Abstract
A novel water soluble ternary copper(ii) complex,—[Cu2(phen)2(3-IAA)2(H2O)](ClO4)2·H2O—(phen: 1,10-phenanthroline, 3-IAA: 3-indoleacetic acid), has been synthesized and characterized by elemental CHN analysis, ESI-TOF, FTIR and single-crystal X-ray diffraction techniques. Interaction of the complex with calf thymus DNA (CT-DNA) has been investigated by absorption spectral titration, ethidium bromide (EB) and Hoechst 33258 displacement assay. The interactions between the complex and bovine serum albumin (BSA) were investigated by electronic absorption and fluorescence spectroscopy methods. The experimental results indicate that the fluorescence quenching mechanism between the complex and BSA is a static quenching process. The Stern–Volmer constants, binding constants, binding sites and the corresponding thermodynamic parameters (ΔG, ΔH, ΔS) of BSA + complex systems were determined at different temperatures. The binding distance between the complex and BSA was calculated according to Förster non-radiation energy transfer theory (FRET). The effect of the complex on the conformation of BSA was also examined using synchronous, two dimensional (2D) and three dimensional (3D) fluorescence spectroscopy. Furthermore, the oxygen radical scavenging activity of the complex was determined in terms of IC50, using the DPPH and H2O2 method, to show that it particularly enables electron loss from radical species. This study highlights the importance of indole and moieties in the development of antioxidant agents.
Graphic abstract
A potent drug candidate novel water soluble ternary copper(II) complex,—[Cu2(phen)2(3-IAA)2(H2O)] (ClO4)2·H2O—(phen: 1,10-phenanthroline, 3-IAA: 3-indoleacetic acid), has been synthesized and characterized by elemental CHN analysis, FTIR, ESI–MS and single-crystal X-ray diffraction techniques. The complex has been tested for in vitro biomacromolecular interactions by spectroscopic methods. Furthermore, radical scavenging activities of the complex were also investigated.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Auxin plays significant roles during the whole life span of a plant. The most abundant naturally occurring auxin is 3-indoleacetic acid (3-IAA) (Strader and Bartel 2011; Kai et al. 2007; Rampey et al. 2004). 3-indoleacetic acid consist of an acetic acid side chain linked by an indole ring. The indole ring is found in compounds as various as the hormone serotonin, the amino acid tryptophan, the psychedelic drug LSD and the antimigrane drug rizatriptan (Lal and Snape 2012; Horton et al. 2003). Indole has important properties because of the presence of an electron rich NH moiety that could interact noncovalently with other molecules by formation of hydrogen bonding in the NH moiety by π–π stacking (Lal and Snape 2012; Shimazakia et al. 2009). The hydrogen bonding donor or acceptor groups of indole are not only facilitative in establishing intermolecular contacts between small molecules and macromolecule such as DNA, protein, enzyme but also bind DNA sequences with some specificity (Pandya et al. 2010). Because of these properties, indole rings are extensively used to advance novel biologically active derivatives and biological activities (antiviral, antifungal, anticancer, antioxidant, antimicrobial, etc.) are assigned to compounds bearing indole moieties (Suzen and Buyukbingol 2000; Suzen et al. 2006; Tiwari et al. 2006; Ryu et al. 2007; Williams et al. 2005). Compounds containing indole are significant and efficient antioxidants, protecting both lipids and proteins from peroxidation, and the indole structure effects the antioxidant efficacy in biological systems (Dannhardt and Kiefer 2001; Brown et al. 1991). Another ligand used in this study, 1,10-phenanthroline and derivatives are an important class of chelating agents and has structural features such as being rigid planar, hydrophobic, and an electron-poor heteroaromatic system. Some metal complexes containing 1,10-phenanthroline and derivatives are known to bind to DNA by an intercalative mode and have interesting anticancer properties (Mital et al. 1989; Mansuri-Torshizi et al. 1991; Skurai et al. 1995). Recently, we have reported the synthesis, characterization, biomolecular interactions and biological activities of some binary and ternary copper(ıı) complexes (Şenel et al. 2019; Kiraz et al. 2019; İnci et al. 2019a, b, 2018a, b, 2017a).
In light of the above attention, our group has focused on the synthesis and characterisation (CHN analysis, FTIR, ESI–MS and single crystal X-ray diffraction techniques) of the complex. The detailed structure of the complex with the formulas [Cu2(phen)2(3-IAA)2(H2O)](ClO4)2·H2O (phen: 1,10-phenanthroline, 3-IAA: 3-indoleacetic acid) has also been determined using single crystal X‐ray analysis (Fig. 1). The binding properties between the complex and DNA/BSA were studied by electronic absorption and fluorescence spectroscopy and the binding mechanism regarding the binding parameters were investigated. The effect of radical scavenging activity of the complex was also evaluated.
Experimental
Materials and measurements
All chemicals were of reagent grade, purchased from different sources, and were used without further purification. 1,10-phenanthroline, 3-indoleacetic acid (3-IAA), Cu(ClO4)2·6H2O, methanol, ethanol, DMSO, CH3CN, DMF, NaCl, tris-(hydroxymethyl)aminomethane-HCl, CT-DNA, BSA, ethidium bromide, Hoescht 33258 (2-(4-hydroxy-phenyl)-5-[5-(4-methylpipera-zine-1-yl) benzimidazo-2-yl]-benzimidazole), hydrogen peroxide, 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (Trolox), 1,1-diphenyl-picrylhydrazyl (DPPH), butylated hydroxytoluene (BHT), and ascorbic acid were provided from Sigma-Aldrich. All solutions were prepared with analytical grade water (R = 18 MΩ) using grade A glassware. CHN elemental analysis were performed using a Costech elemental analyzer at the Technical and Scientific Research Council of Turkey, TUBITAK Bursa Test, and Analysis Laboratory. Infrared spectra were recorded from 4000 to 400 cm−1 with a Thermo–Nicolet 6700 Fourier Transform Infrared Spectrometer using KBr pellets. The spectrophotometric studies were performed using a GBC Cintra 303 UV–Visible spectrophotometer connected with a Peltier thermocell. Emission intensity measurements were carried out using a Jasco FP-750 spectrofluorometer. Electrospray ionization (ESI) mass spectra were recorded using a Bruker Daltonics Microtof II‐ESI‐TOF mass spectrometer at the TUBITAK‐MAM Test and Analysis Laboratory.
General procedure for the synthesis of the complex
[Cu2(phen)2(3-IAA)2(H2O)](ClO4)2·H2O
The complex was prepared by following a procedure in which a mixture of 1.0 mmol phen and 1.0 mmol Cu(ClO4)2·6H2O in 20 mL of CH3OH was added dropwise to 1.0 mmol 3-IAA with stirring for about 2 h. Slow evaporation of the solvent gave crystalline product in 91% yield. The resulting solution was left to evaporate slowly at room temperature and the dark green crystals were obtained.
Yield was 91%. Anal. calcd. for C44 H36 Cl2 Cu2 N6 O14 (1070.77 g mol–1) (%): C, 49.36; H, 3.39; N, 7.85. Found: C, 48.95; H, 3.42; N, 7.81. FTIR (w, weak; m, medium, s, strong): ν(C=N)phen 1418s, ν(C=C)phen 1618s, δ(C–H) 791 and 712, ν(N–H)3-IAA 3373m, νs(COO)3-IAA 1578, νas(COO)3-IAA 1432, (ClO4−) 1070m. UV–Vis (λnm, nm and in 1 × 10–3 M water): 271.6 (55,735.96 M−1 cm−1). ESI‐MS (m/z): 417.05 for [Cu(phen)(3-IAA)]+.
Crystal structure determination
Data for the complex were obtained with Bruker APEX II QUAZAR three-circle diffractometer. Indexing was performed using Bruker (2014a). Data integration and reduction were carried out with Bruker (2013). Absorption correction was performed by multi-scan method implemented in Bruker (2014b). The structure was solved using SHELXT (Sheldrick 2015a) and then refined by full-matrix least-squares refinements on F2 using the SHELXL (Sheldrick 2015b) in Olex2 Software Package (Dolomanov et al. 2009). Crystallographic data and refinement details of the data collection for the complex are given in Table 1. Crystal structure validations and geometrical calculations were performed using Platon software (Spek 2009) and Mercury software (Macrae et al. 2006) was used for visualization of the cif files. Additional crystallographic data with CCDC reference number 1996812 for the complex has been deposited within the Cambridge Crystallographic Data Center via www.ccdc.cam.ac.uk/deposit.
DNA interaction studies
The stock solution of CT-DNA was prepared by dissolving DNA in Tris–HCl buffer (5 mM Tris–HCl/50 mM NaCl buffer at pH 7.2). The stock solutions of the CT-DNA in Tris–HCl buffer gave the ratio of UV absorbance at 260 and 280 nm, A260/A280, of ca. 1.9, indicating that the DNA was sufficiently free of protein. The concentration of CT-DNA (in nucleotide) stock solution was determined by electronic absorption spectroscopy, in a series diluted samples, using the molar absorption coefficient 6600 M−1 cm−1 of a single nucleotide at 260 nm (Reichmann et al. 1954). Concentrated stock solution of the complex was dissolved with water to required concentrations. Electronic absorption spectral titration experiments were performed by keeping the concentration of the complex constant (15 µM) while varying CT-DNA concentration (0–450 µM). After each addition of CT-DNA to the complex, the resulting solution was allowed to equilibrate at 298.2 K for 2 min followed by recording of absorption patterns. The fluorescence spectral method using EB as a reference was used to determine the relative DNA binding properties of the complex to CT-DNA. For fluorescence experiments, CT-DNA (5 μM) was pretreated with EB (5 μM). The emission spectra of the complex (0–500 μM) were recorded in the 520–700 nm range upon excitation at 295 nm in EB displacement assay. Hoechst 33258 displacement assay was also performed under similar conditions to EB assay. CT-DNA (5 μM) was pretreated with Hoechst 33258 (5 μM). The emission spectra of the complex (0–500 μM) were recorded in the 350–700 nm range upon excitation at 341 nm in Hoechst 33258 displacement assay.
BSA interaction studies
All experiments containing BSA were performed in Tris–HCl buffer solution (5 mM Tris–HCl/50 mM NaCl buffer at pH 7.2). Final solutions of BSA and the complex were prepared by dissolving them in the water to required concentrations. The electronic absorption spectra of the BSA and the solution of the complex at 15 μM concentrations were measured in the range of 200–400 nm. In the tryptophan fluorescence quenching experiment, quenching of the tryptophan residues of BSA was done by keeping the constant concentration of BSA (1 μM) while varying the complex concentration (0–18 μM). The fluorescence measurements were performed at three temperatures (298.2, 310.2 and 318.2 K). The range of synchronous scanning were λex = 280 and λem = 298 at 15 nm, λex = 280 and λem = 342 at 60 nm. The three-dimensional (3D) and (2D) countour plot excitation and emission spectra of BSA in the absence and presence of the complex were recorded in the range of 220–350 nm.
Radical scavenging studies
DPPH radical scavenging activity
To determine the DPPH· free radical scavenging capacity of the complex, a solution of DPPH· (0.5 mL, 0.1 mM) was added to 1.5 mL of the complex solutions in ethanol at different concentrations (Gulcin 2006). These solutions were then vortexed thoroughly and incubated in the dark for 30 min. After another 30 min, the absorbance was measured at 517 nm.
Hydrogen peroxide scavenging activity
The principle of the assay is based on a decrease in the absorbance of hydrogen peroxide upon oxidation of hydrogen peroxide (Ruch et al. 1989). A solution of 40 mM hydrogen peroxide was prepared in 0.1 M phosphate buffer (pH 7.4). After different concentrations of the complex in 3.4 mL phosphate buffer were added to 0.6 mL of hydrogen peroxide solution (40 mM), the absorbance of the mixture was recorded at 230 nm. Trolox, ascorbic acid, and BHT were used as reference compounds. The percentage of radical scavenging by the complex and reference compounds was calculated using the following equation:
where A0 and Ac represent the absorbance in the absence and presence of the complex, respectively. The 50% activity (IC50) was calculated using the percentage of activity.
Results and discussion
Description of the crystal structure
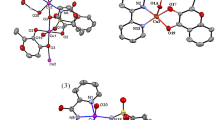
The solid-state structures and geometries of the complex were determined by single-crystal X-ray structural analysis. Crystallographic data and refinement details of the data collection are given in Table 1. The selected bond lengths and bond angles are given in Table 2. Single-crystal X-ray diffraction analysis reveals that the complex crystallizes in the triclinic crystal system with space group P-1. The solid-state structure of the complex is shown in Fig. 2a.
a Crystal structure of [Cu2(phen)2(3-IAA)2(H2O)](ClO4)2·H2O with displacement ellipsoids drawn at the 30% probability level. The uncoordinated chlorate anion and lattice water molecule are omitted for clarity. H-atoms are shown as small spheres of arbitrary radii. b A view of the copper coordination environment in [Cu2(phen)2(IA)2(H2O)]2+. c Perspective view of intermoleculer hydrogen bonding interactions in crystal structure. d, e Illustration of intra- and intermolecular π · π stacking interactions
The structure consists of two Cu(ıı) ion linked by two indole-3-acetate (3-IAA−), one bridged water ligand, two chelating phenanthroline (phen), two uncoordinated perchlorate (ClO4−) as an electron balancing anion, and one lattice water molecule. As can be seen in Fig. 2a, b, the central Cu1 ion has a distorted five-coordinate square pyramidal geometry (CuN3O2), in which the tau‐descriptor (τ5) values for Cu1 atoms is 0.01 (Addision et al. 1984), through two carboxylate oxygen atoms (O1, O3) of 3-IAA, one chelating phen nitrogen atoms (N1, N2), an oxygen atom (O13) of water ligand in the axial position. The other Cu2 centre has almost same coordination arrangement as in Cu1 ion, except for the weak coordination bond (Cu2–O4 = 2.676 (4) Å). In [Cu2(phen)2(3-IAA)2(H2O)](ClO4)2·H2O, equatorial bond lengths (Cu1–N1 = 2.002 (4) Å, Cu1–N2 = 1.987 (4) Å, Cu2–N3 = 2.002 (4) Å, Cu2–N4 = 1.995 (4) Å, Cu1–O1 = 1.933 (4) Å, Cu1–O3 = 1.999 (3) Å, Cu2–O2 = 1.945 (4) Å, Cu2–O3 = 2.010 (3) 2.010 (3)) and weakly bound axial bond distances (Cu1–O13 = 2.411 (4) Å, Cu2–O13 = 2.348 (4) Å) are comparable to Cu(ii) based complexes (İnci et al. 2016, 2017b, 2019b). Crystal structure is mainly stabilized by classical O–H · O, N–H · O and non-classical C–H · O, X–H · Cl (X: N, O, C) hydrogen bonding interactions (Table 3) as existed in many supramolecular systems. The chlorate anion as hydrogen bond acceptor participates three classical hydrogen bonding interactions (O14 · O9 = 2.831 (10) Å, O8 · N5 = 2.996 (6) Å, O6 · O13 = 2.802 (7) Å), which are defined as moderate H-bonds. The lattice water molecule behave as both hydrogen bond donor and acceptor to link perchlorate and complex cation [Cu2(phen)2(3-IAA)2(H2O)]2+ by moderate H-bonds, namely, O14 · O4 = 2.831 (9) Å and O13 · O14 = 2.909 (9) Å as can be seen in Fig. 2c. Additionaly, the weak C–H · O interactions contibuted the stability of crystal structure (Table 3). As shown in Fig. 2d, complex exhibits strong (3.366 and 3.744 Å) and moderate (4.264, 4.316 Å) intramolecular aromatic ring stacking interactions between the aromatic ring of the phen and 3-IAA ligand. In addition to intramolecular aromatic ring stacking, strong and moderate intermolecular π-stacking interactions (3.662, 3.720, and 3.918 Å) are observed in the three-dimensional supramolecular network (Fig. 2e).
Spectral characterization
The FTIR spectrum showed peaks associated with ligands (phen and 3-IAA) coordinated to a copper(ıı) metal ion. The strong absorption peaks at 3373 cm−1 belong to the –NH– group. No significant shifts were observed on the peaks among 3-IAA, suggesting that nitrogen atom in the indole ring did not coordinate with copper(ii) ion. The characteristic peaks in the FTIR spectra were because of asymmetric and symmetric stretching vibrational modes of COO− group. The absorption peaks appearing at 1578 and 1432 cm−1 are assigned to the asymmetric stretching vibrations and symmetric stretching vibrations of the carboxylate groups, while the characteristic bands of asymmetric and symmetric stretching vibrations of the carboxylate groups in free 3-IAA are 1661 and 1467 cm−1, respectively. This results indicates that oxygen atoms on the carboxylate groups of the 3-IAA are involved in coordination with the copper(ii) ion. The value of Δ(COO−) (υa − υs) is equal to 194 cm−1 near to the values reported by several authors in this chelating mode (Viossat et al. 2003; Donnay et al. 1965; Tian et al. 1995). Compared to the free ligands, the bands observed in the 1580–1389 cm−1 range of the ν(C=N) and ν(C = C) groups were shifted to higher frequencies in the range 1620–1419 cm−1 in the complex. The FTIR spectra of the complex showed peaks at 791 and 712 cm−1. These peaks (out-of-plane CH stretching vibrations) were assigned as coordinated νphen skeletal vibrations. This results indicate that the participation with nitrogen atom of the C=N (phenanthroline ring) groups in coordination of the copper(ıı) ion. The broad band centered about 3600–3400 cm−1 confirms the existence of the water molecule. The very strong broad peaks in the ~ 1070 cm−1 region suggest discrete perchlorate anions in the complex.
The complex showed good solubility in water, methanol, CH3CN and DMSO and were found to be stable in both the solid and solution phases. The electronic absorption spectra of the complex recorded indicated two bands which appeared around 272 and 295 nm corresponded to π–π* and n–π* intra-ligand transitions of coordinated 3-IAA and phen (Kamnev et al. 2001; Ambrosek et al. 2010). The ESI‐MS analysis of the complex shows the presence of [Cu(phen)(3-IAA)]+ ion (m/z 417.05) as the most abundant species, representing the original complex with loss of perchlorate ion, coordinated and uncoordinated water molecules (Fig. S1).
DNA binding activities
The interaction of the complex with CT-DNA has been studied with electronic absorption spectroscopy to research the possible binding modes to CT-DNA and determine the binding constants (Kb) to CT-DNA. The results of the electronic absorption spectra of the complex in the absence and presence of CT-DNA are displayed in Fig. 3a. Upon increasing the ratio of the concentration of CT-DNA to those of the complex, the absorption bands of the complex displayed hypochromism and from the absorption data, a plot was drawn of [DNA]/(εa − εf) values versus DNA concentrations for the titration of DNA to the complex using the following equation (Pyle et al. 1989):
a Electronic absorption spectra of the complex upon addition of CT-DNA, Inset: Plot of [DNA]/(εa − εf) versus [DNA] for the titration of DNA to complex. b Effect of addition of the complex on the emission intensity of the CT- DNA bound EB at different concentrations Inset: Stern–Volmer plot of the fluorescence data
[DNA] is the concentration of DNA in terms of base-pairs, εa is the apparent extinction bound form. Each of data, when fitted to the above equation, gave a straight line with a slope of 1/(εb − εf) and a y-intercept of 1/Kb(εb − εf). Kb was calculated from the ratio of the slope to the intercept. The Kb value of the complex was calculated and given in Table 4. The logarithm of the Kb value (log Kb) for the complex was 3.42. Absorption spectra of the complex showed that as the concentration of CT-DNA increases, the curve presents hypochromism. Hypochromic effect is attributed to the intercalative binding mode and also combined with a red shift is considered as an indication of intercalation of small molecules into DNA because of the stacking of the chromophore pairs (Chen et al. 2011). The calculated Kb value for the complex was found to be of lower magnitude than the classical intercalator EB (ethidium bromide) (Kb = 1.23 × 105 M−1, log Kb = 5.09) (Psomas 2008). The Kb values found for the complex could be explained by the presence of an indole ring that reduces partially the binder flexibility (Pandya et al. 2010). Additionally, the indole ring and 1,10-phenanthroline ring shows high electron density favorable to π-stacking interactions CT-DNA nucleotides (Hossain and Kumar 2009; Li et al. 2010; İnci et al. 2019c). The electronic absorption titration studies reveal that the complex could a moderate intercalative binding mode with CT-DNA base pairs.
EB displacement experiments of the complex have also been performed. The fluorescent emission titration of the complex is displayed in Fig. 3b. An increase in the concentration of the complex results in a decrease in the emission intensity of the EB + CT-DNA solution. The apparent binding constant (Kapp) is the concentration of the compound in the medium that reduces the fluorescence intensity of EB by 50% and it could be calculated using Eq. (3) (Karlin et al. 1987). With the addition of the complex to CT-DNA, there was a considerable decrease in the emission intensity because of the replacement of EB by the complex.
where [Complex] is the concentration of the complex at 50% reduction of fluorescence intensity (KEB = 1.0 × 107 M−1, [EB] = 1.3 μM) (Lee et al. 1993). The quenching plots followed the Stern–Volmer relationship of the form (Lakowicz and Weber 1973):
where I0 and I are the fluorescence intensities of the excited CT-DNA + EB in the absence and presence of the complex. Ksv is the Stern–Volmer quenching constant and [Complex] is the concentration of the quencher. The Stern–Volmer quenching constant (Ksv) and Kapp values are illustrated in Table 5. The log Ksv value was calculated to be 3.72 for the complex. The Ksv value of the complex is lower than that observed for classical intercalator EB (107 M−1) (Waring 1965) which imply that the complex bind with CT-DNA relatively less strongly than classical intercalators. This results could be clarified that cationic metal complexes including aromatic planar ligands show moderate binding strength (Cory et al. 1985). There are likely reasons that explaining the decrease of EB + CT-DNA fluorescence intensity on addition of the complex: (i) The binding between EB and the complex could occur and intensity of EB + CT-DNA decreases (ii) the complex competes EB in binding with CT-DNA and excludes intercalated EB from the CT-DNA double helix, and thus the concentration of bound EB is reduced. (iii) The addition of the complex contributes to formation of a novel non fluorescence complex, EB + CT-DNA + complex, which induces the fluorescence quenching of EB + CT-DNA (Bi et al. 2008).
To clarify the intercalative mode of binding of the complex with CT-DNA, Hoechst 33258 displacement assay was also performed. Hoechst 33258 binds to the minor groove of DNA with specificity for AT-rich sequence, and it is used to explore competitive replacement by groove binders (Khajeh et al. 2018). With increasing concentration of the complex, there was no change in the fluorescent intensity of Hoechst 33258 + CT-DNA. For this reason, we could consider that the complex could not be a minor groove binder.
BSA binding activities
The electronic absorption spectra was used to find out the mechanism of quenching process. Figure 4a indicates the electronic absorption spectra of BSA in the absence and presence of the complex. On the addition of the complex, there were evident changes in the absorption intensity and peak position of BSA, suggesting that the quenching process was static quenching, since the formation of BSA + complex in the static quenching caused some changes in the electronic absorption spectrum of BSA, whereas the collision and energy transfer during the dynamic quenching could not result in changes in absorption spectrum of BSA. Therefore, the fluorescence quenching of BSA is induced by the formation of BSA + complex, which displayed a static quenching mechanism (Zhang et al. 2013).
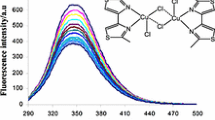
Quenching measurement of BSA fluorescence is a significant method to determine the interactions of the complex with BSA. As known, there were two types of quenching mechanisms of protein fluorescence, static and dynamic quenching (Kang et al. 2004). In static quenching, the complex binds to amino acids residues (Phe, Tyr and Trp) of BSA and a non-fluorescence BSA + complex was formed, resulting in decrease of the fluorescence intensity of BSA (Lakowicz and Weber 1973). The dynamic quenching results from the collision diffusion of the complex and BSAduring the excitation process (Liu et al. 2011). These quenching mechanisms could be distinguished by their different dependence on temperature. In the static quenching, Stern–Volmer quenching constants, Ksv, decrease with increasing temperature because increased temperature result in decreased stability of BSA + complex. On the contrary, in the dynamic quenching, higher temperature results in larger diffusion coefficients, Stern–Volmer quenching constants, Ksv, increase with increasing temperature (Wani et al. 2017). To determine BSA binding behaviors of the complex, quenching experiments were performed at different temperatures (298.2, 310.2 and 318.2 K) and analyzed using classical Stern–Volmer equation (Fig. 4b):
where I0 and I are the fluorescence intensities in the absence and presence of the complex, respectively. [Complex], Kq and τ0 are the total concentration of the complex, the quenching rate constant and the average lifetime of biopolymer in the absence of the quencher (τ0 = 10–8 s) (Lakowicz 1999), respectively. The log Ksv and log Kq is given in Table 5. As shown in Table 5, for the complex, the Stern–Volmer plots were linear and the values of Ksv decreased with increasing temperature. Evidently, the rate constants of BSA quenching initiated by the complex are higher than 2.0 × 1010 M−1 s−1, which is the maximum collision quenching constant of various types of quenchers of biomacromolecules (Ware 1962). These results indicate that the binding of the complex to BSA is responsible for the quenching of its tryptophan fluorescence by the static quenching mechanism. These results also suggested that there was a strong binding between the complex and BSA and BSA + complex might be stored and carried by protein in the body (Jiao et al. 2020).
The modified Stern–Volmer equation was also used to evaluate the association constant, log Ka, for BSA + complexes interaction (Lakowicz 1999):
where Ka is the modified Stern–Volmer association constant, which equals the quotient of an ordinate 1/fa and slope 1/faKa for the accessible fluorophores, and ƒa is the fraction of accessible fluorescence. As shown in Table 5, the decreasing of log Ka with increasing temperature was in accordance with the dependence of log Ksv values on temperature, which is a static type quenching mechanism. In static quenching, the binding constant (KA) and the number of binding sites (n) could be calculated according to the following equation.
where in the present case, KA is the binding constant to a site, and n is the number of binding sites per albumin. The log KA and n for the complex were listed in Table 5. These results displayed that the log KA values decreased as temperature increased for the complexes. This could mean that when temperature is raised hydrogen interactions become weaker, resulting in a decrease of the binding constant values (log KA) between the complex and BSA. Additionally, the value of n was approximately equal to 1, implying that there was one single binding site on BSA for the complex.
The thermodynamic parameters could be calculated on the basis of the van’t Hoff equation:
ΔG is the free energy change, KA binding constant, T is temperature, and R is the gas constant. ΔG, ΔH and ΔS could be determined by the regression curve of ln KA versus 1/T. The values of ΔH, ΔS and ΔG were presented in Table 5. For the complex, the negative values of ΔH and ΔS demonstrated that hydrogen bonds and van der Waals interactions play major roles in the binding process and stabilizes the BSA + complex formation. Furthermore, a negative value of ΔG shows the spontaneity of the interaction. Besides aromaticity and ring shape, hydrogen bonding could play a role in the partitioning of the indole ring (Wimley and White 1996; Persson et al. 1988). Zhang and co-workers conducted a study of the anticancer activity of [Cu(3-IAA)2(phen)] which inhibited the growth of the cells MDA-MB-31 and MCF7 lines at very low concentrations, 4.20 and 5.21 µM, respectively. They performed to analyze the possible interaction between [Cu(3-IAA)2(phen)] and the catalytically active 20S proteasome by molecular docking simulation. The results show that [Cu(3-IAA)2(phen)] forms a few hydrogen bonds with the receptor. It forms a hydrogen bond with sidechain by its indole nitrogen atom and its carboxylic oxygen atom (Zhang et al. 2016). Here, it is significant that 3-IAA having in the structure of the complex containing indole ring plays an active role.
The distance between the acceptor and donor could be determined by FRET (Sklar et al. 1977). Figure 4c displays the overlap between the electronic absorption spectrum of the complex and the fluorescence emission spectrum of BSA. The energy transfer efficiency (E) is obtained using the equation:
where I and I0 are the luminescence intensities of BSA in the presence and absence of the complex (1 μM), r is the distance between the acceptor and the donor and R0 is the critical distance when the transfer efficiency is 50%. It can be given by following equation:
F(λ) is the luminescence intensity of the BSA at the particular wavelength (λ), ε(λ) is the molar absorption coefficient of the complex at the particular wavelength (λ) and its unit is cm−1 M−1 (Wang et al. 2008). E, energy transfer, and overlapping integration values were obtained from Eqs. (10) and (12), respectively. To determine the Förster’s critical distance using Eq. (10), we have used K2 = 2/3, N = 1.336 and ϕ = 0.118 for BSA (Shaikh et al. 2006). The values of r and E % of the BSA + complex system are presented in Table 5. The exact binding location over BSA of the complex might not be detected. However admissible prediction about the binding position could be made from the FRET study which indicates that the distance between BSA and the complex. Found r values is about 0.48 nm for the complex and thus we could recommend that the complex bind near BSA (Cheng et al. 2009; Prasanth et al. 2016). Furthermore, the value of r less than 8 nm indicated that the nonradiative energy transfer and the static quenching interaction between BSA and the complex. The energy transferred to the complex from BSA is 44.71% for the complex (Table 5).
Synchronous fluorescence spectroscopy gives the valuable knowledge about the molecular environment in the surroundings of the tyrosine and tryptophan residues in proteins. As shown in Fig. S2, on addition of the complex, the fluorescence intensity of tyrosine residue at 300 nm decreased of up to 37.94% and the fluorescence intensity of tryptophan residue at 340 nm decreased of up to 72.22%. It might be found that the quenching of fluorescence intensity of Trp residues was stronger than that of Tyr residues, displaying that Trp residues induced more quenching of the intrinsic fluorescence of BSA and as a result the complex was closer to Trp residues compared to Tyr residues (Klajnert and Bryszewska 2002).
Three dimensional fluorescence spectroscopy (3D) analysis and the contour map (2D) could provide much considerable knowledge and detailed conformational changes of BSA. Thus, the 3D spectra of free BSA and BSA + complex system was measured as illustrated in Fig. 5. As given in Fig. 5, “Peak a” (λex = λem) indicates the Rayleigh scattering peak. “Peak b” indicates the spectral behavior of the tryptophan and tyrosine residues. The results indicated that the contour map (2D) of BSA and BSA + the complex was clearly distinct. The fluorescence intensity of “Peak a” and “Peak b” was found to be increased with the addition of the complex. The fluorescence intensity of “Peak a” was found to be increased with the addition of the complex due to the formation of BSA + complex. Thus, the diameter of the BSA increased and resulted in the enhancement of scattering effect (Tian et al. 2010). These experimental results displayed that the binding of the complex with BSA affected conformational and microenvironment of tyrosine and tryptophan residues in BSA.
Radical scavenging activity
To determine whether the complex has radical scavenging activity, we performed DPPH and H2O2 assays. The half minimum inhibitory concentration (IC50) calculated for the complex, asc, trolox and BHT is tabulated (Table 6). The radical scavenging effect of the complex and standards on DPPH radical decreased in the order of the complex > trolox > BHT > ascorbic acid and H2O2 radical decreased in the order of ascorbic acid > BHT > trolox > the complex (Fig. 6). Comparing both methods with each other, it was observed that the complex had better H2O2 scavenging activity. The H2O2 scavenging activity depends on compound ability to lose electron (ET), whereas in the DPPH scavenging activity depends on compound ability to transfer hydrogen atom (HAT) (Wright et al. 2001). Therefore, it could be said that the complex has better ability to induce electron loss according to H2O2 scavenging activity results.
β-Carotene is used as an antioxidant agent. From Table 6, it is evident that the complex demonstrated higher radical svavenging effects on DPPH methods with lower IC50 values when compared with β-carotene (Widowati et al. 2017). Additionally, catechin is also used an antioxidant agent. It is clear that the complex demonstrated higher radical svavenging effects on H2O2 methods with lower IC50 values when compared with catechin (Rahman et al. 2015). An odd electron in copper(ii) ion of the complex decreases the capacity to balance the unpaired electrons and there by arrest the free radicals (İnci et al. 2019a). Shirinzadeh and co-workers found that the indole ring in melatonin indicated as the center responsible for antioxidant properties, because of its high resonance stability and low activation energy barrier in direct reactions against free radicals (Shirinzadeh et al. 2010). Monica and co-workers have synthesized a novel series of indole based tryptophan and tryptamine derivatives and evaluated that their antioxidant activities depend on the substituents on the relative positions of indole ring (Monica Estevao et al. 2010). In another group’s study, a series of substituted 2-arylindoles was synthesized and their antioxidant properties evaluated. They uncovered that fluoro analogues have indicated antioxidant activity comparable to melatonin (Karaaslan et al. 2013).
Conclusions
A novel water soluble copper(ii) complex, [Cu2(phen)2(3-IAA)2(H2O)](ClO4)2·H2O, was synthesized and structurally charactrized. The complex indicated a moderate intercalative CT-DNA binding propensity which was proved by electronic absorption studies, EB and Hoechst 33258 displacement assays. The results also indicated that the complex could bind with BSA, the fluorescence quenching mechanism was static and the binding reaction was spontaneous. The complex indicated free radical scavenging activity when compared to standard antioxidants. The indole ring can be used as a strategy to increase macromolecular interactions (DNA, protein, enzyme, etc.) and radical scvening activity of new complexes. This study also provides a new approach for understanding the nature and site of binding of the complexes with macromolecules.
References
Addision AW, Rao TN, Reedijk J, Rijn JV, Verschoor GC (1984) Synthesis, structure, and spectroscopic properties of copper(ii) compounds containing nitrogen–sulphur donor ligands; the crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]copper(ii) perchlorate. J Chem Soc Dalton Trans 7:1349–1356. https://doi.org/10.1039/DT9840001349
Ambrosek D, Loos PF, Assfeld X, Daniel C (2010) A theoretical study of Ru(ii) polypyridyl DNA intercalators structure and electronic absorption spectroscopy of [Ru(phen)2(dppz)]2+ and [Ru(tap)2(dppz)]2+ complexes intercalated in guanine-cytosine base pairs. J Inorg Biochem 104:893–901. https://doi.org/10.1016/j.jinorgbio.2010.04.002
Bi S, Zhang H, Qiao C, Sun Y, Liu C (2008) Studies of interaction of emodin and DNA in the presence of ethidium bromide by spectroscopic method. Spectrochim Acta A 69:123–129. https://doi.org/10.1016/j.saa.2007.03.017
Brown DW, Graupner PR, Sainsbury M, Shertzer HG (1991) New antioxidants incorporating indole and indoline chromophores. Tetrahedron 25:4383–4408. https://doi.org/10.1016/S0040-4020(01)87108-8
Bruker (2013) SAINT, version V8.34A, Bruker AXS Inc., Madison, Wisconsin, USA
Bruker (2014a) APEX2, version 2014.11-0, Bruker AXS Inc., Madison, Wisconsin, USA
Bruker (2014b) SADABS, version 2014/5, Bruker AXS Inc., Madison, Wisconsin, USA
Chen G-J, Qiao P-Q, Xu G-J, Xu J-Y, Tian J-L, Gu W, Liu X, Yan S-P (2011) Synthesis, DNA binding, photo-induced DNA cleavage, cytotoxicity and apoptosis studies of copper(ii) complexes. J Inorg Biochem 105:119–126. https://doi.org/10.1016/j.jinorgbio.2010.11.008
Cheng XX, Lui Y, Zhou B, Xiao XH, Liu Y (2009) Probing the binding sites and the effect of berbamine on the structure of bovine serum albümin. Spectrochim Acta A Mol Biomol Spectrosc 72:922–928. https://doi.org/10.1016/j.saa.2008.12.003
Cory M, McKee DD, Kagan J, Henry DW, Miller JA (1985) Design, synthesis, and DNA binding properties of bifunctional intercalators. Comparison of polymethylene and diphenyl ether chains connecting phenanthridine. J Am Chem Soc 107:2528–2536. https://doi.org/10.1021/ja00294a054
Dannhardt G, Kiefer W (2001) Cyclooxygenase inhibitors—current status and future prospects. Eur J Med Chem 36(2):109–126. https://doi.org/10.1016/S0223-5234(01)01197-7
Dolomanov OV, Bourhis LJ, Gildea RJ, Howard JAK, Puschmann H (2009) OLEX2: a complete structure solution, refinement and analysis program. J Appl Cryst 42:339–341. https://doi.org/10.1107/S0021889808042726
Donnay G, Donnay JDH, Harding MJC (1965) Crystal data on o-phenanthroline hydrate. Acta Crystallogr 19(4):688–689. https://doi.org/10.1107/S0365110X65004188
Gulcin İ (2006) Antioxidant activity of caffeic acid (3,4-dihydroxycinnamic acid). Toxicology 217(2–3):213–220. https://doi.org/10.1016/j.tox.2005.09.011
Horton DA, Bourne GT, Smythe ML (2003) The combinatorial synthesis of bicyclic privileged structures or privileged substructures. Chem Rev 103:893–930. https://doi.org/10.1021/cr020033s
Hossain M, Kumar GS (2009) DNA binding of benzophenanthridine compounds sanguinarine versus ethidium: comparative binding and thermodynamic profile of intercalation. J Chem Thermodyn 41:764–774. https://doi.org/10.1016/j.jct.2008.12.008
İnci D, Aydin R, Zorlu Y (2016) Affinity of a new copper(ii) complex to DNA/BSA and antioxidant/radical scavenging activities: crystal structure of [Cu(4,7-diphenyl-1,10-phenanthroline)(leucine)(NO3)(H2O)]. J Coord Chem 69:2677–2696. https://doi.org/10.1080/00958972.2016.1213390
İnci D, Aydın R, Vatan Ö, Sevgi T, Yılmaz D, Zorlu Y, Yerli Y, Çoşut B, Demirkan E, Çinkılıç N (2017a) Synthesis and crystal structures of novel copper(II) complexes with glycine and substituted phenanthrolines: reactivity towards DNA/BSA and in vitro cytotoxic and antimicrobial evaluation. J Biol Inorg Chem 22(1):61–85. https://doi.org/10.1007/s00775-016-1408-1
İnci D, Aydin R, Sevgi T, Zorlu Y, Demirkan E (2017b) Synthesis, crystal structure, stability studies, DNA/albumin interactions, and antimicrobial activities of two Cu(ii) complexes with amino acids and 5-nitro-1,10-phenanthroline. J Coord Chem 70:512–543. https://doi.org/10.1080/00958972.2016.1267729
İnci D, Aydın R, Vatan Ö, Zorlu Y, Çinkılıç N (2018a) New binary copper(ii) complexes containing intercalating ligands: DNA interactions, an unusual static quenching mechanism of BSA and cytotoxic activities. J Biomol Struct Dyn 36(15):3878–3901. https://doi.org/10.1080/07391102.2017.1404936
İnci D, Aydın R, Huriyet H, Zorlu Y, Çinkılıç N (2018b) Newly synthesized Cu(ii) pyrazino[2,3-f][1,10]phenanthroline complexes as potential anticancer candidates. Appl Organomet Chem 32:e4309. https://doi.org/10.1002/aoc.4309
İnci D, Aydın R, Huriyet H, Vatan Ö, Zorlu Y, Çoşut B, Çinkılıç N (2019a) Cu(ii) tyrosinate complexes containing methyl substituted phenanthrolines: synthesis, X-ray crystal structures, biomolecular interactions, antioxidant activity, ROS generation and cytotoxicity. Appl Organomet Chem 33(1):e4652. https://doi.org/10.1002/aoc.4652
İnci D, Köseler A, Zeytünlüoğlu A, Aydın R, Zorlu Y (2019b) Interaction of a new copper(ii) complex by bovine serum albumin and dipeptidyl peptidase-IV. J Mol Struct 1177:317–322. https://doi.org/10.1016/j.molstruc.2018.09.086
İnci D, Aydın R, Vatan Ö, Şahin O, Çinkılıç N (2019c) Water-soluble binary and ternary palladium(ıı) complexes containing amino acids and intercalating ligands: synthesis, characterization, biomolecular interactions and cytotoxicities. New J Chem 43(12):4681–4697. https://doi.org/10.1039/C8NJ05934A
Jiao Y-H, Zhang Q, Zou T, Ruan M (2020) Synthesis of a novel p-hydroxycinnamic amide and its interaction with bovine serum albümin. J Mol Struct 1210:127959. https://doi.org/10.1016/j.molstruc.2020.127959
Kai K, Horita J, Wakasa K, Miyagawa H (2007) Three oxidative metabolites of indole-3-acetic acid from Arabidopsis thaliana. Phytochemistry 68:1651–1663. https://doi.org/10.1016/j.phytochem.2007.04.030
Kamnev AA, Shchelochkov AG, Perfiliev YD, Tarantilis PA, Polissiou MG (2001) Spectroscopic investigation of indole-3-acetic acid interaction with iron(iii). J Mol Struct 563–564:565–572. https://doi.org/10.1016/S0022-2860(00)00911-X
Kang J, Liu Y, Xie MX, Li S, Jiang M, Wang YD (2004) Interactions of human serum albumin with chlorogenic acid and ferulic acid. Biochim Biophys Acta 1674(2):205–214. https://doi.org/10.1016/j.bbagen.2004.06.021
Karaaslan C, Kadri H, Coban T, Suzen S, Westwell AD (2013) Synthesis and antioxidant properties of substituted 2-phenyl-1H-indoles. Bioorg Med Chem Lett 23(9):2671–2674. https://doi.org/10.1016/j.bmcl.2013.02.090
Karlin KD, Cohen BI, Hayes JC, Farooq A, Zubieta J (1987) Models for methemocyanin derivatives: structural and spectroscopic comparisons of related azido-coordinated (N3−) mono- and dinuclear copper(ii) complexes. Inorg Chem 26:147–153. https://doi.org/10.1021/ic00248a029
Khajeh MA, Dehghan G, Dastmalchi S, Shaghaghi M, Iranshahi M (2018) Spectroscopic profiling and computational study of the binding of tschimgine: a natural monoterpene derivative, with calf thymus DNA. Spectrochim Acta A Mol Biomol Spectrosc 192:384–392. https://doi.org/10.1016/j.saa.2017.11.042
Kiraz S, İnci D, Aydın R, Vatan Ö, Zorlu Y, Cavaş T (2019) Antiproliferative activity of copper(ii) glutamine complexes with N, N-donor ligands: synthesis, characterization, potentiometric studies and DNA/BSA interactions. J Mol Struct 1194:245–255. https://doi.org/10.1016/j.molstruc.2019.05.086
Klajnert B, Bryszewska M (2002) Fluorescence studies on PAMAM dendrimers interactions with bovine serum albümin. Bioelectrochemistry 55(1):33–35. https://doi.org/10.1016/S1567-5394(01)00170-0
Lakowicz JR (1999) Principles of fluorescence spectroscopy. Plenum Press, New York, NY
Lakowicz JR, Weber G (1973) Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry 12:4161–4170. https://doi.org/10.1021/bi00745a020
Lal S, Snape TJ (2012) 2-arylindoles: a privileged molecular scaffold with potent, broad-ranging pharmacological activity. Curr Med Chem 19:4828–4837. https://doi.org/10.2174/092986712803341449
Lee M, Rhodes AL, Wyatt MD, Forrow S, Hartley JA (1993) GC base sequence recognition by oligoimidazolecarboxamide and C-terminus-modified analogs of distamycin deduced from circular dichroism, proton nuclear magnetic resonance, and methidiumpropylethylenediaminetetraacetate-iron(ii) footprinting studies. Biochemistry 32:4237–4245. https://doi.org/10.1021/bi00067a011
Li X, Wang Q, Qing Y, Lin Y, Zhang Y, Qian X, Cui J (2010) Novel DNA intercalators without basic side chains as efficient antitumor agents: design, synthesis and evaluation of benzo-[c, d]-indol-malononitrile derivatives. Bioorg Med Chem 18:3279–3284. https://doi.org/10.1016/j.bmc.2010.03.017
Liu Y, Liu R, Mou Y, Zhou G (2011) Spectroscopic identification of interactions of formaldehyde with bovine serum albümin. J Biochem Mol Toxic 25:95–100. https://doi.org/10.1002/jbt.20364
Macrae CF, Edgington PR, McCabe P, Pidcock E, Shields GP, Taylor R, Towler M, Van De Streek J (2006) Mercury: visualization and analysis of crystal structures. J Appl Crystallogr 39(3):453–457. https://doi.org/10.1107/S002188980600731X
Mansuri-Torshizi H, Mital R, Srivastava TS, Parekh H, Chitnis MP (1991) Synthesis, characterization, and cytotoxic studies of α-diimine/1,2-diamine platinum(ii) and palladium(ii) complexes of selenite and tellurite and binding of some of these complexes to DNA. J Inorg Biochem 44:239–247. https://doi.org/10.1016/0162-0134(91)84029-9
Mital R, Jain N, Srivatava TS (1989) Synthesis, characterization and cytotoxic studies of diamine and diimine palladium(ii) complexes of diethyldithiocarbamate and binding of these and analogous platinum(ii) complexes with DNA. Inorg Chim Acta 166:135–140. https://doi.org/10.1016/S0020-1693(00)80798-7
Monica Estevao S, Luisa Carvalho C, Ribeiro D, Couto D, Freitas M, Gomes A, Ferreira LM, Fernandes E, Manuel M, Marques B (2010) Antioxidant activity of unexplored indole derivatives: synthesis and screening. Eur J Med Chem 45(11):4869–4878. https://doi.org/10.1016/j.ejmech.2010.07.059
Pandya P, Islam MM, Kumar GS, Jayaram B, Kumar S (2010) DNA minor groove binding of small molecules: experimental and computational evidence. J Chem Sci 122(2):247–257. https://doi.org/10.1007/s12039-010-0029-4
Persson S, Killian JA, Lindblom G (1988) Molecular ordering of interfacially localized tryptophan analogs in ester- and ether-lipid bilayers studied by 2H-NMR. Biophys J 75:1365–1371. https://doi.org/10.1016/S0006-3495(98)74054-8
Prasanth S, Rithesh Raj DR, Vineeshkumar TV, Thomas RK, Sudarsanakumar C (2016) Exploring the interaction of l-cysteine capped CuS nanoparticles with bovine serum albumin (BSA): a spectroscopic study. RSC Adv 6:58288–58295. https://doi.org/10.1039/C6RA03583C
Psomas G (2008) Mononuclear metal complexes with ciprofloxacin: synthesis, characterization and DNA-binding properties. J Inorg Biochem 102:1798–1811. https://doi.org/10.1016/j.jinorgbio.2008.05.012
Pyle AM, Rehmann JP, Meshoyrer R, Kumar CV, Turro NJ, Barton JK (1989) Mixed-ligand complexes of ruthenium(ii): factors governing binding to DNA. J Am Chem Soc 111:3051–3058. https://doi.org/10.1021/ja00190a046
Rahman MM, Islam MB, Biswas M, Alam AHMK (2015) In vitro antioxidant and free radical scavenging activity of different parts of Tabebuia pallida growing in Bangladesh. BMC Res Notes 8:621. https://doi.org/10.1186/s13104-015-1618-6
Rampey RA, LeClere S, Kowalczyk M, Sandberg G, Bartel B (2004) A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol 135:978–988. https://doi.org/10.1104/pp.104.039677
Reichmann ME, Rice SA, Thomas CA, Doty PA (1954) A further examination of the molecular weight and size of desoxypentose nucleic Acid. J Am Chem Soc 76(11):3047–3053. https://doi.org/10.1021/ja01640a067
Ruch RJ, Cheng SJ, Klaunig JE (1989) Prevention of cytotoxicity and inhibition of intercellular communication by antioxidant catechins isolated from Chinese green tea. Carcinogenesis 10(6):1003–1008. https://doi.org/10.1093/carcin/10.6.1003
Ryu CK, Lee JY, Park RE, Ma MY, Nho JH (2007) Synthesis and antifungal activity of 1H-indole-4,7-diones. Bioorg Med Chem Lett 17:127–131. https://doi.org/10.1016/j.bmcl.2006.09.076
Şenel P, İnci D, Aydın R, Huriyet H, Zorlu Y, Çinkılıç N (2019) Methyl substituent effect on one-dimensional copper(ii) coordination polymers containing biologically active ligands: synthesis, characterization, DNA interactions and cytotoxicities. Appl Organomet Chem 33(10):e5122. https://doi.org/10.1002/aoc.5122
Shaikh SMT, Seetharamappa J, Kandagal PB, Ashoka S (2006) Binding of the bioactive component isothipendyl hydrochloride with bovine serum albümin. J Mol Struct 786:46–52. https://doi.org/10.1016/j.molstruc.2005.10.021
Sheldrick GM (2015a) SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr Sect A Found Crystallogr 71:3–8. https://doi.org/10.1107/S2053273314026370
Sheldrick GM (2015b) Crystal structure refinement with SHELXL. Acta Crystallogr Sect C Struct Chem 71:3–8. https://doi.org/10.1107/S2053229614024218
Shimazakia Y, Yajimab T, Talanic M, Yamauchib O (2009) Metal complexes involving indole rings: structures and effects of metal–indole interactions. Coord Chem Rev 253:479–492. https://doi.org/10.1016/j.ccr.2008.04.012
Shirinzadeh H, Eren B, Gurer-Orhan H, Suzen S, Ozden S (2010) Novel indole-based analogs of melatonin: synthesis and in vitro antioxidant activity studies. Molecules 15:2187–2202. https://doi.org/10.3390/molecules15042187
Sklar LA, Hudson BS, Simoni RD (1977) Conjugated polyene fatty acids as fluorescent probes: binding to bovine serum albümin. Biochemistry 16:5100–5108. https://doi.org/10.1021/bi00642a024
Skurai H, Tamura H, Okatani K (1995) Mechanism for a new antitumor vanadium complex: hydroxyl radical-dependent DNA cleavage by 1,10-phenanthroline vanadyl complex in the presence of hydrogen peroxide. Biochem Biophys Res Commun 206:133–137. https://doi.org/10.1006/bbrc.1995.1019
Spek AL (2009) Structure validation in chemical crystallography. Acta Crystallogr Sect D Biol Crystallogr 65(2):148–155. https://doi.org/10.1107/S090744490804362X
Strader LC, Bartel B (2011) Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant 4:477–486. https://doi.org/10.1093/mp/ssr006
Suzen S, Buyukbingol E (2000) Anti-cancer activity studies of indolalthiohydantoin (PIT) on certain cancer cell lines. Il Farmaco 55(4):246–248. https://doi.org/10.1016/S0014-827X(00)00028-8
Suzen S, Bozkaya P, Coban T, Nebioglu D (2006) Investigation of the in vitro antioxidant behaviour of some 2-phenylindole derivatives: discussion on possible antioxidant mechanisms and comparison with melatonin. J Enzyme Inhib Med Chem 21:405–411. https://doi.org/10.1080/14756360500381210
Tian Y-P, Duan C-Y, Xu X-X, You X-Z (1995) Screw-chain structure of 1,10-phenanthroline hydrate, C12H8N2 H2O. Acta Crystallogr C51:2309–2312. https://doi.org/10.1107/S010827019500549X
Tian FF, Jiang FL, Han XL, Xiang C, Ge YS, Li JH, Zhang Y, Li R, Ding XL, Liu Y (2010) Synthesis of a novel hydrazone derivative and biophysical studies of its interactions with bovine serum albumin by spectroscopic, electrochemical, and molecular docking methods. J Phys Chem B 114:14842–14853. https://doi.org/10.1021/jp105766n
Tiwari RK, Singh D, Singh J, Yadav V, Pathak AK, Dabur R, Chhillar AK, Singh R, Sharma GL, Chandra R, Verma AK (2006) Synthesis and antibacterial activity of substituted 1,2,3,4-tetrahydropyrazino [1,2-a] indoles. Bioorg Med Chem Lett 16:413–416. https://doi.org/10.1016/j.bmcl.2005.09.066
Viossat V, Lemoine P, Dayan E, Nguyen-Huy D, Viossat B (2003) Synthesis, crystal structure and IR spectroscopy of MnII(2-IC)2(NC)(DMSO) and [MnII(2-IC)2(phen)(H2O)]·DMA; (2-HIC, indole-2-carboxylic acid; phen, 1,10-phenanthroline; NC, 2,9-dimethyl-1,10-phenanthroline; DMSO, dimethyl sulfoxide; DMA, dimethyl acetamide); catalysts for the disproportionation of hydrogen peroxide. Polyhedron 22:1461–1470. https://doi.org/10.1016/S0277-5387(03)00126-8
Wang F, Huang W, Dai ZX (2008) Spectroscopic investigation of the interaction between riboflavin and bovine serum albümin. J Mol Struct 875:509–514. https://doi.org/10.1016/j.molstruc.2007.05.034
Wani TA, AlRabiah H, Bakheit AH, Kalam MA, Zargar S (2017) Study of binding interaction of rivaroxaban with bovine serum albumin using multi-spectroscopic and molecular docking approach. Chem Cent J 11:134–142. https://doi.org/10.1186/s13065-017-0366-1
Ware WR (1962) Oxygen quenching of fluorescence in solution: an experimental study of the diffusion process. J Phys Chem 66(3):455–458. https://doi.org/10.1021/j100809a020
Waring MJ (1965) Complex formation between ethidium bromide and nucleic acids. J Mol Biol 13:269–282. https://doi.org/10.1016/S0022-2836(65)80096-1
Widowati W, Rani AP, Hamzah RA, Arumwardana S, Afifah E, Kusuma HSW, Rihibiha DD, Nufus H, Amalia A (2017) Antioxidant and antiaging assays of Hibiscus sabdariffa extract and its compounds. Nat Prod Sci 23(3):192–200. https://doi.org/10.20307/nps.2017.23.3.192
Williams JD, Drach JC, Townsend LB (2005) Synthesis and antiviral activity of some 2-substituted 3-formyl-and 3-cyano-5,6-dichloroindole nucleosides. Nucleosides Nucleos Nucleot Nucl Acids 24:1613–1626. https://doi.org/10.1080/15257770500265836
Wimley WC, White SH (1996) Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol 3:842–848. https://doi.org/10.1038/nsb1096-842
Wright JS, Johnson ER, Di Labio GA (2001) Predicting the activity of phenolic antioxidants: theoretical method, analysis of substituent effects, and application to major families of antioxidants. J Am Chem Soc 123(6):1173–1183. https://doi.org/10.1021/ja002455u
Zhang J, Yan Q, Liu J, Lu X, Zhu Y, Wang J, Wang S (2013) Study of the interaction between 5-sulfosalicylic acid and bovine serum albumin by fluorescence spectroscopy. J Lumin 134:747–753. https://doi.org/10.1016/j.jlumin.2012.06.053
Zhang Z, Wang H, Wang Q, Yan M, Wang H, Bi C, Sun S, Fan Y (2016) Anticancer activity and computational modeling of ternary copper(ii) complexes with 3-indolecarboxylic acid and 1,10-phenanthroline. Int J Oncol 49:691–699. https://doi.org/10.3892/ijo.2016.3542
Acknowledgements
This research was financed by Bursa Uludağ University Scientist Training and Development Project (Project Number B.İ.Y.G.P-2018/1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts to declare. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
İnci, D., Aydın, R. & Zorlu, Y. Cu(ii) complex with auxin (3-indoleacetic acid) and an aromatic planar ligand: synthesis, crystal structure, biomolecular interactions and radical scavenging activity. Eur Biophys J 50, 771–785 (2021). https://doi.org/10.1007/s00249-021-01525-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-021-01525-5