Abstract
Hexavalent chromium [Cr(VI)] is a priority pollutant causing serious environmental issues. Microbial reduction provides an alternative strategy for Cr(VI) remediation. The dissimilatory metal-reducing bacterium, Shewanella oneidensis MR-1, was employed to study Cr(VI) reduction and toxicity in this work. To understand the effect of membrane cytochromes on Cr(VI) response, a comparative protein profile analysis from S. oneidensis MR-1 wild type and its mutant of deleting OmcA and MtrC (△omcA/mtrC) was conducted using two-dimensional electrophoresis (2-DE) technology. The 2-DE patterns were compared, and the proteins with abundant changes of up to twofold in the Cr(VI) treatment were detected. Using mass spectrometry, 38 and 45 differentially abundant proteins were identified in the wild type and the mutant, respectively. Among them, 25 proteins were shared by the two strains. The biological functions of these identified proteins were analyzed. Results showed that Cr(VI) exposure decreased the abundance of proteins involved in transcription, translation, pyruvate metabolism, energy production, and function of cellular membrane in both strains. There were also significant differences in protein expressions between the two strains under Cr(VI) treatment. Our results suggest that OmcA/MtrC deletion might result in the Cr(VI) toxicity to outer membrane and decrease assimilation of lactate, vitamin B12, and cystine. When carbohydrate metabolism was inhibited by Cr(VI), leucine and sulfur metabolism may act as the important compensatory mechanisms in the mutant. Furthermore, the mutant may regulate electron transfer in the inner membrane and periplasm to compensate for the deletion of OmcA and MtrC in Cr(VI) reduction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chromium (Cr), one of the most abundant heavy metals, is widely discharged into the environment through industrial activities including electroplating, leather tanning, dyeing, metallurgy, and wood treatment. Due to its mutagenic and carcinogenic properties, Cr leads to severe environmental pollution and poses a significant threat to plants, microorganisms, and human (Narayani and Shetty 2013). Two dominant forms of Cr in the environment are the trivalent [Cr(III)] and hexavalent [Cr(VI)] forms. The adverse effects of Cr are mainly from Cr(VI) because of its solubility, mobility, and high oxidizing potential. On the contrary, Cr(III) is much less toxic and insoluble. A great deal of work has focused on Cr(VI) reduction through various applications of chemical or biological approaches to Cr(III) (Dhal et al. 2013), in which microbe-mediated reduction of Cr(VI) is a promising strategy for Cr detoxification (Ge et al. 2013).
The dissimilatory metal-reducing bacterium Shewanella oneidensis MR-1 is capable of reducing Cr(VI) under both aerobic and anaerobic conditions (Belchik et al. 2011; Brown et al. 2006). The anaerobic reduction of Cr(VI) is associated with electron transfer systems that involve membrane cytochromes (e.g., b and c) (Wang and Shen 1995). In S. oneidensis MR-1, the membrane c-type cytochromes, including CymA, MtrA, MtrB, MtrC, and OmcA, transfer electron from the inner membrane, across the periplasm and outer membrane, to the cell surface (Shi et al. 2012). MtrA and MtrB are present in the periplasm and outer membrane, respectively. They are required to transfer electrons from CymA to MtrC. MtrC and OmcA located on the outer membrane are responsible for electron transfer to extracellular electron acceptors (Shi et al. 2012). By this process, extracellular Cr(VI) can be reduced as the terminal electron acceptor (Belchik et al. 2011).
Bencheikh-Latmani et al. (2005) reported that the cytochrome genes encoding MtrA, MtrB, MtrC, and OmcA in S. oneidensis MR-1 were upregulated under Cr(VI) exposure. A mutant experiment showed that deletion of these cytochrome genes (mtrA, mtrB, mtrC, and omcA) significantly decreased Cr(VI) reduction rates (Belchik et al. 2011; Bencheikh-Latmani et al. 2005). MtrC and OmcA can respectively transfer electrons to extracellular Cr(VI) at the outer membrane, and their cooperation effectively improves Cr(VI) reduction (Wang et al. 2013b). Compared to deleting single MtrC or OmcA, the mutant of MtrC and OmcA that is simultaneously deleted could further decrease Cr(VI) reduction ability (Belchik et al. 2011; Wang et al. 2013b) and may ultimately affect the response of S. oneidensis MR-1 to the Cr(VI) challenge. However, there was a paucity of current knowledge about the effect of MtrC/OmcA on the Cr(VI) response. Cr(VI) reduction and toxicity are two co-existing processes. The change in reduction ability may influence the Cr(VI) toxicity, because both toxicity and resistance level exhibited by S. oneidensis MR-1 are highly dependent on Cr(VI) concentrations (Brown et al. 2006; Thompson et al. 2007). Furthermore, other electron transfer pathways may be induced to be responsible for Cr(VI) reduction after deletion of MtrC and OmcA. A better understanding of how both MtrC and OmcA affect the response of S. oneidensis MR-1 to Cr(VI) can give new insights into microbial remediation mechanisms.
Proteomics is a powerful tool for understanding the cellular molecular networks responsive to environment stress through identifying the differentially expressed proteins. Since two-dimensional electrophoresis (2-DE) was successfully employed as a proteomic technology, a large number of cellular proteomes have been studied. With the application of 2-DE in recent years, the Cr(VI)-responsive proteins identified in S. oneidensis MR-1 provided useful information on the molecular mechanism underlying Cr(VI) reduction and toxicity (Brown et al. 2006; Thompson et al. 2007). Unfortunately, these studies failed to provide the detailed information on the biological effects of OmcA/MtrC on Cr(VI) response. Therefore, in the present study, a comparative proteomic analysis between S. oneidensis MR-1 and its OmcA/MtrC mutant under Cr(VI) treatment was conducted using 2-DE. The differentially expressed proteins in response to Cr(VI) treatment were identified by matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry (MALDI-TOF/TOF-MS). In order to assess the effect of OmcA/MtrC on cellular processes of S. oneidensis MR-1 under Cr(VI) exposure, the identified proteins were further analyzed on the basis of their biological functions. In addition, the transcript expressions of genes encoding the differentially abundant proteins were detected using real-time quantitative polymerase chain reaction (qRT-PCR). The obtained information will shed light on the understanding of the metabolisms of Cr(VI) reduction and toxicity in S. oneidensis MR-1 and facilitate its application in bioremediation.
Materials and methods
Strains and Cr(VI) treatment
S. oneidensis MR-1 (ATCC No. 700550) and the cyt c mutant with deletion of OmcA and MtrC (△omcA/mtrC) have been described in our previous study (Wu et al. 2013). Strains were grown anaerobically in 100-mL serum bottles containing 50-mL Luria-Bertani (LB) medium (pH 7.2). Sodium lactate (10 mM) was added to the medium as the electron donor. The cultures were operated on a shaker at 100 rpm at 30 °C. Anaerobic conditions were provided by nitrogen. When the OD at 600 nm reached 0.45 (mid-exponential phase), Cr(VI) stock solution (100 mM K2CrO4) was added anaerobically to the medium to obtain a final concentration of 0.3 mM. The treatment of heat-killed cells was carried out as the control. The cultures without Cr(VI) were also conducted for proteomic and qRT-PCR analysis. Then, all cultures were allowed to grow at 100 rpm at 30 °C. After exposure for 0, 30, 60, 120, and 240 min, Cr(VI) concentration remaining in the medium was measured using 1,5-diphenylcarbazide method (Urone 1955). Each treatment had three replicates.
After Cr(VI) exposure of 60 min, the cells (wild type and △omcA/mtrC mutant) incubated with or without Cr(VI) were harvested from three independent biological replicates by centrifugation at 5,000 g for 5 min at 4 °C and washed three times with ice-cold sterile distilled water. The operation for harvesting and washing the cells was carried out under anaerobic condition, which was provided by nitrogen in an anaerobic incubator. The washed cells were immediately used for protein and RNA extraction.
Protein extraction
The washed cells were lysed by sonication at 4 °C (Brown et al. 2006) in a lysis buffer (50 mM Tris-HCl, pH 7.5, 250 mM sucrose, 10 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol [DTT], 1 % [w/v] Triton X-100). Total soluble protein was extracted according to the phenol extraction method (Carpentier et al. 2005). The final protein pellet was air-dried and dissolved in a hydration solution (8 M urea, 2 M thiourea, 4 % [w/v] 3[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate [CHAPS], 40 mM DTT, and 1 % [v/v] immobilized pH gradient [IPG] buffer [pH 4-7, GE Healthcare, Piscataway, NJ, USA]). Protein concentration was determined using the Bradford method (Bradford 1976).
2-DE and image analysis
Two-dimensional electrophoresis was carried out according to Bjellqvist et al. (1982). For each replicate, 800 μg of protein was loaded onto an 18-cm IPG strip (pH 4–7, GE Healthcare, Piscataway, NJ, USA) and rehydrated for 18 h at room temperature. Isoelectric focusing was performed using an Ettan IPGphor system (GE Healthcare Amersham Bioscience, Little Chalfont, UK) at 20 °C for a total of 64,000 Vh. The focused strips were first equilibrated in an equilibration solution (50 mM Tris-HCl, pH 8.8, 6 M urea, 30 % [v/v] glycerol, 2 % [w/v] sodium dodecyl sulfate [SDS], and 1 % [w/v] DTT) for 15 min. After that, a same equilibration solution with DTT replaced with 2.5 % (w/v) iodoacetamide was used for another 15 min. The proteins in IPG strips were separated using 12.5 % SDS-polyacrylamide gels. Gels stained with Coomassie brilliant blue R250 were scanned at 600-dpi resolution using a PowerLook 2100XL (UMAX Technologies Inc., Dallas, TX, USA). Gel images were analyzed using PDQuest software (version 8.01, Bio-Rad, Hercules, CA, USA). Protein spots on the gel were detected, background subtracted, matched, and quantified. The reproducibility of the gels among the biological replicates was analyzed using the scatter plot. Statistical differences in the protein abundance from three independent gels for each treatment were defined using the Student’s t test (p < 0.05). The protein spots with an average abundance change of more than twofold and present in all biological replicates for each treatment were identified by MS analysis.
Protein identification
Protein spots excised from the gels were firstly distained in a solution of methanol and 50 mM NH4HCO3 (1:1, v/v) and dried using a vacuum centrifuge. Dried gel spots were digested with 0.1 mg μL−1 of sequencing grade trypsin (Promega, Madison, WI, USA) in 25 mM NH4HCO3 for 16 h at 37 °C. The protein peptides were collected and dried and then dissolved in 1 μL 40 % acetonitrile (ACN)/0.5 % trifluoroacetic acid (TFA). Finally, the mixture of 0.5-μL peptide solution and 0.5-μL matrix solution (α-cyano-4-hydroxycinnamic acid in 50 % ACN/0.1 % TFA) was spotted onto anchor chips (Bruker Daltonics, Bremen, Germany). The peptide mass fingerprint was acquired using MALDI-TOF/TOF-MS analysis (Bruker Daltonics, Bremen, Germany). The mass spectrum was processed with the software FlexAnalysis (version 3.2, Bruker Daltonics, Bremen, Germany). After that, the peptide mass was submitted to the Mascot search engine (www.matrixscience.com) to search the National Center for Biotechnology Information non-redundant (NCBInr) database (37,332,560 entries, downloaded on 6 January 2014). The searched parameters were set: taxonomy, bacteria; no MW/pI restrictions; missed cleavages, 1; enzyme, trypsin; mass tolerance, 100 ppm; the fixed modifications: carbamidomethyl; and significant protein MOWSE score at p < 0.05. The match of the highest Mascot score (≥86) was considered as a criterion for correct identification.
Protein functional classification and hierarchical cluster analysis
The biological functions of the identified proteins were searched in the UniPort database (www.ebi.uniprot.org/index) and NCBI database (www.ncbi.nlm.nih.gov). Hierarchical clustering of protein abundance profiles was carried out using Cluster software (version 3.0), and the linkage clustering was visualized using TreeView software (version 1.6) (Wang et al. 2013a).
RNA extraction and qRT-PCR analysis
Total RNA was extracted with RNAprep pure Cell/Bacteria Kit (Tiangen Biotech Co., Ltd., Beijing, China). The complementary DNA (cDNA) was immediately synthesized using FastQuant RT kit (Tiangen) according to the manufacturer’s instructions. After that, the cDNAs were used as templates for qRT-PCR. The qRT-PCR was performed for the genes encoding differentially abundant proteins detected in the 2-DE gel, and primers for the assayed genes were given in Supplemental Table S1. Bacterial ribosomal DNA (16S rDNA) primer was used as the internal standard. PCR reactions were carried out in an iCycler iQ real-time detection system (Bio-Rad, Hercules, CA, USA) as described by Ram et al. (2008). The relative quantification of each gene was obtained using the 2−ΔΔCt method (Livak and Schmittgen 2001). Three independent replicates were performed for each sample.
Results
Cr(VI) reduction by the wild type and the △omcA/mtrC mutant
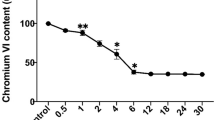
When 0.3 mM Cr(VI) was added to the LB medium, approximately 10 % Cr(VI) was immediately removed by the reducing substances existing in the medium (Fig. 1). Cr(VI) removal by the wild type in 30 and 60 min was 65.9 and 82.7 %, respectively, which were significantly higher than that by the mutant (23.8 and 30.9 %) (Fig. 1). Cr(VI) was completely removed by the wild type in 120 min, whereas 58.4 and 43.9 % of Cr(VI) still remained in the medium for the mutant at 120 and 240 min, respectively. The heat-killed cell did not affect the Cr(VI) concentration (Fig. 1). These results indicate that deletion of MtrC and OmcA significantly decreases the Cr(VI) reduction ability of S. oneidensis MR-1.
Differentially expressed proteins in the wild type and mutant under Cr(VI) treatment
More than 1,100 protein spots were detected reproducibly on each gel for the wild type and the △omcA/mtrC mutant (Fig. 2). Three repeated gels showed a high degree of identity, and the correlation coefficients of the triplicates varied from 0.860 to 0.913 (Supplemental Fig. S1). A total of 58 protein spots exhibited the differentially abundance changes (more than twofold) in response to Cr(VI) exposure (Table 1 and Fig. 2). Figure 2c shows close-up views of several protein spots. Of these 58 proteins, 38 and 45 proteins were differentially expressed in the wild type and the mutant, respectively. In the wild type, the relative abundances of six protein spots were increased, and those of 32 protein spots were decreased (Fig. 3). In the mutant, the number of increased and decreased protein spots was 6 and 39, respectively. Regarding the differentially abundance proteins shared by the two strains, 1 was increased in relative abundance and 24 were decreased in relative abundance (Fig. 3).
Representative 2-DE gels of the protein profiles of S. oneidensis MR-1 wild type (a) and the mutant (b) in response to Cr(VI) exposure. c Close-up views of some differentially abundance proteins. The 58 differentially Cr(VI)-responsive proteins are marked by numbers on the gels, and their characteristics are shown in Table 1
Functional classification and clustering analysis
The 58 protein spots with differentially expressed abundance were identified by MALDI-TOF/TOF-MS, and their detailed information is shown in Table 1. Functional annotations in databases (UniPort or NCBI) existed for the majority of the protein spots, whereas 7 proteins (spots 5, 15, 27, 35, 49, 50, and 56) were unknown or hypothetical function proteins (Table 1). These proteins were classified according to their annotated biochemical functions. As shown in Fig. 4, the most enriched categories in the two strains were metabolism (36.84 % for the wild type, 35.56 % for the mutant). The second enriched categories in the wild type and the mutant were protein biosynthesis (15.79 %) and transport (13.33 %), respectively. Other categories were involved in transcription, biogenesis of cellular components, cell cycle, and unknown functions (Fig. 4). In the wild type, six upregulated proteins were mainly involved in protein biosynthesis, metabolism, and biogenesis of cellular components. For six upregulated proteins in the mutant, the biological functions included transport, metabolism, transcription, and biogenesis of cellular components (Fig. 4).
To comprehensively analyze the abundance dynamics of the identified proteins in the wild type and the mutant, hierarchical clustering was performed. Figure 5 shows that all the identified proteins were divided into two groups, i.e., an increased group and a decreased group. In the increased group, although the protein functions were involved in metabolism, transport, transcription, and protein biosynthesis, they appeared independently in the wild type (spots 4, 6, 35, 37, and 53) or in the mutant (spots 30, 32, 39, 41, and 48). Specifically, the increased proteins related to electron transfer were only found in the mutant, suggesting that some election transfer pathways may be induced by Cr(VI) for the mutant (Fig. 5 and Table 1). The decreased group mainly contained three change patterns. First, the proteins in the first pattern were decreased in relative abundance in both strains. Second, seven proteins (spots 10, 12, 19, 21, 46, 50, and 56), which were related to metabolism and protein biosynthesis, were only identified in the wild type. The third pattern was the proteins statistically changed only in the mutant (Fig. 5); they were involved in transport, metabolism, protein biosynthesis, and cell cycle. The proteins related to outer membrane synthesis were decreased in relative abundance only in the mutant (Fig. 5 and Table 1), indicating that Cr(VI) treatment has a serious impact on the formation of outer membrane in the mutant.
Hierarchical cluster analysis of the identified proteins in the wild type and the mutant under Cr(VI) exposure. Fold changes of protein abundances between Cr(VI) treatment and control are shown in Table 1. Spot numbers are labeled to the right of the corresponding heat map
Expression analysis of Cr(VI)-responsive genes using qRT-PCR
To further confirm the proteomic results, we used qRT-PCR to analyze the messenger RNA (mRNA) levels of nine genes encoding protein spots 9, 11, 20, 29, 30, 32, 38, 39, and 52. Figure 6 shows that the relative abundances of eight protein spots changed in parallel with their mRNA levels in both strains when exposed to Cr(VI) treatment and only the relative transcription of gene encoding protein spot 32 exhibited no significant change in Cr(VI)-treated mutant.
Changes in mRNA (a) and protein (b) levels for selected nine protein spots in both strains with Cr(VI) treatment. The log2 values were used as the relative expression, and the expression change with a <0.5 or >2 was calculated and normalized to <−1 or >1 via the fit of log2. Spot 9, pyruvate formate-lyase PflB; spot 11, TonB-dependent vitamin B12 receptor BtuB; spot 20, succinate dehydrogenase flavoprotein subunit; spot 29, dTDP-glucose 4,6-dehydratase RfbB; spot 30, Na-translocating NADH-quinone reductase subunit A NqrA; spot 32, aldehyde oxidoreductase; spot 38, d-alanyl-d-alanine carboxypeptidase DacA; spot 39, isovaleryl-CoA dehydrogenase LiuA; spot 52, l-lactate dehydrogenase complex protein LldG
Discussion
Deletion of OmcA and MtrC aggravates Cr(VI) toxicity to outer membrane
During bioremediation of Cr(VI), microbes also suffer from the toxic effect of Cr(VI) in a dose-dependent manner (Brown et al. 2006). In this study, more Cr(VI) ions were remained in the medium for culturing the mutant (Fig. 1), indicating that the mutant should be subjected to more serious toxicity of Cr(VI) than the wild type at the same time of Cr(VI) treatment. The outer membrane of bacteria is the first site in response to outer stress (Nikaido 1994). Here, we observed that Cr(VI) treatment led to the significant reduction in the abundances of four proteins related to outer membrane synthesis (Table 1, spots 1, 29, 38, and 51). Interestingly, these proteins were only identified in the mutant. Spots 1, 38, and 51 are involved in the peptidoglycan synthesis of bacterial outer membrane. For example, UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate ligase (spot 1) catalyzes the assembly of the peptidoglycan pentapeptide (McCoy and Maurelli 2006), and glycosyl transferase (spot 51) catalyzes the formation of the carbohydrate chains from disaccharide subunits in the peptidoglycan synthesis (Lovering et al. 2007). The thymidine diphosphate (dTDP)-4-dehydro-6-deoxy-d-glucose as product of the enzyme dTDP-glucose 4,6-dehydratase (spot 29) participates in the outer membrane lipopolysaccharide production (Liu and Thorson 1994). Decreased abundances of these proteins indicate that Cr(VI) interfered the outer membrane syntheses of the mutant. A previous study in S. oneidensis MR-1 also found that prolonged Cr(VI) exposure disturbed outer membrane synthesis and further led to the abnormally cell morphologies (Chourey et al. 2006).
As the components of outer membrane, cytochromes OmcA and MtrC are considered as two important electron transporters. In this study, the growth of wild type was slightly better than the mutant under anaerobic condition (Fig. S2 in the Supplementary Material). Deletion of OmcA and MtrC decreased electron transfer from periplasm to extracellular electron acceptors (Belchik et al. 2011); it may result in negative effect on growth. In addition, their absence also affects surface composition of outer membrane. Our previous study showed that the deletion of both MtrC and OmcA decreased surface N content, protein, and polysaccharide content and increased lipid content in S. oneidensis MR-1 (Wu et al. 2013). Peptidoglycan consists of amino acids and polysaccharide and can modulate its structure and composition to respond to environmental challenge (Horcajo et al. 2012). The decreased content of N and polysaccharide in the mutant may affect the function of peptidoglycan in the outer membrane. This should attribute to Cr(VI) susceptibility, probably to facilitate Cr(VI) toxicity to outer membrane in the mutant.
Deletion of OmcA and MtrC disturbs transcription and translation processes under Cr(VI) exposure
Cr(VI) exposure results in an inevitable impact on transcription and translation processes of bacteria (Bencheikh-Latmani et al. 2005). Here, Cr(VI) decreased the abundances of three transcription-related proteins (spots 13, 33, and 43) in the two strains. AraC family transcriptional regulator (spot 41), which can regulate the expression of some genes responded to stress and virulence (Martin and Rosner 2001), increased in relative abundance only in the mutant under Cr(VI) treatment. AraC family transcriptional regulators function as monomers to regulate gene transcription. For example, MarA, a member of the AraC family transcriptional regulators, could facilitate bacteria resistance to antibiotics, organic solvents, and oxidative stress agents by controlling expression of the mar regulon (Alekshun and Levy 1997). This result indicates that AraC family transcriptional regulator may play positive role in defending Cr(VI) toxicity in the mutant.
Under Cr(VI) exposure, three (spots 12, 28, and 58) and five (spots 7, 18, 28, 36, and 58) translation-related proteins decreased in their abundances in the wild type and the mutant, respectively. Cr(VI) treatment might inhibit translation elongation and glycine-rich protein synthesis due to the decreased abundances of elongation factor G (spots 7 and 58) and glycyl-transfer RNA (tRNA) synthetase (spot 28) in the two strains. Additionally, the toxic effect of Cr(VI) on the mutant also showed the decreased abundance of SurA (spot 36), a chaperone involved in the folding and assembly of outer membrane proteins (Rouviere and Gross 1996). This supports the above result that deletion of MtrC and OmcA increased the toxic effect of Cr(VI) on the synthesis of outer membrane.
Three proteins, which were involved in phenylalanine-rich protein synthesis (phenylalanyl-tRNA synthetase, spot 4), hydrolysis of peptide bond from proline residues (peptidase S9 prolyl oligopeptidase, spot 6), and phosphorylation of serine or threonine residues of proteins (serine/threonine protein kinase, spot 37), significantly increased in their abundances only in the wild type, but not in the mutant. In contrast, the abundance of serine protein kinase (spot 18) showed negative change in the mutant. Serine/threonine protein kinase widely functions in bacteria development and is required for phosphorylation of proteins, which is an important signal of environmental stress (Yang et al. 1996). These results imply that the wild type might adapt Cr(VI) stress through regulating the synthesis and function of specific proteins, whereas they were not associated with the mutant.
Different metabolism pathways between the two strains in response to Cr(VI)
Carbohydrate metabolism is primarily responsible for providing substrates and energy to biological processes. In our study, a large number of proteins involved in carbohydrate metabolism were identified (Table 1 and Fig. 4). Firstly, we focused on the proteins associated to pyruvate turnover. Pyruvate kinase (spot 26) and phosphoenolpyruvate synthase (spot 2) are enzymes involved in the conversion of phosphoenolpyruvate (PEP) to pyruvate in the glycolysis and gluconeogenesis, respectively (Imanaka et al. 2006). PEP-protein phosphotransferase (spot 42) catalyzes PEP to pyruvate for sugar phosphorylation which participates in carbohydrate transport (Barabote and Saier 2005). The abundances of these three proteins decreased in the two strains when exposed to Cr(VI). Formate metabolism is an important anaerobic carbohydrate metabolism pathway. In cells, formate is produced from the non-oxidative cleavage of pyruvate via pyruvate formate-lyase and is oxidized by formate dehydrogenase to CO2 (Leonhartsberger et al. 2002). Here, the proteins related to pyruvate formate-lyase (formate acetyltransferase) (spots 8, 9, and 10) and formate dehydrogenase (spot 30) decreased in their abundances in the two strains, which were consistent with their mRNA changes (Fig. 6). These results suggest that carbohydrate metabolism involving pyruvate may be one of the Cr(VI) toxicity mechanism and that pyruvate may be an important target of Cr(VI) toxicity.
Most notably, the decreased abundance of l-lactate dehydrogenase (spot 52) was only found in the mutant. In the present experiment, lactate was added as an important carbon source and is converted to pyruvate by l-lactate dehydrogenase with concomitant production of NADH (Dong et al. 1993). Consistent with our results, Thompson et al. (2010) also reported that when lactate was used as the sole carbon source, the abundance of lactate dehydrogenase obviously decreased in Cr(VI)-treated Pseudomonas putida F1. Hence, we proposed that Cr(VI) inhibited the expression of l-lactate dehydrogenase in the mutant, thereby decreasing utilization of lactate.
Some compensatory mechanisms may be induced to be responsible for Cr(VI) adaptation. Under Cr(VI) treatment, glycerol-3-phosphate dehydrogenase (spot 53) was observed to increase in abundance only in the wild type. This enzyme catalyzes the production of glycerol from the glycerol 3-phosphate during anaerobic growth (Rawls et al. 2011). The glycerol production can maintain cytosolic redox balance and compensate for cellular reactions through producing NADH under anaerobic condition (Gonzalez et al. 2008). This should be an adaptive strategy employed by the wild type in response to Cr(VI).
In the △omcA/mtrC mutant, amino acid and sulfur metabolism can be regulated by Cr(VI), and they may be the important compensatory mechanisms. When carbohydrate metabolism is restrained, some amino acids are produced from protein breakdown to metabolize for respiratory (Aubert et al. 1996). In our study, the relative abundance of isovaleryl-CoA dehydrogenase (IVD, spot 39), which is responsible for the conversion of isovaleryl-CoA to 3-methylcrotonyl-CoA in the leucine catabolism pathway, increased in protein and mRNA levels only in the mutant exposed to Cr(VI). IVD is also a membrane flavoenzyme, which transfers the electrons generated into the respiratory chain through the ubiquinol pool (Binder et al. 2007). Previous study has showed that leucine might participate in Cr(VI) reduction (Gnanamani et al. 2010). Besides amino acid, some sulfur compounds including sulfoacetate, sulfolactate, taurine, isethionate, and ethane-1,2-disulfonate can be utilized as sulfur or as carbon and energy sources, and all are converted to sulfoacetaldehyde, which is catalyzed by sulfoacetaldehyde acetyltransferase (XSC) to acetyl phosphate with desulfonation. The acetyl phosphate is subsequently converted to acetyl CoA for the TCA cycle (Cook and Denger 2002). It should be noted that the abundance of XSC (spot 48) increased only in the mutant after Cr(VI) treatment. In agreement with our result, the increased abundances of Cr(VI)-responsive proteins involved in sulfur metabolism were also detected in other studies about S. oneidensis MR-1 (Brown et al. 2006) and P. putida F1 (Thompson et al. 2010). Although carbohydrate metabolism was inhibited by Cr(VI), leucine and sulfur metabolism could be induced to provide substrate and energy for the △omcA/mtrC mutant.
Change of substance and electron transport-related proteins in cell membrane
The interaction between bacteria and external environment requires transportation function of cell membrane, and these processes consume energy. In bacteria, cellular energy is produced in the inner membrane. In this study, membrane transportation-related protein (spot 55) and energy generation-related proteins (spots 23, 45, and 47) exhibited obvious decrease in their abundances in both strains. Twin-arginine translocation (Tat) system (spot 55) is responsible for the transportation of proteins from cytoplasm to the periplasm, outer membrane, or medium (Robinson et al. 2011). Spots 23 and 45 are directly associated with ATP synthesis. Additionally, TonB2 energy transduction system (spot 47), a member of TonB system, transmits the energy produced in the inner membrane to the outer membrane for substrate transportation (Postle and Larsen 2007). These observations suggest that Cr(VI) repressed energy production and substrate transportation into and out of the cell, thereby decreasing interaction with external environment.
TonB system has been well studied in iron and vitamin B12 transport (Fischer et al. 1989). Vitamin B12 plays a significant role in affecting the DNA synthesis, fatty acid synthesis, and energy production (Rodionov et al. 2003). The relative abundance of TonB-dependent vitamin B12 receptor BtuB (spot 11) involved in the transportation of vitamin B12 into cells decreased in protein and mRNA levels only in the mutant, revealing that Cr(VI) may decrease assimilation of vitamin B12 in the mutant. ATP-binding cassette (ABC) transporters are transmembrane proteins that utilize energy to carry substrates into the cells (Bulut et al. 2012). The decreased abundance of cystine ABC transporter (spot 40) in the mutant might reduce the uptake of cystine that is an important sulfur-containing amino acid for microbial growth (Cantoni et al. 1995). These results suggest that the toxic effect of Cr(VI) on the mutant might also exhibit the decreased utilization of vitamin B12 and cystine in cells.
The anaerobic Cr(VI) reduction is linked to the electrons transferred from cellular internal to cell surface. This process is directly related to electron transport chains, which are present in the cellular membrane (Belchik et al. 2011). The flavoprotein, an important component of the electron transport chains, is involved in electron transfer from NADH to the ferredoxin in the inner membrane of anaerobic bacteria (Herrmann et al. 2008). Here, the decreased abundances of three flavoprotein subunits (spots 20, 21, and 44) suggest that Cr(VI) exposure may reduce electron production from NADH in the two strains. However, in the mutant, Na-translocating NADH-quinone reductase (NQR) may exhibit functional compensation for this process. In our study, the abundance of Na-translocating NQR subunit A (spot 30) increased in protein and mRNA levels only in Cr(VI)-treated mutant. Na-translocating NQR is a Na+-dependent redox enzyme which drives primary Na+ pump in the inner membrane and further improves the activity of the membrane-bound NADH oxidase (Hayashi et al. 2001). Additionally, flavin as the cofactor of Na+-translocating NQR can transfer electrons from the inner membrane to the periplasm through reducing ubiquinone-1 to ubiquinol (Hayashi et al. 2001). A previous study has reported that Na+-translocating NQR participated in the extracellular reduction of Fe(III) and azo compound in S. oneidensis MR-1 (Wang et al. 2010).
Besides, another protein related to membrane electron transfer, aldehyde oxidoreductase (spot 32), was also induced only in the mutant. The aldehyde oxidoreductase belongs to the xanthine oxidase family which is molybdenum-containing enzyme. As its cofactors, two [2Fe-2S] clusters are involved in intramolecular electron transfer in the periplasm (Neumann et al. 2009). Cr(VI), as a soluble ion, is also transported into the cytoplasm and periplasm to be reduced (Belchik et al. 2011). This result suggests that aldehyde oxidoreductase should play an important role in Cr(VI) reduction in the mutant, through transporting electrons in the periplasm, further to the outer membrane.
The mutant incompletely inhibited the reduction ability of S. oneidensis MR-1 to Cr(VI) (Fig. 1), suggesting that OmcA and MtrC are not the only outer membrane c-Cyts that function as terminal reductases. For example, MtrF, an MtrC homolog, is another decaheme c-type cytochrome located on the surface of the cell and involved in Fe(III) oxide reduction (Clarke et al. 2011). MtrF can also compensate for the loss of MtrC with respect to flavin reduction (Coursolle and Gralnick 2010). Due to deletion of both OmcA and MtrC, MtrF may play a key role in Cr(VI) reduction in the △omcA/mtrC mutant. The function of MtrF in Cr(VI) reduction requires further investigation. In this study, the outer membrane proteins involved in electron transfer chains were not detected, implying that Cr(VI) has less effects on these decaheme c-type cytochromes in S. oneidensis MR-1.
Based on these data, we proposed a schematic model of the responses of S. oneidensis MR-1 wild type and the mutant to Cr(VI) (Fig. 7). The differences between the two strains were attributable to the deletion of OmcA and MtrC. In the model, Cr(VI)-responsive proteins were involved in transcription, translation, metabolism, and the synthesis and function of cellular membrane. Under Cr(VI) treatment, OmcA and MtrC affect the cell responses in these biological processes. Deletion of OmcA and MtrC may aggravate Cr(VI) toxicity via decreasing the abundances of outer membrane synthesis-related proteins and reducing the abundances of proteins involved in assimilation of substrates. Notably, OmcA/MtrC deletion affects electron transport chains in the cellular membrane. The mutant could regulate the expressions of Na-translocating NQR and aldehyde oxidoreductase to improve electron transfer in the inner membrane and periplasm, which might be useful for Cr(VI) reduction. This study would be helpful to understand the biological functions of OmcA and MtrC in the bioremediation of Cr(VI) pollution.
Schematic model of the wild type and the △omcA/mtrC mutant in response to Cr(VI) exposure. The protein spots in the figure are the same as those in the Fig. 2 and Table 1. The increased and decreased proteins are marked by ↑ and ↓, respectively. The red arrows represent the differentially change shared by both the wild type and the mutant; the green and blue arrows represent the change independently in the wild type and the mutant, respectively
References
Alekshun MN, Levy SB (1997) Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother 41:2067–2075
Aubert S, Alban C, Bligny R, Douce R (1996) Induction of β-methylcrotonyl-coenzyme A carboxylase in higher plant cells during carbohydrate starvation: evidence for a role of MCCase in leucine catabolism. FEBS Lett 383:175–180
Barabote RD, Saier MH Jr (2005) Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol Mol Biol Rev 69:608–634
Belchik SM, Kennedy DW, Dohnalkova AC, Wang YM, Sevinc PC, Wu H, Lin YH, Lu HP, Fredrickson JK, Shi L (2011) Extracellular reduction of hexavalent chromium by cytochromes MtrC and OmcA of Shewanella oneidensis MR-1. Appl Environ Microbiol 77:4035–4041
Bencheikh-Latmani R, Williams SM, Haucke L, Criddle CS, Wu LY, Zhou JZ, Tebo BM (2005) Global transcriptional profiling of Shewanella oneidensis MR-1 during Cr(VI) and U(VI) reduction. Appl Environ Microbiol 71:7453–7460
Binder S, Knill T, Schuster J (2007) Branched-chain amino acid metabolism in higher plants. Physiol Plant 129:68–78
Bjellqvist B, Ek K, Righetti PG, Gianazza E, Gorg A, Westermeier R, Postel W (1982) Isoelectric focusing in immobilized pH gradients: principle, methodology and some applications. J Biochem Biophys Methods 6:317–339
Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem 72:248–254
Brown SD, Thompson MR, VerBerkmoes NC, Chourey K, Shah M, Zhou JZ, Hettich RL, Thompson DK (2006) Molecular dynamics of the Shewanella oneidensis response to chromate stress. Mol Cell Proteomics 5:1054–1071
Bulut H, Moniot S, Licht A, Scheffel F, Gathmann S, Saenger W, Schneider E (2012) Crystal structures of two solute receptors for L-cystine and L-cysteine, respectively, of the human pathogen Neisseria gonorrhoeae. J Mol Biol 415:560–572
Cantoni O, Brandi G, Albano A, Cattabeni F (1995) Action of cystine in the cytotoxic response of Escherichia coli cells exposed to hydrogen peroxide. Free Radic Res 22:275–283
Carpentier SC, Witters E, Laukens K, Deckers P, Swennen R, Panis B (2005) Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 5:2497–2507
Chourey K, Thompson MR, Morrell-Falvey J, VerBerkmoes NC, Brown SD, Shah M, Zhou JZ, Doktycz M, Hettich RL, Thompson DK (2006) Global molecular and morphological effects of 24-hour chromium(VI) exposure on Shewanella oneidensis MR-1. Appl Environ Microbiol 72:6331–6344
Clarke TA, Edwards MJ, Gates AJ, Hall A, White GF, Bradley J, Reardon CL, Shi L, Beliaev AS, Marshall MJ, Wang ZM, Watmough NJ, Fredrickson JK, Zachara JM, Butt JN, Richardson DJ (2011) Structure of a bacterial cell surface decaheme electron conduit. Proc Natl Acad Sci U S A 108:9384–9389
Cook AM, Denger K (2002) Dissimilation of the C2 sulfonates. Arch Microbiol 179:1–6
Coursolle D, Gralnick JA (2010) Modularity of the Mtr respiratory pathway of Shewanella oneidensis strain MR-1. Mol Microbiol 77:995–1008
Dhal B, Thatoi HN, Das NN, Pandey BD (2013) Chemical and microbial remediation of hexavalent chromium from contaminated soil and mining/metallurgical solid waste: a review. J Hazard Mater 250:272–291
Dong JM, Taylor JS, Latour DJ, Iuchi S, Lin ECC (1993) Three overlapping lct genes involved in L-lactate utilization by Escherichia coli. J Bacteriol 175:6671–6678
Fischer E, Gunter K, Braun V (1989) Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J Bacteriol 171:5127–5134
Ge SM, Zhou MH, Dong XJ, Lu Y, Ge SC (2013) Distinct and effective biotransformation of hexavalent chromium by a novel isolate under aerobic growth followed by facultative anaerobic incubation. Appl Microbiol Biotechnol 97:2131–2137
Gnanamani A, Kavitha V, Radhakrishnan N, Rajakumar GS, Sekaran G, Mandal AB (2010) Microbial products (biosurfactant and extracellular chromate reductase) of marine microorganism are the potential agents reduce the oxidative stress induced by toxic heavy metals. Colloids Surf B 79:334–339
Gonzalez R, Murarka A, Dharmadi Y, Yazdani SS (2008) A new model for the anaerobic fermentation of glycerol in enteric bacteria: trunk and auxiliary pathways in Escherichia coli. Metab Eng 10:234–245
Hayashi M, Nakayama Y, Unemoto T (2001) Recent progress in the Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. BBA-Bioenergetics 1505:37–44
Herrmann G, Jayamani E, Mai G, Buckel W (2008) Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol 190:784–791
Horcajo P, de Pedro MA, Cava F (2012) Peptidoglycan plasticity in bacteria: stress-induced peptidoglycan editing by noncanonical D-amino acids. Microb Drug Resist 18:306–313
Imanaka H, Yamatsu A, Fukui T, Atomi H, Imanaka T (2006) Phosphoenolpyruvate synthase plays an essential role for glycolysis in the modified Embden-Meyerhof pathway in Thermococcus kodakarensis. Mol Microbiol 61:898–909
Leonhartsberger S, Korsa I, Bock A (2002) The molecular biology of formate metabolism in enterobacteria. J Mol Microbiol Biotechnol 4:269–276
Liu HW, Thorson JS (1994) Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu Rev Microbiol 48:223–256
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25:402–408
Lovering AL, de Castro LH, Lim D, Strynadka NCJ (2007) Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science 315:1402–1405
Martin RG, Rosner JL (2001) The AraC transcriptional activators. Curr Opin Microbiol 4:132–137
McCoy AJ, Maurelli AT (2006) Building the invisible wall: updating the chlamydial peptidoglycan anomaly. Trends Microbiol 14:70–77
Narayani M, Shetty KV (2013) Chromium-resistant bacteria and their environmental condition for hexavalent chromium removal: a review. Crit Rev Environ Sci Technol 43:955–1009
Neumann M, Mittelstädt G, Iobbi-Nivol C, Saggu M, Lendzian F, Hildebrandt P, Leimkühler S (2009) A periplasmic aldehyde oxidoreductase represents the first molybdopterin cytosine dinucleotide cofactor containing molybdo-flavoenzyme from Escherichia coli. FEBS J 276:2762–2774
Nikaido H (1994) Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382–388
Postle K, Larsen RA (2007) TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 20:453–465
Ram S, Vajpayee P, Shanker R (2008) Rapid culture-independent quantitative detection of enterotoxigenic Escherichia coli in surface waters by real-time PCR with molecular beacon. Environ Sci Technol 42:4577–4582
Rawls KS, Martin JH, Maupin-Furlow JA (2011) Activity and transcriptional regulation of bacterial protein-like glycerol-3-phosphate dehydrogenase of the haloarchaea in Haloferax volcanii. J Bacteriol 193:4469–4476
Robinson C, Matos CFRO, Beck D, Ren C, Lawrence J, Vasisht N, Mendel S (2011) Transport and proofreading of proteins by the twin-arginine translocation (Tat) system in bacteria. BBA-Biomembranes 1808:876–884
Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS (2003) Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem 278:41148–41159
Rouviere PE, Gross CA (1996) SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Gene Dev 10:3170–3182
Shi L, Rosso KM, Clarke TA, Richardson DJ, Zachara JM, Fredrickson JK (2012) Molecular underpinnings of Fe(III) oxide reduction by Shewanella oneidensis MR-1. Front Microbiol 3:50
Thompson MR, VerBerkmoes NC, Chourey K, Shah M, Thompson DK, Hettich RL (2007) Dosage-dependent proteome response of Shewanella oneidensis MR-1 to acute chromate challenge. J Proteome Res 6:1745–1757
Thompson DK, Chourey K, Wickham GS, Thieman SB, VerBerkmoes NC, Zhang B, McCarthy AT, Rudisill MA, Shah M, Hettich RL (2010) Proteomics reveals a core molecular response of Pseudomonas putida F1 to acute chromate challenge. BMC Genomics 11:311
Urone PF (1955) Stability of colorimetric reagent for chromium, s-diphenylcarbazide, in various solvents. Anal Chem 27:1354–1355
Wang YT, Shen H (1995) Bacterial reduction of hexavalent chromium. J Ind Microbiol 14:159–163
Wang B, Xu MY, Sun GP (2010) Comparative analysis of membranous proteomics of Shewanella decolorationis S12 grown with azo compound or Fe(III) citrate as sole terminal electron acceptor. Appl Microbiol Biotechnol 86:1513–1523
Wang CY, Shen RF, Wang C, Wang W (2013a) Root protein profile changes induced by Al exposure in two rice cultivars differing in Al tolerance. J Proteome 78:281–293
Wang YM, Sevinc PC, Belchik SM, Fredrickson J, Shi L, Lu HP (2013b) Single-cell imaging and spectroscopic analyses of Cr(VI) reduction on the surface of bacterial cells. Langmuir 29:950–956
Wu R, Cui L, Chen L, Wang C, Cao C, Sheng G, Yu H, Zhao F (2013) Effects of bio-Au nanoparticles on electrochemical activity of Shewanella oneidensis wild type and △omcA/mtrC Mutant. Sci Rep 3:3307
Yang XF, Kang CM, Brody MS, Price CW (1996) Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Gene Dev 10:2265–2275
Acknowledgments
This research was supported financially by the National Natural Science Foundation of China (21322703, 21177122), China Postdoctoral Science Foundations (2012M520411), and Science and Technology Innovation and Collaboration Team Project of the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Additional information
Chao Wang and Juan Chen contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 491 kb)
Rights and permissions
About this article
Cite this article
Wang, C., Chen, J., Hu, WJ. et al. Comparative proteomics reveal the impact of OmcA/MtrC deletion on Shewanella oneidensis MR-1 in response to hexavalent chromium exposure. Appl Microbiol Biotechnol 98, 9735–9747 (2014). https://doi.org/10.1007/s00253-014-6143-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6143-3