Abstract
Expression at the mRNA level of six methionine biosynthesis genes in Corynebacterium glutamicum cells under oxygen-deprived conditions was repressed by supplementation of medium with methionine. The repression was not observed in a mutant deficient in the TetR-type transcriptional repressor McbR. Analysis of transcriptional start sites of the methionine biosynthesis genes confirmed that McbR binding motifs exist in the promoter regions of all genes repressed by methionine supplementation. Furthermore, electrophoretic mobility shift assays revealed that not only S-adenosylhomocysteine but also S-adenosylmethionine affects binding of McbR to the promoter region of metY, suggesting that both of these methionine metabolites are involved in the regulation of methionine biosynthesis genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Corynebacterium glutamicum, a gram-positive soil bacterium widely used for the industrial production of amino acids (Kumagai 2000), excretes significant amounts of organic acids in oxygen-deprived conditions under which cellular growth is arrested (Inui et al. 2004b). These characteristics have enabled development of a high-productivity bioprocess for lactate or succinate and its further tailoring for ethanol production from recombinant C. glutamicum (Inui et al. 2004a; Okino et al. 2005, 2008). However, the mechanism of maintaining metabolic activity in the growth-arrested cells under oxygen-deprived conditions is still unknown. The elucidation of this mechanism of metabolic control is important for further development of the high-productivity bioprocess.
Using global DNA microarrays based on the C. glutamicum R genome (Yukawa et al. 2007), transcriptional profiles of densely packed cells under oxygen-deprived conditions in minimal medium compared to those under normal aeration conditions in nutrient medium at physiological cell densities revealed a total of 161 genes that showed a greater than twofold increase in expression and 221 that were transcribed at levels less than half those observed under aerobic conditions (Inui et al. 2007). The National Center for Biotechnology Information Cluster of Orthologous Groups database groups the upregulated and downregulated genes as genes involved in transcription, translation, DNA replication, transport, or metabolism. Of these, expression of several genes involved in methionine biosynthesis was upregulated under oxygen-deprived conditions.
There are two major pathways for methionine biosynthesis in microorganisms. In Escherichia coli, it occurs through the transsulfuration pathway, whereas Saccharomyces cerevisiae and Pseudomonas aeruginosa utilize the direct sulfhydrylation pathway. The transsulfuration pathway involves cystathionine as an intermediate and utilizes cysteine as the sulfur source, but the direct sulfhydrylation pathway bypasses cystathionine and uses inorganic sulfur instead of cysteine. While most microorganisms synthesize methionine via either one of these pathways, C. glutamicum utilizes both (Hwang et al. 2002). The set of genes involved in l-methionine biosynthesis in C. glutamicum is found on the genome sequence. The hom gene is repressed at the transcriptional level and the enzymatic activities of metY and metB gene products are repressed in cells grown in the presence of methionine (Follettie et al. 1988; Hwang et al. 2002). Proteomic analysis have also revealed that the levels of MetY, MetK, and Hom protein are reduced by the addition of l-methionine to the growth medium (Rey et al. 2003). However, regulation mechanisms of the methionine biosynthesis genes in response to methionine in C. glutamicum remain unclear. In E. coli and Bacillus subtilis, S-adenosylmethionine (SAM), a derivative of methionine, is the major effector controlling regulation of methionine biosynthesis genes at the level of transcription initiation (Belfaiza et al. 1987; Kromer et al. 2006b; Rodionov et al. 2004; Saint-Girons et al. 1986; Smith et al. 1985; Usuda and Kurahashi 2005). Since SAM is used as a methyl donor in many transmethylation reactions of biomolecules such as proteins, DNA, RNA, and phospholipids, biosynthesis of SAM via methionine must be important for coordination of the relevant cell functions.
McbR, a TetR-type transcriptional regulator, is involved in regulation of the metabolic network directing the synthesis of l-methionine in C. glutamicum (Rey et al. 2003, 2005). A systematic search for the potential effector substance modulating the function of McbR revealed that only S-adenosylhomocysteine, which is produced from SAM in the SAM-dependent methyltransferase reactions, prevented the binding of McbR to its target sequence (Rey et al. 2005). However, involvement of SAH and McbR in downregulation of methionine biosynthesis genes in response to methionine in culture medium is not fully understood, although repression of the MetY and Hom protein levels by methionine was not detected in a mcbR mutant (Rey et al. 2003, 2005).
In this study, we show that McbR is involved in regulation of methionine biosynthesis genes in response to methionine supplementation in C. glutamicum growth-arrested cells under oxygen-deprived conditions. Furthermore, DNA binding of McbR is shown to be affected by not only SAH but also SAM.
Materials and methods
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table 1.
Media and growth conditions
For genetic manipulations, E. coli strains were grown at 37°C in Luria-Bertani (LB) medium and C. glutamicum strains were grown at 33°C in A medium (2 g l−1 yeast extract, 7 g l−1 casamino acid, 2 g l−1 urea, 7 g l−1 (NH4) 2SO4, 0.5 g l−1 KH 2PO4, 0.5 g l−1 K2H2PO4, 0.5 g l−1 MgSO4–7H2O, 6 mg l−1 Fe2SO4–7H2O, 4.2 mg l−1 Mn2SO4–H2O, 0.2 mg l−1 biotin, 0.2 mg l−1 thiamine) with 4% glucose on a rotary shaker at 200 rpm. When appropriate, media were supplemented with antibiotics. The final antibiotic concentrations for E. coli were 50 μg ampicillin per milliliter and 50 μg chloramphenicol per milliliter.
For analysis of gene expression C. glutamicum R cells were cultivated at 33°C, pH 7.6, for 13 h in 500 ml A medium containing 4% glucose in a 1-l jar fermenter (BMJ01PI, Biott). The aeration was set at 1 l min−1 and the agitation speed was 1,000 rpm. The culture was harvested by centrifugation at 6,500×g and 4°C for 15 min. The cell pellet was subsequently washed once with minimal medium (BT medium), which differs from the A medium by the absence of yeast extract and casamino acids. For incubation under oxygen-deprived conditions, the washed cells were resuspended at a final cell concentration of 50 g wet cell weight per liter with 80 ml BT medium containing 200 mM glucose and 400 mM bicarbonate. When indicated, methionine was added at a final concentration of 0.75 mM. The cell suspension was incubated at 33°C with constant agitation without aeration in a lidded 100-ml medium bottle. The pH of the cell reaction was maintained at 8 using a pH controller (DT-1023, Biott) to supplement the medium with NH3.
Quantitative RT-PCR
Total RNA was isolated from cells using the RNeasy Kit (QIAGEN). Total RNA of 20 ng was used as template for analysis of the methionine biosynthesis genes and 0.4 ng was used for analysis of the 16S rRNA to generate cDNA and for the subsequent polymerase chain reaction (PCR) reaction. Each PCR reaction consisted of the amount of total RNA 12.5-μl Power SYBR Green PCR Master Mix, 0.5-μM forward primer, 0.5-μM reverse primer, 8-U RNase inhibitor, and 5-U MuLV reverse transcriptase in a total volume of 20 μl. PCR parameters were 50°C for 30 min, 95°C for 10 min, and 40 cycles at 95°C for 15 s and 60°C for 60 s. The result for 16S rRNA was used as an internal control.
Primer extension
Total RNA (100 μg) and 1.5-pmol primer were mixed and annealed at 80°C for 90 min, at 60°C for 90 min and 30°C for 90 min, using a GeneAmp PCR System 9700 instrument (Applied Biosystems). cDNA was synthesized at 42°C for 1 h using AMV Reverse Transcriptase (Promega). The reaction was terminated by adding EDTA (pH 8.0) at a final concentration of 250 mM and DNase-free RNase A at a final concentration of 3 ng μl−1 to trigger degradation of the RNA templates. The resulting cDNA was treated by phenol–chloroform extraction and ethanol precipitation. Upon centrifugation, the precipitated DNA pellet was resuspended in IR2 Stop Solution (LI-COR). The primer extension products were treated at 95°C for 2 min and placed on ice for 5 min and separated on 3.7% KBPlus Gel Matrix (LI-COR) using a LI-COR 4300 DNA analyzer. The migration position of each primer extension product was determined by comparing a sequencing ladder generated from a DNA fragment corresponding to the same chromosomal region, using the same primers and DYEnamic Direct Cycle Sequencing kit with 7-deaza-dGTP (Amersham Biosciences).
Purification of His-tagged McbR protein in E. coli
Overexpression of McbR with an amino-terminal six-His tag was achieved by using pCold I. E. coli cells harboring pCRB207 were grown at 37°C in 100 ml of LB medium to OD610 of 0.5. Cultures were incubated at 15°C for 30 min and then IPTG was added at a final concentration of 0.1 mM and shaken for 24 h. Cells were harvested by centrifugation and suspended in 900 μl of His-binding buffer (0.5 M NaCl, 20 mM Tris–HCl, 5 mM imidazole, pH 7.9; Novagen), 100 μl of Fast Break Cell Lysis Reagent (Promega), and 0.2 mg of lysozyme. The mixture was incubated for 15 min at room temperature and centrifuged for 5 min at 12,000 × g and the supernatant was pooled. His6-McbR was purified using His Bind Resin and Buffer Kit (Novagen) according to the procedure specified by the manufacturer.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were carried out in a total volume of 20 μl of binding buffer (20 mM Tris–HCl, pH 7.9, 100 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT, 5% (w/v) glycerol, and 50 μg ml−1 bovine serum albumin). The metY promoter DNA fragment was amplified by PCR using primers listed in Table 2. The DNA fragments containing nucleotide substitutions at putative McbR binding sites were prepared as follows.
The DNA fragments were incubated with the purified McbR at 33°C for 10 min. The mixture was fractionated by electrophoresis on a native 5% (w/v) polyacrylamide gel containing 5% (w/v) glycerol in 1/2 TBE (45 mM Tris–borate, pH 8.3, 1 mM EDTA) at room temperature. The DNA bands were stained with ethidium bromide and visualized by UV irradiation.
Results
Methionine biosynthesis genes are upregulated under oxygen-deprived conditions
Expression levels of methionine biosynthesis genes in C. glutamicum cells under oxygen-deprived conditions were compared to those under aerobic conditions using quantitative reverse transcription (RT)-PCR. C. glutamicum cells cultivated in a nutrient-rich A medium under aerobic conditions were collected and resuspended to a high density in BT minimal medium. The resuspended cells were incubated under oxygen-deprived conditions where they produced lactate and succinate from glucose at a high yield even though cellular growth was arrested. Total RNA was prepared from the cells grown under aerobic conditions (exponentially growing cells) and after 2-h incubation under oxygen deprivation. The expression levels of metX, metB, metY, metE, metH, and metK genes under oxygen-deprived conditions were significantly higher than those under aerobic conditions (Fig. 1). This result is consistent with our previous microarray analyses (Inui et al. 2007).
Characteristics of the promoter regions in methionine biosynthesis genes
The transcriptional start sites of eight C. glutamicum methionine biosynthesis genes, metX, metY, metE, metH, metK, metF, aecD, and metB, were determined by primer extension. Two transcriptional start sites of metY were found, while a single transcriptional start site each was detected for the other seven genes (Fig. 2). Based on the findings on promoter regions of C. glutamicum (Patek et al. 2003), the characteristics of the −10 box and −35 box regions located upstream of the methionine biosynthesis genes (Fig. 3), the consensus McbR binding sequence was located in the vicinity of the −10 and −35 boxes of metX, metY, metH, metK, and metB genes. A putative McbR binding site is located 217 bases upstream of the transcriptional start site of metE. However, another binding site in less agreement with the consensus sequence site occurs 16 bases downstream of the transcriptional start site. No probable McbR binding site was found in the promoter regions of metF and aecD genes. It is noteworthy that the putative McbR binding sites are found in the promoter regions of metX, metY, metH, metK, metB, and metE, whose expression was enhanced under oxygen-deprived conditions. The expression of metF and aecD, which have no McbR binding motifs in their promoter regions, was not enhanced under the conditions (Fig. 1).
Primer extension analyses of methionine biosynthesis genes under oxygen-deprived conditions. As for each sequencing gel analysis of primer extension, high-resolution electrophoresis of 5’-labeled cDNA products are shown aligned with a sequencing ladder generated from DNA from the same region, using the same primer. Dideoxy sequencing marker lanes A, C, T, and G are indicated and cDNA synthesis from the RNA samples is shown in the next lane (PE). The migration position of the primer extension product is indicated with an arrow. A portion of the DNA sequence is shown on the left, and the asterisk indicates the putative transcriptional start site
Sequence alignment of promoter regions with potential McbR binding site of methionine biosynthesis genes. The ATG start codon of each gene is indicated in bold italics letters. The experimentally determined sites of transcription initiation are given in bold letters. The deduced extended −10 and −35 boxes for each promoter are boxed. McbR binding motifs (TAGAC-N6-GTCTA) were underlined
Methionine biosynthesis genes in mcbR-deficient strain are bit subject to methionine repression
One of the major advantages of bioprocesses for organic acid production based on growth-arrested C. glutamicum cells under oxygen-deprived conditions is the high productivity in the absence of complex natural nutrients such as yeast extract conventionally used to optimize cell growth. Therefore, the induction of expression of the C. glutamicum methionine biosynthesis genes under oxygen-deprived conditions may be ascribed to the absence of methionine in the culture medium. To confirm this, effects of supplementation of medium with methionine on expression of the genes was examined by quantitative RT-PCR. The results revealed mRNA levels of metE, metH, metB, metY metK, and metX that were significantly lower in the presence of methionine than in its absence (Table 3). It should be noted that these genes are regulated in response to methionine even in the growth-arrested cells under oxygen-deprived conditions.
To examine the role of McbR on methionine response of methionine biosynthesis genes under oxygen-deprived conditions, expression of the genes in an mcbR-deficient mutant was analyzed by quantitative RT-PCR. Aerobic growth was significantly delayed by deletion of mcbR, as reported previously (Kromer et al. 2006a). Under aerobic conditions, metE, metH, metB, metY, metK, and metX mRNA levels were higher in the mcbR mutant than in the wild type (data not shown). The repression of the six genes by methionine observed in the wild-type strain was not observed in the mcbR-deficient mutant strain (Table 3). It should be noted that the mRNA levels of metF and aecD genes which do not possess an McbR binding motif in their promoter regions remained unchanged irrespective of changes in culture conditions.
McbR repressor binding to the metY promoter region is promoted by SAM, inhibited by SAH, but not affected by methionine
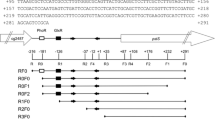
The mRNA level of metY was most markedly increased by disruption of mcbR (data not shown). Although two McbR binding motifs were identified in its promoter region, binding of McbR to the motifs was not verified experimentally yet. EMSA using McbR-His tag fusion protein and each of four DNA probes (Y1, Y2, N, and M) derived from the promoter region of metY (Fig. 4a) was therefore performed. Each DNA probe was incubated with different amounts of the purified McbR-His tag fusion protein and the mixture subjected to gel electrophoresis. The presence of McbR protein caused the band to shift and separate with DNA fragment Y1 containing two sets of the McbR binding motif. One band shift was observed with the Y2 fragment containing only one McbR binding motif. On the other hand, no band shift was observed with an arbitrary fragment N containing no McbR binding motif. Furthermore, mutation of the McbR binding motif in the Y2 fragment (M) lost the binding to McbR (Fig. 4b). These results indicated that both motifs were involved in the McbR binding.
Electrophoretic mobility shift assays using the metY promoter region and a purified His-tagged McbR protein. a The promoter region of the metY gene. metY is indicated with a white arrow. DNA fragments (Y1, 127 bp; Y2, 89 bp; M, 89 bp; and N, 235 bp) used in the EMSAs are indicated with bars. A magnification shows the location of the putative McbR site with arrows (TAGAC-N6-GTCTA). Mutations introduced in the putative McbR binding motif, which are indicated with bold letters, are shown. b Binding of the His-tagged McbR protein to the metY promoter region. Y1, Y2, N, and M at the indicated concentration were incubated with different concentrations of His-tagged McbR protein. c Effect of the binding of the McbR protein to the metY promoter region in the presence of methionine, S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH). Y2 DNA fragment at 100 nM was incubated with the His-tagged McbR protein at 680 nM in the presence of methionine, SAM, and SAH at concentrations indicated in each lane. The DNA–protein complex is indicated by an arrow
It is likely that McbR binds to the promoter region and represses the expression of the methionine biosynthesis genes in cells grown in the presence of methionine. Therefore, methionine or its metabolites may act as a positive effector for McbR binding although a methionine metabolite SAH is rather reported to act as a negative effector for the binding of McbR to the promoter region of hom (Rey et al. 2005). The effects of methionine and its metabolites, SAM and SAH, on McbR binding to the promoter region of metY was examined by EMSAs. While addition of methionine showed minimal effects on the McbR binding to its target DNA, the binding was promoted by addition of SAM. In contrast, the binding was inhibited by SAH, which is consistent with the analyses using the promoter region of hom (Rey et al. 2005; Fig. 4c).
Discussion
In this study, it was shown that the mRNA level of metX, metY, metE, metH, metK, and metB was repressed by methionine in C. glutamicum cells under oxygen-deprived conditions. Although it has been reported that expression of some methionine biosynthesis genes is repressed in the presence of methionine in C. glutamicum, changes in the transcript level were shown only for hom, and changes in the protein level were shown for metY, metK, cysK, and metB (Follettie et al. 1988; Rey et al. 2003). Methionine is not only required for protein synthesis but is also metabolized to SAM, which is a universal methyl donor for various cellular methylation reactions (Kramer et al. 1987; Macintyre et al. 2001). Thus, the intracellular SAM concentration seems to be strictly regulated in response to the intracellular and/or extracellular conditions. SAM also plays an important role in regulation of cell division. A mutant with low levels of SAM exhibited defects in cell division (Newman et al. 1998; Wang et al. 2005; Wei and Newman 2002). The gene encoding SAM-dependent methyltransferase is located in the cell-division-related gene cluster on the E. coli chromosome, and therefore methyltransferase might be involved in cell division. Similarly, the cluster of cell division related genes in the C. glutamicum genome contains a putative SAM-dependent methyltransferase-encoding gene. C. glutamicum cells remain able to excrete, in significant amounts, several metabolites, despite the cessation of cellular growth under oxygen-deprived conditions. The induction of methionine biosynthesis genes under these conditions may suggest the important role of SAM in the high metabolic activity of growth-arrested cells.
In this study, the transcriptional start site of eight genes involved in methionine biosynthesis was determined, indicating that the consensus sequence of the McbR binding site (Rey et al. 2005) is located in the vicinity of the −10 and −35 boxes of six of these genes (metX, metY, metE, metH, metK, and metB), which is consistent with the earlier finding that this type of regulator binds to its operator region to repress transcription (Ramos et al. 2005). In the mcbR mutant, no repression of the six genes expression by methionine was observed. EMSAs confirmed that McbR binds to the McbR binding motif in the promoter region of metY, as well as the corresponding site derived from the upstream region of hom (Rey et al. 2005). These results suggest that McbR acts as a repressor of the methionine biosynthesis genes in the presence of methionine. The binding of the McbR protein was inhibited by SAH, which is a derivative of the methylation agent SAM, as reported for the hom promoter (Rey et al. 2005). It was suggested that the McbR protein senses the cellular SAH level that directly reflects the consumption of SAM, as both metabolites are physiologically linked by the rate of transmethylation reactions (Rey et al. 2005). However, the effect of SAH alone cannot explain the repression of gene expression by addition of methionine. EMSAs in this study suggested that SAM functions as a corepressor, enhancing the binding of McbR to the operator sequence. Therefore, the repression of the McbR regulon by addition of methionine may be due to an increase of the intracellular SAM concentration. The mechanism of regulation of methionine biosynthesis genes at the transcriptional level has been well studied in E. coli and B. subtilis (Auger et al. 2002; Belfaiza et al. 1987; Saint-Girons et al. 1986; Usuda and Kurahashi 2005). In these bacteria, transcription of methionine biosynthesis genes is regulated through a different feedback mechanism by SAM (Rodionov et al. 2004; Usuda and Kurahashi 2005). Distinctively, in C. glutamicum, the regulation of methionine biosynthesis genes via McbR may be modulated by the ratio of SAM to SAH in the cell. The regulatory system of gene expression in response to methionine under oxygen-deprived conditions demonstrated in this study will be applied to control gene expression for improvement of high-productivity bioprocesses under these conditions.
References
Auger S, Danchin A, Martin-Verstraete I (2002) Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J Bacteriol 184:5179–5186
Belfaiza J, Guillou Y, Margarita D, Perrin D, Saint Girons I (1987) Operator-constitutive mutations of the Escherichia coli metF gene. J Bacteriol 169:670–674
Follettie MT, Shin HK, Sinskey AJ (1988) Organization and regulation of the Corynebacterium glutamicum hom-thrB and thrC loci. Mol Microbiol 2:53–62
Hwang BJ, Yeom HJ, Kim Y, Lee HS (2002) Corynebacterium glutamicum utilizes both transsulfuration and direct sulfhydrylation pathways for methionine biosynthesis. J Bacteriol 184:1277–1286
Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H (2004a) Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J Mol Microbiol Biotechnol 8:243–254
Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H (2004b) Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol 7:182–196
Inui M, Suda M, Okino S, Nonaka H, Puskás LG, Vertès AA, Yukawa H (2007) Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology 153:2491–2504
Kramer DL, Sufrin JR, Porter CW (1987) Relative effects of S-adenosylmethionine depletion on nucleic acid methylation and polyamine biosynthesis. Biochem J 247:259–265
Kromer JO, Heinzle E, Schroder H, Wittmann C (2006a) Accumulation of homolanthionine and activation of a novel pathway for isoleucine biosynthesis in Corynebacterium glutamicum McbR deletion strains. J Bacteriol 188:609–618
Kromer JO, Wittmann C, Schroder H, Heinzle E (2006b) Metabolic pathway analysis for rational design of l-methionine production by Escherichia coli and Corynebacterium glutamicum. Metab Eng 8:353–369
Kumagai H (2000) Microbial production of amino acids in Japan. Adv Biochem Eng Biotechnol 69:71–85
Macintyre G, Atwood CV, Cupples CG (2001) Lowering S-adenosylmethionine levels in Escherichia coli modulates C-to-T transition mutations. J Bacteriol 183:921–927
Newman EB, Budman LI, Chan EC, Greene RC, Lin RT, Woldringh CL, D’Ari R (1998) Lack of S-adenosylmethionine results in a cell division defect in Escherichia coli. J Bacteriol 180:3614–3619
Okino S, Inui M, Yukawa H (2005) Production of organic acids by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 68:475–480
Okino S, Suda M, Fujikura K, Inui M, Yukawa H (2008) Production of d-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 78:449–454
Patek M, Nesvera J, Guyonvarch A, Reyes O, Leblon G (2003) Promoters of Corynebacterium glutamicum. J Biotechnol 104:311–323
Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R (2005) The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356
Rey DA, Puhler A, Kalinowski J (2003) The putative transcriptional repressor McbR, member of the TetR-family, is involved in the regulation of the metabolic network directing the synthesis of sulfur containing amino acids in Corynebacterium glutamicum. J Biotechnol 103:51–65
Rey DA, Nentwich SS, Koch DJ, Ruckert C, Puhler A, Tauch A, Kalinowski J (2005) The McbR repressor modulated by the effector substance S-adenosylhomocysteine controls directly the transcription of a regulon involved in sulphur metabolism of Corynebacterium glutamicum ATCC 13032. Mol Microbiol 56:871–887
Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS (2004) Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res 32:3340–3353
Saint-Girons I, Belfaiza J, Guillou Y, Perrin D, Guiso N, Barzu O, Cohen GN (1986) Interactions of the Escherichia coli methionine repressor with the metF operator and with its corepressor, S-adenosylmethionine. J Biol Chem 261:10936–10940
Smith AA, Greene RC, Kirby TW, Hindenach BR (1985) Isolation and characterization of the product of the methionine-regulatory gene metJ of Escherichia coli K-12. Proc Natl Acad Sci USA 82:6104–6108
Usuda Y, Kurahashi O (2005) Effects of deregulation of methionine biosynthesis on methionine excretion in Escherichia coli. Appl Environ Microbiol 71:3228–3234
Wang S, Arends SJ, Weiss DS, Newman EB (2005) A deficiency in S-adenosylmethionine synthetase interrupts assembly of the septal ring in Escherichia coli K-12. Mol Microbiol 58:791–799
Wei Y, Newman EB (2002) Studies on the role of the metK gene product of Escherichia coli K-12. Mol Microbiol 43:1651–1656
Yukawa H, Omumasaba CA, Nonaka H, Kos P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y, Vertès AA, Inui M (2007) Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058
Acknowledgements
We thank Crispinus A. Omumasaba (RITE) for critical reading of the manuscript. This work was financially supported in part by the New Energy and Industrial Technology Development Organization, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Suda, M., Teramoto, H., Imamiya, T. et al. Transcriptional regulation of Corynebacterium glutamicum methionine biosynthesis genes in response to methionine supplementation under oxygen deprivation. Appl Microbiol Biotechnol 81, 505–513 (2008). https://doi.org/10.1007/s00253-008-1694-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1694-9