhibit nitrite oxidation. The suspension was incubated at 25 °C for 24 h in the dark, and the nitrite was extracted with 5 ml of 2 M KCl and reacted with N-(1-naphthyl)ethylenediamine dihydrochloride. The optical density was determined spectrophotometrically at 543 nm (SpectraMax M5, Molecular Devices, USA). A standard curve produced from KNO2 in a range of 0–100 μg l−1 was used to calculate the PNR in the samples.
Statistical analysis
The significant difference level of AOA abundance along the elevation gradient was measured by ANOVA using the software SPSS version 19.0 (IBM Inc., USA). To visualize the shift of AOA community structure, the relative abundance of all T-RFLP peaks was used for canonical correspondence analysis using CANOCO 5 (Microcomputer Power, Ithaca, NY). Absolute abundance of each T-RF, which was used to examine the relationships with elevations, was calculated by multiplying the relative abundance of the T-RF and total AOA gene abundance. To examine the correlations of AOA community structure with soil properties and geographical distance, a Mantel test was employed in vegan package in R (version 3.2.2). The spatial geographical coordinates and elevations at each sampling site were recorded by a GPS (eTrex Venture, Garmin, USA), and the pairwise geographical distance between sites was calculated using the Vegan package (dist() function) in R. Variation partition analysis (VPA) was performed to determine the individual contribution of each environmental factor by partitioning the effect of co-correlated factors on the microbial community variation using CANOCO 5 (Guo et al. 2015). Diversity (inverse Simpson index), evenness (Simpson index), and richness were calculated from T-RFLP profiles in R; species richness was indicated by the number of T-RFs.
Results
Soil properties and AOA abundance along the elevation gradient
Soil properties substantially changed with increasing elevations and have been detailed in our previous study (Guo et al. 2015). Briefly, soil pH (ranging from 5.1 to 7.1), soil temperature (5.7 to 9.7 °C), and nitrate concentration (0.24–0.98 mg kg−1 dry soil) decreased with elevations, whereas total organic carbon (TOC, 16–120 g kg−1 dry soil), ammonium (2.0–30.0 mg kg−1 dry soil), and soil water content (10–85%) increased.
Abundant AOA were observed in all the soils (Fig. 1) and gradually but significantly decreased from 4400 m (4.2 × 108 copies g−1 dry soil) to the grassline elevation at 5100 m (1.6 × 108 copies g−1 dry soil), and the highest decrease occurred at the transition from 4800 to 4950 m (2.4-fold). The AOA abundance positively correlated with soil temperature and pH and negatively correlated with TOC, ammonium, and water content (P < 0.001, Table 1).
AOA diversity and evenness along the elevation gradient
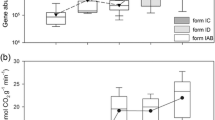
A total of seven distinct T-RFs were detected across all the meadow soils, including T-RFs of lengths 73, 136, 329, 391, 403, 421, and 443 bp (Fig. S4). The AOA diversity, evenness, and richness indexes were calculated based on all detected T-RFs. Diversity exhibited a monotonic increase pattern with elevation (Fig. 2a), and evenness followed the elevational diversity pattern (Fig. 2b). In contrast, richness indicated by the number of T-RFs was relatively stable along the elevations, except a peak at the mid-elevation 4800 m (Fig. 2c). Diversity and evenness significantly correlated with all the measured soil properties (R = 0.1047, P = 0.039, Fig. S1A) rather than geographical distance (Fig. S1B). In contrast, richness did not show any significant correlations with either soil properties or geographical distance. To further elucidate how the AOA diversity and evenness were driven by individual soil properties, Pearson correlations were calculated. The results showed that both the diversity and evenness positively correlated with soil TOC, water content, and ammonium and negatively correlated with soil temperature and pH (Table 1 and Fig. S2).
AOA community structure shift along the elevation gradient
The AOA community structure based on all the detected T-RFs was analyzed using canonical correspondence analysis and revealed three separate community clusters along the elevation gradient (Fig. 3). The three separate clusters were the AOA communities at 4400 m, the communities at 4650 and 4800 m, and the communities at 4950 and 5100 m. Greater shifts occurred at the transitions from 4400 to 4650 m and from 4800 to 4950 m. The Mantel test revealed that the AOA community structure significantly correlated with both soil physicochemical properties (pH, TOC, ammonium, nitrate, soil water, and temperature) (R = 0.59, P = 0.001, Fig. S3A) and geographical distance (R = 0.56, P = 0.001, Fig. S3B). VPA further illuminated that soil pH, water content, and temperature could individually explain the AOA community variation by 8.1, 5.3, and 3.1%, respectively, which were much lower than the combined explanation of all the soil properties (32.6%, Table 2).
The T-RFs of length 329, 391, 421, and 443 bp were detected in all soil samples, while the T-RF 136 bp was only detected at the lower elevations (4400 to 4800 m), and the T-RF 403 bp was detected at middle and higher elevations from 4650 to 5100 m (Fig. S4). Significant linear correlations were observed between six specific T-RFs and the elevation (Fig. S5), confirming that the AOA community structure variation was caused by the shift of specific T-RFs along the elevation gradient (Fig. 3). Among the six specific T-RFs, four T-RFs (136, 329, 391, 421 bp) significantly decreased with increasing elevations, whereas T-RFs 403 and 443 bp significantly increased. Particularly, the T-RF 421 bp exhibited a sharp decrease from 2 × 108 gene copies/g dry soil at 4400 m to 5 × 107 gene copies/g dry soil at 4650 m and the T-RF 136 was absent in the soils of two highest elevations (4950 and 5100 m). The four T-RFs (136, 329, 391, and 421 bp) exhibiting a monotonic decrease pattern were identified to be affiliated with the phylotypes retrieved from lake sediments and soils in Yunnan Plateau, Southern China, or Arizona, which are originally warmer than the yearly-cold Tibetan Plateau (Fig. 4). The two T-RFs (403 and 443 bp) exhibiting the monotonic increase pattern were identified to be affiliated with the phylotypes retrieved from yearly-cold indigenous Kobresia meadow soils on the Tibetan Plateau. Thus, the overall shift of AOA community structure was attributed to the two elevational patterns of specific AOA phylotypes, which were named warm- and cold-adapted, respectively. Pearson correlations showed that the abundances of three warm-adapted AOA phylotypes, including the specific T-RFs 134, 391, and 423 bp, positively correlated with soil temperature and pH (P < 0.01, Table S1). In contrast, the abundances of the two cold-adapted AOA phylotypes T-RFs positively correlated with soil ammonium concentration and TOC (P < 0.01).
Neighbor-joining phylogenetic tree of archaeal amoA gene fragment sequences (635 bp) retrieved from alpine meadow soils. The GenBank accession number was listed in brackets and was followed by specific terminal restriction fragment. Bootstrap values (>50) were shown at branch points. Bar, 0.05 substitutions per nucleotide position
Phylogenetic analysis of archaeal amoA gene
A total of 20 OTUs were retrieved from soils at two elevations (4400 and 5100 m), and all shared high sequence similarity with Nitrososphaera gargensis, which was originally isolated from soil (Fig. 4). These sequences, assigned into four clusters, all fell within soil and sediment group I.1b, one of the two typical known AOA groups. OTU1 belonging to cluster 2 dominated to the AOA community at 4400 m (60% in relative frequency) and shared high sequence similarity to paddy soil clones originally observed in warmer regions (Fig. S6). In contrast, OTU2 belonging to cluster 1 dominated the AOA community at 5100 m (40% in relative frequency) and shared high sequence similarity to clones originally observed in yearly-cold alpine meadow soil on the Tibetan Plateau. Comparison of cloning libraries retrieved from the soils at the two different elevations demonstrated that the OTUs distributed at 5100 m were more cold-adapted or cold-tolerant than those at 4400 m. Therefore, the clone libraries further confirmed the substantial shift of AOA phylotypes along the elevation gradient as described above.
Soil PNR along the elevation gradient
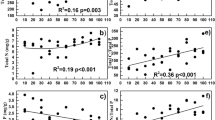
PNR was unchanged from 4440 to 4650 m and then significantly increased along the increasing elevations, and the highest occurred at 5100 m (Fig. S7A). Pearson correlations between PNR and archael amoA gene abundance and their community indexes were calculated. The results showed that PNR did not correlate with either archael amoA gene abundance (Fig. S7B) or diversity (Fig. S7C). Among all the detected T-RFs, PNR positively correlated with absolute abundance of the specific T-RF 403 bp (P < 0.05, Figs. 5 and S8), which was identified to be affiliated with phylotypes in yearly-cold Tibetan Plateau soils.
Discussion
Our findings indicated that the AOA diversity and evenness monotonically increased along the increasing elevations, but their richness kept relatively stable except the peak at the mid-elevation 4800 m. Diversity incorporates richness and evenness; thus, the increase of AOA diversity was attributed to the evenness increase along the elevation gradient. Soils at higher elevations exhibited lower soil temperature and pH, which inhibited the growth of specific AOA phylotypes, such as T-RFs 136, 329, 391, and 421 bp, but facilitated the growth of the other two AOA phylotypes (T-RFs 403 and 443 bp). The opposite response of the AOA phylotypes observed here indicated AOA niche separation (Gubry-Rangin et al. 2015; Okie et al. 2015; Stempfhuber et al. 2015). Given that AOA are well adapted to low-pH pressure in acidic soils (He et al. 2012; Hu et al. 2014), low temperature likely played a more important role than pH in driving AOA growth and their niche separation. Our sequencing data confirmed the affiliation of the decreased AOA phylotypes with increasing elevations (T-RFs 136, 329, 391, and 421 bp), with those retrieved from soils and lake sediments in Yunnan Plateau, Southern China, or Arizona (Li et al. 2015; Liu et al. 2015b), where it is warmer than the yearly-cold Tibetan Plateau. The abundance of warm-adapted AOA phylotypes positively correlated with soil temperature (P < 0.01, Table S1) indicated that their growth was inhibited by lower temperature at higher elevations. In contrast, the AOA phylotypes whose abundances increased with increasing elevations (T-RFs 403 and 443 bp) were affiliated with those retrieved from yearly-cold meadow soils at the elevation 5013 m (Mount Mila) on the Tibetan Plateau, suggesting their adaption or tolerance to low temperature. The abundance of cold-adapted AOA phylotypes positively correlated with soil ammonium concentration and TOC (P < 0.01, Table S1), indicating that their growth was facilitated by richer nutrients (NH4 + and TOC) at higher elevations. Therefore, the evenness increase at higher elevations resulted from the growth inhibition of warm-adapted AOA phylotypes by lower temperature and the growth facilitation of cold-adapted AOA phylotypes by soil nutrients. The elevational evenness pattern of AOA is similar to the arbuscular mycorrhizal fungi, whose evenness demonstrated a monotonic increase pattern along an elevation gradient (Shi et al. 2014). These findings suggest that low temperature might play an important role in the growth and niche separation of AOA, and the cold-adapted AOA phylotypes could benefit from rich nutrients in the yearly-cold meadow soils.
The AOA community structure was driven by both soil properties and geographical distance. The combined effect of all soil properties contributed more (32.6%) to the AOA community structure variation than individual soil properties (3–8% of the total variation). This is consistent with a previous study showing that AOA community structure was mainly driven by multiple environmental factors (Yao et al. 2013). Here, we showed that pH individually explained 8.1% of the total AOA community variation and was the key environmental factor driving AOA community, consistent with previous reports on bacterial elevational variations (Shen et al. 2013; Siles and Margesin 2016; Yashiro et al. 2016) and soil AOA community variations (Gubry-Rangin et al. 2011; Hu et al. 2015; Li et al. 2015). Soil water content and temperature individually explained 5.4 and 3.1% of the total AOA community variation, respectively. Previous findings indicated that that soil water availability and temperature both significantly altered AOA community physiological characteristics and ecological niche (Gleeson et al. 2010; Peralta et al. 2014; Tourna et al. 2008). It has to be mentioned that ammonium availability, which was previously regarded as a main driver (Verhamme et al. 2011), only explained 1.6% of the total variation. This could be attributed to the small ammonium variation range (2–30 mg kg−1) in this study, or some AOA phylotypes were dependent on ammonia arising from mineralization of organic matters rather than inorganic nitrogen in high organic matter soils (Stopnisek et al. 2010). The environmental drivers of AOA community structure are not consistent with the general bacterial community structure in the same location, which was driven by precipitation rather than pH and temperature (Yuan et al. 2014). The reported correlation between bacteria and precipitation could be explained by the fact that the access of soil bacteria to soil organic matter was accelerated by soil moisture at the pore scale (Schimel and Schaeffer 2012). In contrast, AOA are typical autotrophs, which gain energy from ammonia oxidization process and are independent of soil organic matter (Prosser and Nicol 2012).
The archaeal amoA gene abundance significantly decreased along the elevation, indicating that lower soil temperature and pH inhibited the total AOA abundance, although the increasing soil TOC, ammonium, and water content facilitated the growth of specific cold-adapted AOA phylotypes. Soil PNR positively correlated with and the cold-adapted phylotype T-RFs (403 bp), suggesting that they could play a key role in the soil nitrification process in yearly-cold meadow soils on the Tibetan Plateau. The AOA communities at 4400 m were dominated by the phylotypes detected in warmer soil or sediments, whereas those at 5100 m were dominated by indigenous phylotypes detected in Mount Mila meadow soils at the elevation 5013 m on the Tibetan Plateau. Thus, the cloning and sequencing further independently confirmed the AOA niche separation. The cold-adapted AOA phylotypes may be unique or specific to the high-elevation soils (≥4800 m). All the sequences were affiliated with the soil isolate N. gargensis (Tourna et al. 2011). N. gargensis was also detected in alkaline alpine soils in the same regions (Zhang et al. 2009), suggesting that they could inhabit diverse yearly-cold alpine soils.
In conclusion, the elevational gradient, ranging from 4400 m to the grassline at 5100 m, provides a unique model to test microbial elevational diversity pattern and their driving factors. The abundance decrease (T-RFs 136, 329, 391, and 421 bp) and increase (T-RFs 403 and 443 bp) resulted in the monotonic increase of both AOA diversity and evenness as well as the shift of overall AOA community structure along the elevation gradient. Lower temperature likely played an important role in AOA phylotype growth and their niche separation at higher elevations. The positive correlation between cold-adapted phylotype (T-RF 443 bp) and soil PNR suggested a potential negative effect of climate warming on soil nitrification process by altering AOA community compositions, structure, and diversity in the Tibetan region. We have to mention that the AOA elevational diversity pattern and their community compositions/structure are attributed to long-term evolutionary and historical mechanisms. The effects of future climate warming on AOA community structure and diversity could be short-term and more complicated than expected.
References
Alves RJE, Wanek W, Zappe A, Richter A, Svenning MM, Schleper C, Urich T (2013) Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J 7(8):1620–1631. doi:10.1038/ismej.2013.35
Bertuzzo E, Carrara F, Mari L, Altermatt F, Rodriguez-Iturbe I, Rinaldo A (2016) Geomorphic controls on elevational gradients of species richness. Proc Natl Acad Sci U S A 113(7):1737–1742. doi:10.1073/pnas.1518922113
Bryant JA, Lamanna C, Morlon H, Kerkhoff AJ, Enquist BJ, Green JL (2008) Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc Natl Acad Sci U S A 105:11505–11511. doi:10.1073/pnas.0801920105
Chen YL, Hu HW, Han HY, Du Y, Wan SQ, Xu ZW, Chen BD (2014) Abundance and community structure of ammonia-oxidizing archaea and bacteria in response to fertilization and mowing in a temperate steppe in Inner Mongolia. FEMS Microbiol Ecol 89(1):67–79. doi:10.1111/1574-6941.12336
Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, He JZ (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2(9):621–624. doi:10.1038/ngeo613
Fierer N, McCain CM, Meir P, Zimmermann M, Rapp JM, Silman MR, Knight R (2011) Microbes do not follow the elevational diversity patterns of plants and animals. Ecology 92(4):797–804. doi:10.1890/10-1170.1
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102(41):14683–14688. doi:10.1073/pnas.0506625102
Gleeson DB, Mueller C, Banerjee S, Ma W, Siciliano SD, Murphy DV (2010) Response of ammonia oxidizing archaea and bacteria to changing water filled pore space. Soil Biol Biochem 42(10):1888–1891. doi:10.1016/j.soilbio.2010.06.020
Gubry-Rangin C, Nicol GW & Prosser JI (2010) Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol 74:566–574
Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, Schloter M, Griffiths RI, Prosser JI, Nicol GW (2011) Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci U S A 108(52):21206–21211. doi:10.1073/pnas.1109000108
Gubry-Rangin C, Kratsch C, Williams TA, McHardy AC, Embley TM, Prosser JI, Macqueen DJ (2015) Coupling of diversification and pH adaptation during the evolution of terrestrial Thaumarchaeota. Proc Natl Acad Sci U S A 112(30):9370–9375. doi:10.1073/pnas.1419329112
Guo G, Kong W, Liu J, Zhao J, Du H, Zhang X, Xia P (2015) Diversity and distribution of autotrophic microbial community along environmental gradients in grassland soils on the Tibetan Plateau. Appl Microbiol Biotechnol 99(20):8765–8776. doi:10.1007/s00253-015-6723-x
He J-Z, Hu H-W, Zhang L-M (2012) Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils. Soil Biol Biochem 55:146–154. doi:10.1016/j.soilbio.2012.06.006
Heger TJ, Derungs N, Theurillat JP, Mitchell EAD (2016) Testate amoebae like it hot: species richness decreases along a subalpine-alpine altitudinal gradient in both natural calluna vulgaris litter and transplanted minuartia sedoides cushions. Microb Ecol 71(3):725–734. doi:10.1007/s00248-015-0687-3
Herzog SK, Kessler M, Bach K (2005) The elevational gradient in Andean bird species richness at the local scale: a foothill peak and a high-elevation plateau. Ecography 28(2):209–222. doi:10.1111/j.0906-7590.2005.03935.x
Hu HW, Xu ZH, He JZ (2014) Ammonia-oxidizing archaea play a predominant role in acid soil nitrification. In: Sparks DL (ed) Advances in agronomy, Vol 125. Advances in agronomy, vol 125, pp 261–302
Hu HW, Zhang LM, Yuan CL, Zheng Y, Wang JT, Chen D, He JZ (2015) The large-scale distribution of ammonia oxidizers in paddy soils is driven by soil pH, geographic distance, and climatic factors. Front Microbiol 6:938. doi:10.3389/fmicb.2015.00938
Ives AR, Woody ST, Nordheim EV, Nelson C, Andrews JH (2004) The synergistic effects of stochasticity and dispersal on population densities. Am Nat 163(3):375–387. doi:10.1086/381942
Jarvis SG, Woodward S, Taylor AFS (2015) Strong altitudinal partitioning in the distributions of ectomycorrhizal fungi along a short (300 m) elevation gradient. The New phytologist 206(3):1145–1155. doi:10.1111/nph.13315
Jia Z, Conrad R (2009) Bacteria rather than archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol 11(7):1658–1671. doi:10.1111/j.1462-2920.2009.01891.x
Koerner C (2007) The use of ‘altitude’ in ecological research. Trends Ecol Evol 22(11):569–574. doi:10.1016/j.tree.2007.09.006
Kong W & Nakatsu CH (2010) optimization of RNA extraction for PCR quantification of aromatic compound degradation genes. Appl Environ Microbiol 76:1282–1284
Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55:485–529. doi:10.1146/annurev.micro.55.1.485
Kurola J, Salkinoja-Salonen M, Aarnio T, Hultman J, Romantschuk M (2005) Activity, diversity and population size of ammonia-oxidising bacteria in oil-contaminated landfarming soil. FEMS Microbiol Lett 250(1):33–38. doi:10.1016/j.femsle.2005.06.057
Li H, Weng BS, Huang FY, Su JQ, Yang XR (2015) pH regulates ammonia-oxidizing bacteria and archaea in paddy soils in southern China. Appl Microbiol Biotechnol 99(14):6113–6123. doi:10.1007/s00253-015-6488-2
Liu L, Hart MM, Zhang J, Cai X, Gai J, Christie P, Li X, Klironomos JN (2015a) Altitudinal distribution patterns of AM fungal assemblages in a Tibetan alpine grassland. FEMS Microbiol Ecol 91(7):fiv078. doi:10.1093/femsec/fiv078
Liu Y, Zhang J, Zhao L, Li Y, Dai Y, Xie S (2015b) Distribution of sediment ammonia-oxidizing microorganisms in plateau freshwater lakes. Appl Microbiol Biotechnol 99(10):4435–4444. doi:10.1007/s00253-014-6341-z
Lomolino MV (2001) Elevation gradients of species-density: historical and prospective views. Glob Ecol Biogeogr 10(1):3–13. doi:10.1046/j.1466-822x.2001.00229.x
Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA (2009) Ammonia oxidation kinetics determine niche separation of nitrifying archaea and bacteria. Nature 461(7266):976–U234. doi:10.1038/nature08465
Matsuoka S, Mori AS, Kawaguchi E, Hobara S, Osono T (2016) Disentangling the relative importance of host tree community, abiotic environment and spatial factors on ectomycorrhizal fungal assemblages along an elevation gradient. FEMS Microbiol Ecol 92(5):fiw044. doi:10.1093/femsec/fiw044
McGradySteed J, Harris PM, Morin PJ (1997) Biodiversity regulates ecosystem predictability. Nature 390(6656):162–165
Meng H, Li K, Nie M, Wan JR, Quan ZX, Fang CM, Chen JK, Gu JD, Li B (2013) Responses of bacterial and fungal communities to an elevation gradient in a subtropical montane forest of China. Appl Microbiol Biotechnol 97(5):2219–2230. doi:10.1007/s00253-012-4063-7
Novillo A, Ojeda RA (2014) Elevation patterns in rodent diversity in the dry Andes: disentangling the role of environmental factors. J Mammal 95(1):99–107. `doi:10.1644/13-mamm-a-086.1
Okie JG, Van Horn DJ, Storch D, Barrett JE, Gooseff MN, Kopsova L, Takacs-Vesbach CD (2015) Niche and metabolic principles explain patterns of diversity and distribution: theory and a case study with soil bacterial communities. Proc R Soc B-Biol Sci 282(1809):20142630. doi:10.1098/rspb.2014.2630
Peralta AL, Matthews JW, Kent AD (2014) Habitat specialization along a wetland moisture gradient differs between ammonia-oxidizing and denitrifying microorganisms. Microb Ecol 68(2):339–350. doi:10.1007/s00248-014-0407-4
Prosser JI, Nicol GW (2008) Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10(11):2931–2941. doi:10.1111/j.1462-2920.2008.01775.x
Prosser JI, Nicol GW (2012) Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol 20(11):523–531. doi:10.1016/j.tim.2012.08.001
Schimel JP, Schaeffer SM (2012) Microbial control over carbon cycling in soil. Front Microbiol 3:348. doi:10.3389/fmicb.2012.00348
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541. doi:10.1128/aem.01541-09
Shen C, Xiong J, Zhang H, Feng Y, Lin X, Li X, Liang W, Chu H (2013) Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol Biochem 57:204–211. doi:10.1016/j.soilbio.2012.07.013
Shen C, Liang W, Shi Y, Lin X, Zhang H, Wu X, Xie G, Chain P, Grogan P, Chu H (2014) Contrasting elevational diversity patterns between eukaryotic soil microbes and plants. Ecology 95(11):3190–3202
Shi Z, Wang F, Zhang K, Chen Y (2014) Diversity and distribution of arbuscular mycorrhizal fungi along altitudinal gradients in Mount Taibai of the Qinling Mountains. Can J Microbiol 60(12):811–818. doi:10.1139/cjm-2014-0416
Siles JA, Margesin R (2016) Abundance and diversity of bacterial, archaeal, and fungal communities along an altitudinal gradient in alpine forest soils: what are the driving factors? Microb Ecol 72(1):207–220. doi:10.1007/s00248-016-0748-2
Singh D, Takahashi K, Kim M, Chun J, Adams JM (2012) A hump-backed trend in bacterial diversity with elevation on Mount Fuji, Japan. Microb Ecol 63(2):429–437. doi:10.1007/s00248-011-9900-1
Singh D, Takahashi K, Park J, Adams JM (2016) Similarities and contrasts in the archaeal community of two Japanese mountains: Mt. Norikura compared to Mt. Fuji. Microb Ecol 71(2):428–441. doi:10.1007/s00248-015-0681-9
Stempfhuber B, Engel M, Fischer D, Neskovic-Prit G, Wubet T, Schoening I, Gubry-Rangin C, Kublik S, Schloter-Hai B, Rattei T, Welzl G, Nicol GW, Schrumpf M, Buscot F, Prosser JI, Schloter M (2015) pH as a driver for ammonia-oxidizing archaea in forest soils. Microb Ecol 69(4):879–883. doi:10.1007/s00248-014-0548-5
Stopnisek N, Gubry-Rangin C, Hoefferle S, Nicol GW, Mandic-Mulec I, Prosser JI (2010) Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl Environ Microbiol 76(22):7626–7634. doi:10.1128/aem.00595-10
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30(12):2725–2729. doi:10.1093/molbev/mst197
Tourna M, Freitag TE, Nicol GW, Prosser JI (2008) Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol 10(5):1357–1364. doi:10.1111/j.1462-2920.2007.01563.x
Tourna M, Stieglmeier M, Spang A, Koenneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108(20):8420–8425. doi:10.1073/pnas.1013488108
Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C (2005) Novel genes for nitrite reductase and amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol 7(12):1985–1995. doi:10.1111/j.1462-2920.2005.00906.x
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu DY, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304(5667):66–74. doi:10.1126/science.1093857
Verhamme DT, Prosser JI, Nicol GW (2011) Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J 5(6):1067–1071. doi:10.1038/ismej.2010.191
Wang Z, Luo T, Li R, Tang Y, Du M (2013) Causes for the unimodal pattern of biomass and productivity in alpine grasslands along a large altitudinal gradient in semi-arid regions. J Veg Sci 24(1):189–201. doi:10.1111/j.1654-1103.2012.01442.x
Wang J-T, Cao P, Hu H-W, Li J, Han L-L, Zhang L-M, Zheng Y-M, He J-Z (2015) Altitudinal distribution patterns of soil bacterial and archaeal communities along Mt. Shegyla on the Tibetan Plateau. Microb Ecol 69(1):135–145. doi:10.1007/s00248-014-0465-7
Yang Y, Gao Y, Wang S, Xu D, Yu H, Wu L, Lin Q, Hu Y, Li X, He Z, Deng Y, Zhou J (2014) The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J 8(2):430–440. doi:10.1038/ismej.2013.146
Yao H, Campbell CD, Chapman SJ, Freitag TE, Nicol GW, Singh BK (2013) Multi-factorial drivers of ammonia oxidizer communities: evidence from a national soil survey. Environ Microbiol 15(9):2545–2556. doi:10.1111/1462-2920.12141
Yashiro E, Pinto-Figueroa E, Buri A, Spangenberg JE, Adatte T, Niculita-Hirzel H, Guisan A, van der Meer JR (2016) Local environmental factors drive divergent grassland soil bacterial communities in the western Swiss alps. Appl Environ Microbiol 82(21):6303–6316. doi:10.1128/aem.01170-16
Yuan Y, Si G, Wang J, Luo T, Zhang G (2014) Bacterial community in alpine grasslands along an altitudinal gradient on the Tibetan Plateau. FEMS Microbiol Ecol 87(1):121–132. doi:10.1111/1574-6941.12197
Yuan Y, Si G, Li W, Zhang G (2015) Altitudinal distribution of ammonia-oxidizing archaea and bacteria in alpine grassland soils along the south-facing slope of Nyqentangula Mountains, central Tibetan Plateau. Geomicrobiol J 32(1):77–88. 10.1080/01490451.2014.925013
Zhang LM, Wang M, Prosser JI, Zheng YM, He JZ (2009) Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everest. FEMS Microbiol Ecol 70(2):208–217. doi:10.1111/j.1574-6941.2009.00775.x
Zhang LM, Hu HW, Shen JP, He JZ (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6(5):1032–1045. doi:10.1038/ismej.2011.168
Acknowledgements
The authors greatly thank Dr. Cindy Nakatsu at Purdue University for thoughtful discussions and constructive suggestions. This project was financially supported by Chinese Academy of Sciences (XDB15010203 and KZZD-EW-TZ-14 to WK) and National Natural Science Foundation of China (41471054 to WK and 41401287 to GG).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 954 kb)
Rights and permissions
About this article
Cite this article
Zhao, K., Kong, W., Khan, A. et al. Elevational diversity and distribution of ammonia-oxidizing archaea community in meadow soils on the Tibetan Plateau. Appl Microbiol Biotechnol 101, 7065–7074 (2017). https://doi.org/10.1007/s00253-017-8435-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8435-x