Abstract
Congenital heart disease (CHD) is the most common birth defect and is the most prevalent non-infectious cause of infant death. Aggregating evidence demonstrates that genetic defects are involved in the pathogenesis of CHD. However, CHD is genetically heterogeneous and the genetic determinants for CHD in an overwhelming majority of patients remain unknown. In this study, the coding regions and splice junctions of the NKX2.6 gene, which encodes a homeodomain transcription factor crucial for cardiovascular development, were sequenced in 210 unrelated CHD patients. As a result, a novel heterozygous NKX2.6 mutation, p.K152Q, was identified in an index patient with ventricular septal defect (VSD). Genetic analysis of the proband’s available family members showed that the mutation cosegregated with VSD transmitted as an autosomal dominant trait with complete penetrance. The missense mutation was absent in 400 control chromosomes and the altered amino acid was completely conserved evolutionarily across species. Due to unknown transcriptional targets of NKX2.6, the functional characteristics of the identified mutation at transcriptional activity were analyzed by using NKX2.5 as a surrogate. Alignment between human NKX2.6 and NKX2.5 proteins displayed that K152Q-mutant NKX2.6 was equivalent to K158Q-mutant NKX2.5, and introduction of K158Q into NKX2.5 significantly reduced its transcriptional activating function when compared with its wild-type counterpart. This study firstly links NKX2.6 loss-of-function mutation with increased susceptibility to isolated VSD, providing novel insight into the molecular mechanism underpinning VSD and contributing to the development of new preventive and therapeutic strategies for this common form of CHD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Congenital heart disease (CHD) represents the most common form of human birth defect, affecting nearly 1 % of all live births worldwide, and remains a leading cause of morbidity and mortality in childhood, accounting for approximately 10 % of all infant deaths [15, 17]. Congenital cardiovascular abnormalities are clinically classified into at least 21 different categories with specific anatomic lesions, including ventricular septal defect (VSD), atrial septal defect, tetralogy of Fallot, patent ductus arteriosus, aortic stenosis, pulmonary atresia, endocardial cushion defect, double outlet right ventricle, anomalous coronary arteries, abnormal pulmonary venous return, aorticopulmonary window, truncus arteriosus, transposition of great arteries, coronary artery anomaly, and hypoplastic left heart syndrome, of which VSD is the most prevalent type of CHD in children while in adults atrial septal defect is the most frequently seen developmental malformation [17, 63]. Various kinds of CHDs may occur singly or in combination with each other, resulting in degraded quality of life, limited exercise capacity, delayed brain development, pulmonary hypertension, anomalous lung function, impaired muscle function, autonomic dysfunction, aortic aneurysm or dissection, heart failure, infective endocarditis, thromboembolism, arrhythmias, and even cardiac death [1, 3, 5, 12–14, 16, 21, 27, 32–35, 37, 39–41, 43, 49, 65]. Although great advances made in surgical and medical treatment of children born with CHD have led to a growing number of adult survivors, unfortunately, the incidence of late complications and mortality rate are significantly increased in the survivals [19, 35, 48, 50, 51]. Hence, CHD has imposed a vast economic burden on patients and health care systems, and the socioeconomic burden is projected to increase in the future with increasing CHD adults [52]. Despite the high prevalence and pronounced clinical significance, the molecular mechanism underlying CHD remains poorly understood.
In vertebrates, the heart is the first functional organ that develops during embryogenesis. Cardiac morphogenesis is a complex dynamic biological process that requires the accurate spatial and temporal cooperation of cardiac cell commitment, differentiation, proliferation, and migration, and both environmental and genetic risk factors may disturb this process of cardiogenesis, leading to a large spectrum of CHDs [4, 11, 15, 18, 29, 38]. Recently, a growing body of evidence highlights the genetic basis for CHD, and a great number of mutations in more than 60 genes have been implicated in the pathogenesis of CHD [2, 4, 8, 10, 15, 24, 25, 28, 42, 44, 53–56, 62, 64]. Among these well-established CHD-associated genes, most encode cardiac transcription factors and show autosomal dominant inheritance with incomplete disease penetrance, and the cardiac transcription factor genes NKX2.5 and GATA4 are most frequently associated with non-syndromic CHD [9, 23, 31, 57, 59, 60]. Nevertheless, mutations in these genes only explain the CHDs in a minority of cases and in an overwhelming majority of patients, the genetic components responsible for CHDs are still to be identified.
As a member of the cardiac NK-2 family of transcription factors, which are the vertebrate orthologs of the Drosophila tinman gene, NKX2.5 is crucial to normal cardiac development across vertebrate species, and mutations or altered expression of NKX2.5 may lead to abnormal heart formation in vertebrates [6]. In humans, a long list of mutations in NKX2.5 have been causally linked to a great variety of congenital cardiovascular deformities, including atrial septal defect, ventricular septal defect, and tetralogy of Fallot, as well as atrial fibrillation [9, 22, 23, 31, 58, 61]. NKX2.6 is another member of the NK-2 family of transcription factors and its temporal and spatial expression patterns and functional roles are similar to those of NKX2.5 during cardiovascular development [6]. In mice with targeted deletion of Nkx2.6, it has been revealed that the redundant activity of Nkx2.5 is required for proper cardiac morphogenesis [20]. Therefore, it is warranted to screen NKX2.6 as a preferred candidate gene for human CHD.
Materials and Methods
Ethics
This study was conducted in accordance with the ethical principles of the revised Declaration of Helsinki (Somerset West, Republic of South Africa, 1996). The research protocol was reviewed and approved by the local institutional ethics committee of Tongji Hospital, Tongji University (the ethical approval number for cases and controls: LL(H)-09-07; the date for the approval: July 27, 2009) and written informed consent was obtained from all participants or their guardians prior to commencement of investigation.
Study Subjects
A total of 210 unrelated patients affected with non-syndromic CHD were recruited. The available relatives of the index patient harboring an identified Nkx2.6 mutation were also included. All patients underwent a comprehensive clinical evaluation, including individual and familial histories, medical records, complete physical examination, 12-lead electrocardiogram, and two-dimensional transthoracic echocardiography with color flow Doppler. Cardiac catheterization, angiography, chest X-ray, and cardiac magnetic resonance imaging were performed only if there was a strong clinical indication. Medical records were also reviewed in the case of deceased or unavailable relatives. CHD was confirmed by imaging and/or direct view during cardiac surgery. The patients with known chromosomal abnormalities or syndromic cardiovascular defects, such as Down syndrome, Turner syndrome, Marfan syndrome, Di George syndrome, and Holt–Oram syndrome, were excluded from the study. The controls comprised 200 non-CHD individuals from the same geographic area, who were matched to the CHD patients in race and gender. All the study subjects were of Chinese Han ethnicity. The ethnic origin of the participants was ascertained by a combination of self-reported ethnicity and a personal questionnaire about the birthplace, language, religion, and ancestry. Approximately 0.5–2 ml of peripheral venous blood sample was collected from each study participant. The clinical studies were performed with investigators blinded to the results of genotypes.
Genetic Screening of NKX2.6
Genomic DNA was extracted from each participant’s peripheral blood leukocytes with Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). The coding exons and flanking exon–intron boundaries of the NKX2.6 gene were sequenced in 210 unrelated patients with CHD. The available relatives of the index patient carrying an identified NKX2.6 mutation and 200 unrelated control individuals were subsequently genotyped for NKX2.6. The referential genomic DNA sequence of NKX2.6 was derived from GenBank (accession no. NG_030636), a gene sequence database at the National Center for Biotechnical Information (NCBI; http://www.ncbi.nlm.nih.gov/). With the aid of the online Primer 3 program (http://frodo.wi.mit.edu/), the primer pairs used to amplify the coding regions and splice junction sites of NKX2.6 by polymerase chain reaction (PCR) are designed as shown in Table 1. The PCR was carried out using HotStar Taq DNA Polymerase (Qiagen GmbH, Hilden, Germany) on a Veriti Thermal Cycler (Applied Biosystems, Foster, CA, USA), with standard conditions and concentrations of reagents. Both strands of each PCR product were sequenced with a BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) under an ABI PRISM 3130 XL DNA Analyzer (Applied Biosystems). The sequencing primers were the same as those designed for specific region amplifications. The DNA sequences were analyzed with the DNA Sequencing Analysis Software v5.1 (Applied Biosystems). The identified NKX2.6 variant was confirmed by resequencing an independent PCR-generated amplicon from the same case. Additionally, an identified sequence variation was queried in the single nucleotide polymorphism (SNP) database at NCBI (http://www.ncbi.nlm.nih.gov/), the human gene mutation (HGM) database (http://www.hgmd.org/), and the 1000 Genome Project (1000 GP) database (http://www.1000genomes.org/) to confirm its novelty.
Alignment of NKX2.6 Amino Acid Sequences Among Various Species
Conservation of the amino acid altered by missense mutation was estimated by aligning human NKX2.6 to chimpanzee, monkey, dog, cattle, mouse, rat, fowl, zebrafish, and frog NKX2.6 using the HomoloGene and Show Multiple Alignment links on the NCBI’s website (http://www.ncbi.nlm.nih.gov/homologene).
Prediction of the Causative Potential of a Novel NKX2.6 Sequence Variation
The disease-causing potential of a novel NKX2.6 sequence variation was predicted by MutationTaster (an online program at http://www.mutationtaster.org), which automatically gave a probability for the variation to be either a deleterious mutation or a benign polymorphism. Notably, the p value used here is the probability of the correct prediction rather than the probability of error as used in t test statistics (i.e., a value close to 1 indicates high accuracy of the prediction). Additionally, another online program PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) was also used to evaluate the possible pathogenic effect of an amino acid substitution on the structure and function of NKX2.6 protein.
Plasmids and Site-Directed Mutagenesis
The recombinant expression plasmid NKX2.5-pEFSA and the ANF–luciferase (ANF–Luc) reporter vector, which contains the 2600-bp 5′-flanking region of the ANF gene, were kindly provided by Dr. Ichiro Shiojima from Chiba University School of Medicine, Japan. Owing to unknown downstream genes of NKX2.6, NKX2.5 was used as a surrogate in transcriptional analysis to assess functional consequences of the NKX2.6 homeodomain substitution as previously described [20]. Alignment between human NKX2.6 and NKX2.5 proteins displayed that K152Q-mutant NKX2.6 was equivalent to K158Q-mutant NKX2.5 (as shown in Fig. 1). The c.472A>C transition, which was predicted to generate the p.K158Q mutation, was introduced into the wild-type NKX2.5 using a QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) with a complementary pair of primers. The mutant was sequenced to confirm the desired mutation and to exclude any other sequence variations.
Reporter Gene Analysis of Mutant NKX2.5
COS-7 cells were routinely maintained in Dulbecco’s modified Eagle’s medium supplemented with 10 % fetal calf serum and seeded in 12-well plates (1 × 105) before transfection. The ANF(-2600)–Luc reporter construct and an internal control reporter plasmid pGL4.75 (hRluc/CMV, Promega) were used in transient transfection assay to assess the transcriptional activity of the NKX2.5 mutant. Twenty-four hours after plating, the COS-7 cells at about 70 % confluence were transfected with 0.4 μg of wild-type or mutant NKX2.5-pEFSA, 1.0 μg of ANF(-2600)–Luc reporter construct, and 0.04 μg of pGL4.75 using PolyFect Transfection Reagent (Qiagen). For co-transfection experiments, 0.2 μg of wild-type NKX2.5-pEFSA together with 0.2 μg of mutant NKX2.5-pEFSA or 0.2 μg of empty vector were used in the presence of 0.4 μg of ANF(-2600)–Luc and 0.04 μg of pGL4.75. Firefly luciferase and Renilla luciferase activities were measured with the Dual-Glo luciferase assay system (Promega) 48 h after transfection. The activity of the ANF promoter was presented as fold activation of Firefly luciferase relative to Renilla luciferase. At least three independent experiments were conducted in triplicate for wild-type and mutant NKX2.5.
Statistical Analysis
Data are expressed as means ± standard deviations. Continuous variables were tested for normal distribution and Student’s unpaired t test was used to compare the numeric variables between two groups. Comparison of the categorical variables between two groups was completed using Pearson’s χ 2 test or Fisher’s exact test when appropriate. A two-tailed p value less than 0.05 was considered to indicate statistical difference.
Results
Clinical Characteristics of the Study Subjects
A cohort of 210 unrelated patients with CHD was clinically evaluated in contrast to a total of 200 unrelated non-CHD control individuals. All the study subjects had no established environmental risk factors for CHD, such as maternal illness and drug use in the first trimester of pregnancy, parental smoking, and long-term exposure to toxicants and ionizing radiation. The baseline clinical characteristics of the 210 unrelated CHD patients are summarized in Table 2.
Identification of a Novel Nkx2.6 Mutation
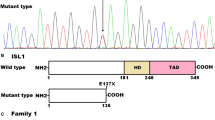
By sequence analysis of the coding exons and flanking introns of NKX2.6, a heterozygous sequence variation was identified in 1 of 210 unrelated CHD patients, with a mutational prevalence of about 0.48 %. Specifically, a substitution of cytosine for adenine in the first nucleotide of codon 152 (c.454A>C), predicting the transition of lysine into glutamine at amino acid position 152 (p.K152Q), was identified in an index patient with VSD. The sequence electropherograms showing the identified heterozygous NKX2.6 variation in contrast to its corresponding control sequence are shown in Fig. 2. The schematic diagrams of NKX2.6 and NKX2.5 proteins showing the structural domains and location of the mutation identified in this study are presented in Fig. 3. The variation was neither observed in 400 control chromosomes nor found in the SNP, HGM, and 1000 GP databases, which were consulted again on August 9, 2014, indicating a novel mutation.
Sequence electropherograms showing the NKX2.6 mutation in contrast to its corresponding control. The arrow indicates the heterozygous nucleotides of A/C in the proband from family 1 (mutant); or the homozygous nucleotides of A/A in the corresponding control individual (wild-type). The rectangle means the nucleotides constituting a codon of NKX2.6
Schematic diagrams of human NKX2.6 and NKX2.5 protein structures with the mutation responsible for congenital ventricular defect shown. The mutation associated with congenital ventricular defect is shown above the structural domains. NH 2 amino-terminus, TN tinman domain, HD homeodomain, NK nucleotide kinase domain, and COOH carboxyl-terminus
Genetic screening of the proband’s relatives showed that the mutation was present in all the affected living family members, but absent in unaffected family members who were examined. Analysis of the pedigree demonstrated that the mutation cosegregated with VSD transmitted in an autosomal dominant pattern and with complete penetrance. The pedigree structure of the family is shown in Fig. 4. The phenotypic characteristics and status of NKX2.6 mutation of the affected family members are listed in Table 3.
Pedigree structure of the family with congenital ventricular defect. Family is designated as family 1. Family members are identified by generations and numbers. Square indicates male family member, circle female member, closed symbol affected member, open symbol unaffected member, symbol with a slash the deceased member, arrow proband, “+” carrier of the heterozygous mutation, and “−” non-carrier
Multiple Alignments of NKX2.6 Protein Sequences Among Various Species
As shown in Fig. 5, a cross-species alignment of multiple NKX2.6 protein sequences displayed that the affected amino acid of p.K152 was completely conserved evolutionarily, suggesting that the amino acid is functionally important.
Disease-Causing Potential of the Identified NKX2.6 Sequence Variation
The NKX2.6 sequence variation of c.454A>C was predicted to be pathogenic, with a p value of 0.956766. The amino acid substitution of p.K152Q was also predicted to be probably damaging by another software PolyPhen-2, with a score of 0.964 (sensitivity: 0.62; specificity: 0.92), implying that mutated NKX2.6 contributes to the development of VSD in these mutation carriers.
Diminished Transcriptional Activity of the Mutant NKX2.5
As shown in Fig. 6, the wild-type NKX2.5 and the K158Q-mutant NKX2.5 activated the ANF promoter by ~11- and ~3-fold, respectively. When wild-type NKX2.5 was co-expressed with the same amount of K158Q-mutant NKX2.5, the induced activation of the ANF promoter was ~fivefold. These results reveal that the K158Q-mutant NKX2.5 is associated with significantly diminished transactivational activity compared with its wild-type counterpart.
Functional impairment of K158Q-mutant NKX2.5. Activation of the ANF–Luc reporter in cultured COS-7 cells by wild-type NKX2.5 (WT) or mutant (K158Q), alone or in combination, showed significantly decreased transcriptional activity by mutant protein. Experiments were performed in triplicate and mean and standard deviations are given. **p < 0.001 and *p < 0.005, respectively, when compared with wild-type NKX2.5
Discussion
In the present study, a novel heterozygous mutation in NKX2.6, p.K152Q, was detected in a family with VSD. The mutant allele was present in all affected family members available but absent in unaffected relatives examined and 400 referral chromosomes from an ethnically matched control population. A cross-species alignment of multiple NKX2.6 protein sequences showed that the altered amino acid was completely conserved evolutionarily. The p.K152Q variation was predicted to be pathogenic by both MutationTaster and PolyPhen-2, and the functional analysis indicated that the homeobox substitution resulted in functional impairment. Therefore, it is very likely that mutant NKX2.6 predisposes these mutation carriers to VSD. To our knowledge, this is the first to associate NKX2.6 loss-of-function mutation with enhanced susceptibility to isolated VSD in humans.
The official name of NKX2.6 is NK-2 transcription factor-related locus 6. This term refers to the grouping of homeodomain-containing proteins into 20 different classes, one of which is the NK-2 class. The abbreviation NK refers to Nirenberg and Kim, who completed a search for Drosophila genes containing homeodomain-encoding motifs. The “X” in NKX was added to designate vertebrate members of the family. Among the NK-2 genes expressed in vertebrates, a subset, referred to as the “cardiac group”, includes Nkx2.3, Nkx2.5, Nkx2.6, Nkx2.7, Nkx2.8, and Nkx2.10 [6]. The human NKX2.6 gene maps to chromosome 8p21.2 and consists of two exons encoding a protein of 301 amino acids, which is expressed in early embryonic heart progenitor cells, playing an important role in proper cardiovascular development [7, 46]. The NKX2.6 protein contains an evolutionarily conserved homeodomain that recognizes and binds to a consensus DNA motif, AAGTG. The homeodomain is centrally located at amino acid positions 132–191 and is predominantly involved in target DNA binding, nuclear translocation as well as interaction with other transcription factors [20, 36]. The NKX2.6 mutation of p.K152Q identified in this study is located in homeodomain, and thus may be anticipated to exert influence on the transcriptional activity of NKX2.6 by interfering with its nuclear distribution or DNA-binding ability.
In order to determine the functional consequence of the K152Q homeodomain substitution in NKX2.6, NKX2.5 was chosen as a surrogate mainly based on the following reasons. Firstly, neither a target gene nor a binding site recognition sequence for NKX2.6 has been verified. Secondly, both NKX2.5 and NKX2.6 belong to members of the NK family, and they share highly conserved structural motifs, especially in the homeodomain. Thirdly, biochemical analyses of NKX2.5 have been well performed repeatedly. Finally, there are in vivo studies substantiating that NKX2.5 may compensate for the lack of NKX2.6 during embryogenesis, and therefore implying that some DNA binding and transcriptional regulatory activities are shared [7, 20, 46]. Consequently, functional assays of K158Q-mutant NKX2.5 instead of K152Q-mutant NKX2.6 revealed that the homeodomain substitution significantly reduced transcriptional activity on a target gene, ANF. These findings suggest that haploinsufficiency or dominant-negative effect caused by NKX2.6 mutation is potentially an alternative pathological mechanism of VSD. Nevertheless, the direct molecular mechanism by which mutant NKX2.6 predisposes to VSD needs to be explored in order to overcome the limitation to the current indirect functional analysis.
Previously NKX2.6 mutations were associated with truncus arteriosus, another type of CHD. Heathcote et al. made a genetic analysis of a large consanguineous family with truncus arteriosus, and identified an F151L mutation in the homeodomain of NKX2.6. Due to unknown transcriptional targets of NKX2.6, the functional characteristics of the F151L-mutant NKX2.6 were investigated by using F157L-mutant NKX2.5 as a surrogate, and introduction of F157L into human NKX2.5 substantially reduced its transcription activating function, its synergism with partners at the ANF promoter and its specific DNA-binding affinity [20]. Ta-Shma et al. performed a whole exome analysis of a three-patient family with complex CHD, including the proband with VSD and aortic arch hypoplasia, his dead brother with truncus arteriosus, stenosis of right pulmonary artery and right aortic arc, and his sister with truncus arteriosus and right aortic arc, and discovered a homozygous insertion mutation of c.453_454insT in NKX2.6, which was predicted to generate a truncated protein with only N-terminal 152 amino acids, namely p.K152X mutation. However, functional analysis of this truncation mutation was not made [47]. Khetyar et al. screened the coding regions and splice signal sequences of the NKX2.6 gene in 12 unrelated CHD patients, but found no non-synonymous mutation [26]. The discrepancy in the mutational prevalence of these reports including the present study may be partially explained by different sample size and ethnicity as well as great genetic heterogeneity. Taken together, these findings support that NKX2.6 plays a key role in human heart development.
Association of genetically defective Nkx2.6 with enhanced susceptibility to CHD has been revealed in experimental animals. In mice, targeted deletion of Nkx2.5 led to abnormal heart morphogenesis and embryonic lethality; although a linear heart tube formed normally in mutant embryos, the looping morphogenesis was not initiated [30]. In contrast, Nkx2.6-null mice were viable with no obvious abnormalities in the heart [45]. Nevertheless, in mutant embryos homozygous for Nkx2.6, Nkx2.5 mRNA expression expanded to the pharyngeal pouch endoderm, suggesting functional compensation for loss of Nkx2.6 in the pharyngeal pouches of these embryos [45]. Furthermore, overlapping functions for Nkx2.5 and Nkx2.6 have been demonstrated in the Nkx2.5 and Nkx2.6 double-knockout mouse embryos, which lacked pharyngeal pouches and had less advanced atrium, mainly due to enhanced apoptosis and reduced proliferation of pharyngeal endodermal cells [46]. Given the high degree of homology between Nkx2.5 and Nkx2.6 proteins over the homeodomain and their overlapping expression patterns, these data indicate that Nkx2.5 acts at a subset of Nkx2.6 target genes [20].
In conclusion, this study firstly associates functionally compromised NKX2.6 with increased vulnerability to isolated VSD, highlighting the essential role of NKX2.6 in human cardiovascular development, and implying the potential implications for genetic counseling and clinical care of the patients with VSD.
References
Agha H, El Heinady F, El Falaky M, Sobih A (2014) Pulmonary functions before and after pediatric cardiac surgery. Pediatr Cardiol 35:542–549
Al Turki S, Manickaraj AK, Mercer CL, Gerety SS, Hitz MP, Lindsay S, D’Alessandro LC, Swaminathan GJ, Bentham J, Arndt AK, Low J, Breckpot J, Gewillig M, Thienpont B, Abdul-Khaliq H, Harnack C, Hoff K, Kramer HH, Schubert S, Siebert R, Toka O, Cosgrove C, Watkins H, Lucassen AM, O’Kelly IM, Salmon AP, Bu’lock FA, Granados-Riveron J, Setchfield K, Thornborough C, Brook JD, Mulder B, Klaassen S, Bhattacharya S, Devriendt K, Fitzpatrick DF; UK10 K Consortium, Wilson DI, Mital S, Hurles ME (2014) Rare variants in NR2F2 cause congenital heart defects in humans. Am J Hum Genet 94:574–585
Alonso-Gonzalez R, Borgia F, Diller GP, Inuzuka R, Kempny A, Martinez-Naharro A, Tutarel O, Marino P, Wustmann K, Charalambides M, Silva M, Swan L, Dimopoulos K, Gatzoulis MA (2013) Abnormal lung function in adults with congenital heart disease: prevalence, relation to cardiac anatomy, and association with survival. Circulation 127:882–890
Andersen TA, Troelsen Kde L, Larsen LA (2014) Of mice and men: molecular genetics of congenital heart disease. Cell Mol Life Sci 71:1327–1352
Bang JS, Jo S, Kim GB, Kwon BS, Bae EJ, Noh CI, Choi JY (2013) The mental health and quality of life of adult patients with congenital heart disease. Int J Cardiol 170:49–53
Bartlett H, Veenstra GJ, Weeks DL (2010) Examining the cardiac NK-2 genes in early heart development. Pediatr Cardiol 31:335–341
Biben C, Hatzistavrou T, Harvey RP (1998) Expression of NK-2 class homeobox gene Nk2–6 in foregut endoderm and heart. Mech Dev 73:125–127
Chang SW, Mislankar M, Misra C, Huang N, Dajusta DG, Harrison SM, McBride KL, Baker LA, Garg V (2013) Genetic abnormalities in FOXP1 are associated with congenital heart defects. Hum Mutat 34:1226–1230
Costa MW, Guo G, Wolstein O, Vale M, Castro ML, Wang L, Otway R, Riek P, Cochrane N, Furtado M, Semsarian C, Weintraub RG, Yeoh T, Hayward C, Keogh A, Macdonald P, Feneley M, Graham RM, Seidman JG, Seidman CE, Rosenthal N, Fatkin D, Harvey RP (2013) Functional characterization of a novel mutation in NKX2-5 associated with congenital heart disease and adult-onset cardiomyopathy. Circ Cardiovasc Genet 6:238–247
Cowan J, Tariq M, Ware SM (2014) Genetic and functional analyses of ZIC3 variants in congenital heart disease. Hum Mutat 35:66–75
Cresci M, Foffa I, Ait-Ali L, Pulignani S, Kemeny A, Gianicolo EA, Andreassi MG (2013) Maternal environmental exposure, infant GSTP1 polymorphism, and risk of isolated congenital heart disease. Pediatr Cardiol 34:281–285
Demir M (2013) The relationship between atrial septal aneurysm and autonomic dysfunction. Exp Clin Cardiol 18:104–106
Dimitropoulos A, McQuillen PS, Sethi V, Moosa A, Chau V, Xu D, Brant R, Azakie A, Campbell A, Barkovich AJ, Poskitt KJ, Miller SP (2013) Brain injury and development in newborns with critical congenital heart disease. Neurology 81:241–248
Dimopoulos K, Wort SJ, Gatzoulis MA (2014) Pulmonary hypertension related to congenital heart disease: a call for action. Eur Heart J 35:691–700
Fahed AC, Gelb BD, Seidman JG, Seidman CE (2013) Genetics of congenital heart disease: the glass half empty. Circ Res 112:707–720
Garcia Guerra G, Joffe AR, Robertson CM, Atallah J, Alton G, Sauve RS, Dinu IA, Ross DB, Rebeyka IM, Western Canadian Complex Pediatric Therapies Follow-up Group (2014) Health-related quality of life experienced by children with chromosomal abnormalities and congenital heart defects. Pediatr Cardiol 35:536–541
Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Judd SE, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Mackey RH, Magid DJ, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER 3rd, Moy CS, Mussolino ME, Neumar RW, Nichol G, Pandey DK, Paynter NP, Reeves MJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Wong ND, Woo D, Turner MB; American Heart Association Statistics Committee and Stroke Statistics Subcommittee (2014) Heart disease and stroke statistics—2014 update: a report from the American Heart Association. Circulation 129:e28–e292
Gorini F, Chiappa E, Gargani L, Picano E (2014) Potential effects of environmental chemical contamination in congenital heart disease. Pediatr Cardiol 35:559–568
Greutmann M, Tobler D (2012) Changing epidemiology and mortality in adult congenital heart disease: looking into the future. Future Cardiol 8:171–177
Heathcote K, Braybrook C, Abushaban L, Guy M, Khetyar ME, Patton MA, Carter ND, Scambler PJ, Syrris P (2005) Common arterial trunk associated with a homeodomain mutation of NKX2.6. Hum Mol Genet 14:585–593
Hoffmann A, Chockalingam P, Balint OH, Dadashev A, Dimopoulos K, Engel R, Schmid M, Schwerzmann M, Gatzoulis MA, Mulder B, Oechslin E (2010) Cerebrovascular accidents in adult patients with congenital heart disease. Heart 96:1223–1226
Huang RT, Xue S, Xu YJ, Zhou M, Yang YQ (2013) A novel NKX2.5 loss-of-function mutation responsible for familial atrial fibrillation. Int J Mol Med 31:1119–1126
Huang W, Meng H, Qiao Y, Pang S, Chen D, Yan B (2013) Two novel and functional DNA sequence variants within an upstream enhancer of the human NKX2-5 gene in ventricular septal defects. Gene 524:152–155
Huang RT, Xue S, Xu YJ, Zhou M, Yang YQ (2014) Somatic GATA5 mutations in sporadic tetralogy of Fallot. Int J Mol Med 33:1227–1235
Jiang JQ, Li RG, Wang J, Liu XY, Xu YJ, Fang WY, Chen XZ, Zhang W, Wang XZ, Yang YQ (2013) Prevalence and spectrum of GATA5 mutations associated with congenital heart disease. Int J Cardiol 165:570–573
Khetyar M, Tinworth L, Syrris P, Abushaban L, Abdulazzaq Y, Silengo M, Carvalho J, Carter N (2008) NKX2.5/NKX2.6 mutations are not a common cause of isolated type 1 truncus arteriosus in a small cohort of multiethnic cases. Genet Test 12:467–469
Kröönström LA, Johansson L, Zetterström AK, Dellborg M, Eriksson P, Cider Å (2014) Muscle function in adults with congenital heart disease. Int J Cardiol 170:358–363
Lahm H, Deutsch MA, Dreßen M, Doppler S, Werner A, Hörer J, Cleuziou J, Schreiber C, Böhm J, Laugwitz KL, Lange R, Krane M (2013) Mutational analysis of the human MESP1 gene in patients with congenital heart disease reveals a highly variable sequence in exon 1. Eur J Med Genet 56:591–598
Lee LJ, Lupo PJ (2013) Maternal smoking during pregnancy and the risk of congenital heart defects in offspring: a systematic review and metaanalysis. Pediatr Cardiol 34:398–407
Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP (1995) Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev 9:1654–1666
McCulley DJ, Black BL (2012) Transcription factor pathways and congenital heart disease. Curr Top Dev Biol 100:253–277
Mondésert B, Abadir S, Khairy P (2013) Arrhythmias in adult congenital heart disease: the year in review. Curr Opin Cardiol 28:354–359
Moutafi AC, Manis G, Dellos C, Tousoulis D, Davos CH (2014) Cardiac autonomic nervous activity in adults with coarctation of the aorta late after repair. Int J Cardiol 173:566–568
Mulkey SB, Swearingen CJ, Melguizo MS, Schmitz ML, Ou X, Ramakrishnaiah RH, Glasier CM, Bradley Schaefer G, Bhutta AT (2013) Multi-tiered analysis of brain injury in neonates with congenital heart disease. Pediatr Cardiol 34:1772–1784
Müller J, Engelhardt A, Fratz S, Eicken A, Ewert P, Hager A (2014) Improved exercise performance and quality of life after percutaneous pulmonary valve implantation. Int J Cardiol 173:388–392
Nam HJ (2012) Crystal structure of the human NKX2.5 homeodomain in complex with DNA target. Biochemistry 51:6312–6319
O’Byrne ML, Mercer-Rosa L, Ingall E, McBride MG, Paridon S, Goldmuntz E (2013) Habitual exercise correlates with exercise performance in patients with conotruncal abnormalities. Pediatr Cardiol 34:853–860
Patel SS, Burns TL (2013) Nongenetic risk factors and congenital heart defects. Pediatr Cardiol 34:1535–1555
Perry JC (2012) Sudden cardiac death and malignant arrhythmias: the scope of the problem in adult congenital heart patients. Pediatr Cardiol 33:484–490
Rushani D, Kaufman JS, Ionescu-Ittu R, Mackie AS, Pilote L, Therrien J, Marelli AJ (2013) Infective endocarditis in children with congenital heart disease: cumulative incidence and predictors. Circulation 128:1412–1419
Sakata M, Hayabuchi Y, Inoue M, Onishi T, Kagami S (2013) Left atrial volume change throughout the cardiac cycle in children with congenital heart disease associated with increased pulmonary blood flow: evaluation using a novel left atrium-tracking method. Pediatr Cardiol 34:105–111
Sanchez-Castro M, Gordon CT, Petit F, Nord AS, Callier P, Andrieux J, Guérin P, Pichon O, David A, Abadie V, Bonnet D, Visel A, Pennacchio LA, Amiel J, Lyonnet S, Le Caignec C (2013) Congenital heart defects in patients with deletions upstream of SOX9. Hum Mutat 34:1628–1631
Schuck R, Abd El Rahman MY, Rentzsch A, Hui W, Weng Y, Alexi-Meskishvili V, Lange PE, Berger F, Abdul-Khaliq H (2014) Altered right ventricular function in the long-term follow-up evaluation of patients after delayed aortic reimplantation of the anomalous left coronary artery from the pulmonary artery. Pediatr Cardiol 35:530–535
Shi LM, Tao JW, Qiu XB, Wang J, Yuan F, Xu L, Liu H, Li RG, Xu YJ, Qian Wang Q, Zheng HZ, Li X, Wang XZ, Qu XK, Yang YQ (2014) GATA5 loss-of-function mutations associated with congenital bicuspid aortic valve. Int J Mol Med 33:1219–1226
Tanaka M, Yamasaki N, Izumo S (2000) Phenotypic characterization of the murine Nkx2.6 homeobox gene by gene targeting. Mol Cell Biol 20:2874–2879
Tanaka M, Schinke M, Liao HS, Yamasaki N, Izumo S (2001) Nkx2.5 and Nkx2.6, homologs of Drosophila tinman, are required for development of the pharynx. Mol Cell Biol 21:4391–4398
Ta-Shma A, El-lahham N, Edvardson S, Stepensky P, Nir A, Perles Z, Gavri S, Golender J, Yaakobi-Simhayoff N, Shaag A, Rein AJ, Elpeleg O (2014) Conotruncal malformations and absent thymus due to a deleterious NKX2-6 mutation. J Med Genet 51:268–270
Tutarel O, Kempny A, Alonso-Gonzalez R, Jabbour R, Li W, Uebing A, Dimopoulos K, Swan L, Gatzoulis MA, Diller GP (2014) Congenital heart disease beyond the age of 60: emergence of a new population with high resource utilization, high morbidity, and high mortality. Eur Heart J 35:725–732
Ueda A, Adachi I, McCarthy KP, Li W, Ho SY, Uemura H (2013) Substrates of atrial arrhythmias: histological insights from patients with congenital heart disease. Int J Cardiol 168:2481–2486
Valente AM, Gauvreau K, Assenza GE, Babu-Narayan SV, Evans SP, Gatzoulis M, Groenink M, Inuzuka R, Kilner PJ, Koyak Z, Landzberg MJ, Mulder B, Powell AJ, Wald R, Geva T (2013) Rationale and design of an International Multicenter Registry of patients with repaired tetralogy of Fallot to define risk factors for late adverse outcomes: the INDICATOR cohort. Pediatr Cardiol 34:95–104
van der Bom T, Zomer AC, Zwinderman AH, Meijboom FJ, Bouma BJ, Mulder BJ (2011) The changing epidemiology of congenital heart disease. Nat Rev Cardiol 8:50–60
Verheugt CL, Uiterwaal CS, van der Velde ET, Meijboom FJ, Pieper PG, Sieswerda GT, Plokker HW, Grobbee DE, Mulder BJ (2010) The emerging burden of hospital admissions of adults with congenital heart disease. Heart 96:872–878
Wang J, Wang LJ, Luo XJ, Liu ZM, Xu WJ, Wang Q, Zheng HZ, Huang RT, Xue S, Yang YQ (2014) GATA5 loss-of-function mutation underlies double outlet right ventricle. Exp Clin Cardiol 20:856–882
Wei D, Bao H, Liu XY, Zhou N, Wang Q, Li RG, Xu YJ, Yang YQ (2013) GATA5 loss-of-function mutations underlie tetralogy of Fallot. Int J Med Sci 10:34–42
Wei D, Bao H, Zhou N, Zheng GF, Liu XY, Yang YQ (2013) GATA5 loss-of-function mutation responsible for the congenital ventriculoseptal defect. Pediatr Cardiol 34:504–511
Wei D, Gong XH, Qiu G, Wang J, Yang YQ (2014) Novel PITX2c loss-of-function mutations associated with complex congenital heart disease. Int J Mol Med 33:1201–1208
Xiang R, Fan LL, Huang H, Cao BB, Li XP, Peng DQ, Xia K (2014) A novel mutation of GATA4 (K319E) is responsible for familial atrial septal defect and pulmonary valve stenosis. Gene 534:320–323
Xie WH, Chang C, Xu YJ, Li RG, Qu XK, Fang WY, Liu X, Yang YQ (2013) Prevalence and spectrum of Nkx2.5 mutations associated with idiopathic atrial fibrillation. Clinics (Sao Paulo) 68:777–784
Yang YQ, Gharibeh L, Li RG, Xin YF, Wang J, Liu ZM, Qiu XB, Xu YJ, Xu L, Qu XK, Liu X, Fang WY, Huang RT, Xue S, Nemer G (2013) GATA4 loss-of-function mutations underlie familial tetralogy of Fallot. Hum Mutat 34:1662–1671
Yang YQ, Wang J, Liu XY, Chen XZ, Zhang W, Wang XZ (2013) Mutation spectrum of GATA4 associated with congenital atrial septal defects. Arch Med Sci 9:976–983
Yu H, Xu JH, Song HM, Zhao L, Xu WJ, Wang J, Li RG, Xu L, Jiang WF, Qiu XB, Jiang JQ, Qu XK, Liu X, Fang WY, Jiang JF, Yang YQ (2014) Mutational spectrum of the NKX2–5 gene in patients with lone atrial fibrillation. Int J Med Sci 11:554–563
Yuan F, Zhao L, Wang J, Zhang W, Li X, Qiu XB, Li RG, Xu YJ, Xu L, Qu XK, Fang WY, Yang YQ (2013) PITX2c loss-of-function mutations responsible for congenital atrial septal defects. Int J Med Sci 10:1422–1429
Yuksel S, Meric M, Soylu K, Gulel O, Zengin H, Demircan S, Yilmaz O, Sahin M (2013) The primary anomalies of coronary artery origin and course: a coronary angiographic analysis of 16,573 patients. Exp Clin Cardiol 18:121–123
Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, Carriero NJ, Cheung YH, Deanfield J, DePalma S, Fakhro KA, Glessner J, Hakonarson H, Italia MJ, Kaltman JR, Kaski J, Kim R, Kline JK, Lee T, Leipzig J, Lopez A, Mane SM, Mitchell LE, Newburger JW, Parfenov M, Pe’er I, Porter G, Roberts AE, Sachidanandam R, Sanders SJ, Seiden HS, State MW, Subramanian S, Tikhonova IR, Wang W, Warburton D, White PS, Williams IA, Zhao H, Seidman JG, Brueckner M, Chung WK, Gelb BD, Goldmuntz E, Seidman CE, Lifton RP (2013) De novo mutations in histone-modifying genes in congenital heart disease. Nature 498:220–223
Zomer AC, Vaartjes I, van der Velde ET, de Jong HM, Konings TC, Wagenaar LJ, Heesen WF, Eerens F, Baur LH, Grobbee DE, Mulder BJ (2013) Heart failure admissions in adults with congenital heart disease; risk factors and prognosis. Int J Cardiol 168:2487–2493
Acknowledgments
The authors are greatly thankful to the participants for their devotion to the study. This work was supported in part by Grants from the National Natural Science Fund of China (81270161 and 81271927), the Key Program for Basic Research of Shanghai, China (14JC1405500), the Experimental Animal Program of Shanghai, China (12140902800), the Fundamental Research Fund for Central University of Ministry of Education, China (15072), the Scientific Research Fund of Health Bureau, Shanghai, China (2010119), and the Excellent Discipline Leader Training Foundation of Pudong New Strict, Shanghai, China (PWRd2012-05).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Juan Wang, Jian-Hui Mao, and Ke-ke Ding have contributed equally to the work.
Rights and permissions
About this article
Cite this article
Wang, J., Mao, JH., Ding, KK. et al. A Novel NKX2.6 Mutation Associated with Congenital Ventricular Septal Defect. Pediatr Cardiol 36, 646–656 (2015). https://doi.org/10.1007/s00246-014-1060-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00246-014-1060-x