Abstract
Detection of species fraud in meat products is very important in order to protect consumers from undesirable adulteration, as well as for the economic, religious and health aspects. The most important reason for verification of the labeling statements is to detect fraudulent substitution of expensive meat components with other cheaper animals or mislabeling. The aim of this study was to develop a multiplex PCR that could be used in the simultaneous identification of multiple meat species. In this study, ten sausages with a minimum beef content of 55 %, from ten different manufacturing companies, and five samples of cow, chicken, goat, camel and donkey raw meats, for the purpose of positive control, were collected from food markets in Tehran, Iran. Total DNA was extracted from each sausage and the raw meats. Primers were selected in different regions of mitochondrial DNA (12S rRNA, cytochrome b and NADH dehydrogenase subunits 2) for identification of meat species. 12S rRNA and NADH dehydrogenase subunits 2 primers generated specific fragments of 183 and 145 bp length, for chicken and donkey, respectively. Three different specific primers were used for amplification of cytochrome b gene in goat, camel and cattle species and amplified species-specific DNA fragments of 157, 200 and 274 bp, respectively. The results proved that half of the specimens were contaminated with chicken meat, and this was greater than the proportion of beef stated on the label, while the other half only had chicken residuals, and no beef content. No contamination was found with goat, donkey or camel meats. These findings showed that molecular methods, such as multiplex PCR, is a potentially reliable, sensitive and accurate assay for the detection of adulterated meat species in mixed meat products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Meat is a major source of protein and it is also rich in micronutrients such as magnesium, zinc and iron, which are essential for human health. Usually red raw meat such as beef and sheep is more expensive than the other red meat products such as hamburger, sausages and cold cut because of lower content of costly meat in these products. So, some people prefer to use ready meat products instead of pure red meat. Therefore, meat products can only partially meet the nutritional needs of the community [1, 2]. On the other hand, the maximum potential for fraud occurs in the production of meat products compared to other foods because the raw materials, once uniformly mixed, are no longer distinguishable in appearance from one another. A common fraud in the production of meat products such as hamburger, sausages and kebab is mixing beef with cheaper meats such as chicken and chicken residuals. Currently, the authenticity of meat products is a serious concern for food industry officials [3]. Increasing consumer awareness about the ingredients in meat products requires accurate monitoring of the origins of the meats used in these products, because the incorrect labeling of meat products indicates commercial fraud [4]. Since detecting the different ingredients is important, attention has been drawn toward the labeling of food products, especially processed foods in recent years. Furthermore, due to various crises in the food industry, including crises associated with public health such as outbreaks of bovine spongiform encephalopathy (BSE), avian influenza virus, swine flu and dioxin contamination, the trend toward the accurate identification of animal species in meat products has also increased in recent years [5, 6]. One of the most important principles in maintaining food safety is the supervision of the health and safety of food products supplied to the market. Given all of these concerns, it is important to verify the declared claims on labels, both for the consumer and food industry officials. Thus, to support the accurate supervision of food products, various techniques have been developed to analyze food ingredients, which include analytical methods based on protein and DNA analysis [4]. Protein-based techniques include electrophoresis [7], chromatography [8] and immunological methods [9–11]. These methods are not usually suitable for processed mixed food products like sausages because they cannot distinguish between closely-related meat species in the processed foods, furthermore, they are also time consuming and expensive [12–14]. As an alternative, molecular authentication or molecular detection of meat species through DNA proliferation using a PCR method has been developed in the last 15 years [15, 16]. This method is preferred due to its speed, simplicity, high sensitivity, specificity and high stability of DNA, compared to protein-based methods, especially in cooked or autoclaved meat products [17, 18]. Common DNA-based methods are randomly amplified polymorphic DNA-PCR (RAPD-PCR) [19], hybridization [20–23], PCR sequencing or PCR–restriction fragment length polymorphism (PCR–RFLP) [24–28], PCR–single-strand conformation polymorphism (PCR–SSCP) [29, 30], multiplex PCR [31–33] and specific species PCR [34]. Given the increasing consumption of ready-made foods and meat products, this study aims to support consumer rights against frauds that may occur in meat products like sausages. In this study, the multiplex PCR method was used for the amplification of various mitochondrial genes to concurrently identify meat species in cooked sausages.
Materials and methods
Samples
The raw meats were selected from different species (camel, cow, donkey, goat and chicken) as a positive control and industrial meat products (sausages) were collected from 10 different companies in Tehran, Iran (randomly coded as A–J), and these were stored at 4 °C until used for DNA extraction.
DNA extraction
Total cellular DNA was extracted from samples according to the cetyltrimethylammonium bromide (CTAB) method with some modifications [35]. In brief, 0.06 g of chopped tissues was mixed with 1,000 µl 2× CTAB lysis buffer (100 mM Tris–HCl, 1.4 M NaCL, 20 mM EDTA, 2 g CTAB, pH 8.0), and 30 µl proteinase K in 1.5 ml microtubes, then incubated at 65 °C for 3 h. Microtubes were inverted every 15 min to produce a good solution of the crushed tissues with the buffer. A total of 600 µl phenol–chloroform–isoamyl alcohol (25:24:1) mixture was added, followed by vigorous shaking for 10 min and centrifugation at 12,000 rpm for 5 min. The upper aqueous phase was separated without disturbing the interphase which contained cell debris and proteins. This step was repeated once with a fresh aliquot of phenol, chloroform and isoamyl alcohol mixture (25:24:1) and once with a chloroform isoamyl alcohol mixture (24:1). The aqueous phase of each tube was transferred to a new 1.5 ml microcentrifuge tube DNA in the supernatant was precipitated with 600 μl of cold (−20 °C) absolute ethanol followed by mild inversion for 15 min. The resultant mixture was centrifuged at 12,000 rpm for 10 min and the upper aqueous phase was removed. The DNA pellet was washed with cold 70 % ethanol followed by 15-min mild inversion at room temperature and centrifugation at 12,000 rpm at 4 °C for 10 min. The washed DNA pellet was dried by leaving the tubes at 37 °C for 40 min. The DNA sample was dissolved in 50 μl TE buffer (10 mM Tris–HCl, 0.5 mM EDTA, pH 8.0) [36].
Genomic DNA purity and quantity was assessed with a NanoDrop™ ND-2000 spectrophotometry. By measuring the A260/230 and A260/280 absorbance ratios, the DNA concentration, purity and protein contamination of samples were determined. The quality of extracted DNA was analyzed by electrophoresis pattern of samples in a 0.7 % agarose gel stained with ethidium bromide and visualized under ultraviolet light.
Oligonucleotide primers
Species-specific primers were selected for amplification of different regions of the mitochondrial genome is presented in Table 1.
Simplex PCR
In a preliminary phase of this investigation, primer specificity was assessed with DNA extracted from raw meats. For detection of cross-reaction, primer set of each species was analyzed by another DNA species in five separate simplex PCRs. Simplex PCRs were carried out in a final volume of 25 μl containing 2.5 µl of 1× PCR buffer (50 mM Kcl, 20 mM Tris, pH 8.4), 1.5 U smart Taq DNA Polymerase (Fermentas, Germany), 200 µM each of dNTPs (Fermentas), 1.5 mM MgCl2, 0.4, 0.4, 0.2, 0.36 and 0.36 µM of donkey, poultry, cattle, goat and camel forward primers, respectively, 0.4 µM of all reverse primers and 20 ng of DNA template. Amplification was carried out in Thermal Cycler (Biorad). After an initial denaturation step at 95 °C for 1 min, 30 cycles were programmed as follows: strand denaturation at 94 °C for 1 min, primer annealing at 60 °C for 1 min, primer extension at 72 °C for 90 s and final extension at 72 °C for 5 min. PCR products were determined by visualization of amplicons on 2 % agarose gels stained with ethidium bromide.
Multiplex PCR
In order to simultaneously identify each animal species, all primer sets were used to develop a one-step reaction. Amplifications were developed in a final volume of 50 µl containing 5 µl of 10× PCR buffer (500 mM Kcl, 100 mM Tris, pH 8.8), 2.5 U smart Taq DNA Polymerase (Fermentas, Germany), 200 µM each of dNTPs (Fermentas), 1.5 mM MgCl2, 0.8, 0.8, 0.4, 0.72 and 0.72 µM of donkey, poultry, cattle, goat and camel forward primers, respectively, 0.8 µM of all reverse primers and 40 ng of DNA template. Amplification was carried out in a Biorad Thermal Cycler. After an initial denaturation step at 95 °C for 2 min, 30 cycles were programmed as follows: 94 °C for 1 min, 60 °C for 1 min, 72 °C for 90 s and final extension at 72 °C for 5 min.
Specificity and sensitivity of PCR
The specificity of each species-specific primer was confirmed by amplification of 20 ng DNA of other meat species used in this study (cow, chicken, camel, donkey and goat meats genomic DNAs). In the determination of the detection limit of specific primers, serial 1:10 (50, 5, 0.5, 0.05 and 0.005 ng DNA/µl water) dilutions of raw cow, chicken, camel, donkey and goat DNAs were prepared and each dilution was added separately to the reaction mixtures.
Results
DNA extraction
The quantity of DNA in the solution was calculated from the absorbance of 260 nm (A260) which revealed high concentrations of extracted DNA, and the purity was calculated with the ratio A260/A280. The adsorption rate of the extracted DNA was found to be in the range of 1.7–2, which indicated a high quality of DNA extraction. In fact, results of the extracted DNA from the raw meat samples (control) and ten brands of sausages showed the suitability of the extracted DNA for PCR amplification.
Specific simplex PCR
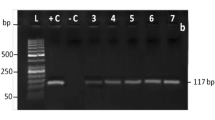
Simplex PCR with species-specific primers was optimized to confirm the specificity of the primers. To that end, a simplex PCR with a species-specific primer using DNA from other meat species (camel, cattle, donkey, goat and chicken) was performed. In the initial stage of this study, simplex PCR on the DNA of raw meats (control) was optimized with species-specific primers. Species-specific primers of donkey, goat, chicken, camel and cattle produced amplified 145-, 157-, 183-, 200- and 274-bp fragments, respectively. To identify cross-reactions between the DNAs of other species and species-specific primers, simplex PCR was performed with nontarget DNA of the species, proving the specificity of the primer of each species. No cross-reaction was observed (Fig. 1). This test was repeated several times with the same results each time, indicating repeatability of the test. Then, simplex PCR with each species-specific primer was performed on the DNAs extracted from ten sausage brands. Specific simplex PCR for the ten sausage brands with species-specific primers of camel, cattle and goat, for the amplification of the cytochrome b gene, the poultry-specific primer for the 12S rRNA gene, and the donkey specific primer for the NADH mitochondrial subunit 2 gene, are presented in Fig. 2. The results obtained from amplification of the 200-bp fragment for the mitochondrial cytochrome b gene in the sausage samples on 2 % agarose gel, revealed no camel meat in any of the samples (Fig. 2a). The specific 183-bp fragment from the 12S rRNA mitochondrial gene region was amplified in all samples, thus indicating the presence of chicken in every sample (Fig. 2b). Lack of amplification of the 145-bp fragment from the mitochondrial NADH subunit 2 gene revealed that there was no donkey meat in the samples (Fig. 2c). Amplification of the 274-bp fragment of cattle mitochondrial cytochrome b gene is shown in samples A, C, D, G and I (Fig. 2d). Results obtained from a lack of amplification of the 157-bp fragment of the mitochondrial cytochrome b gene in the sausage samples revealed no goat meat in any sample (Fig. 2e).
Specificity of simplex PCR of DNA from 10 sausages brands with camel primer (a), poultry primer (b), donkey primer (c), cattle primer (d), goat primer (e). Lane 1 sausage (A), lane 2 sausage (B), lane 3 sausage (C), lane 4 sausage (D), lane 5 sausage (E), lane 6 sausage (F), lane 7 sausage (G), lane 8 sausage (H), lane 9 sausage (I), lane 10 sausage (J), lane 11 positive control with target meat DNA in each figure, C− negative control, M 100 bp DNA ladder
Multiplex PCR
After confirmation of specificity in the primers of each animal species, a multiplex PCR was optimized for simultaneous identification of species with the control samples. As expected, no cross-reaction in multiplex PCR was observed between species-specific primers and the DNA from the other control meats. Then, a multiplex PCR was performed with DNA extracted from ten sausage brands (A–J), and their results are presented in Fig. 3. Results of the multiplex PCR were in complete agreement with those of the simplex PCR. As can be seen in Fig. 3, all of the brands in this study (A–J) contained chicken ingredients, which is inconsistent with the label on the meat product supplied to the market, because all of these brands had red meat labels on them, and according to the food regulation, they must not contain any chicken ingredients. This clearly showed fraud in all of the sausages studied. Some beef meat was found only in samples A, C, D, G, and I.
Multiplex PCR on 10 sausages brands on 2 % agarose gel, lanes 1–10 sausage (A), (B), (C), (D), (E), (F), (G), (H), (I), (J), respectively, C− negative control; lane M 100 bp DNA ladder, M1 multiplex PCR of raw cattle, camel, chicken, goat and donkey meats DNAs, lanes 11–15 positive control with raw cattle meat DNA, camel meat DNA, chicken meat DNA, goat meat DNA, donkey meat DNA, respectively
Sensitivity of the species-specific primers
The detection limits of specific PCR assay were determined by PCR amplification of DNA extracted from each species. The sensitivity of the method was determined as 0.05 ng DNA for each species (Fig. 4a–g).
The results of PCR sensitivity test for donkey primer (a), poultry primer (b), goat primer (c), cattle primer (d) and camel primer (e) with raw meats. Sensitivity results of simplex PCR of different dilution of DNA from sausages with cattle primer (f) and chicken primer (g). Lane 1 50 ng, lane 2 5 ng, lane 3: 0.5 ng, lane 4 0.05 ng, lane 5 0.005 ng, lane 6 0.0005 ng, lane 7 0.00005 ng, C− negative control, M 100 bp DNA ladder
Discussion
Detecting animal species in terms of consumer preference and for supervisory purposes is often necessary. In a country such as Iran, identifying animal species in meat products is particularly important because of the cultural and religious issues from a consumer standpoint. The first step in recognizing the type of meat used, involves DNA extraction followed by a PCR specific reaction.
Quantitative test results of extracted DNA by NanoDrop indicated that the adsorption rate of DNA solution was in the range of 1.7–2, which indicates a high purity of extracted DNA. In fact, results of the extracted DNA from raw meat (control) and ten brands of sausages indicated that the quality of extracted DNA for amplification of the PCR was good. As a result of the various processes such as seasoning with salt, drying, smoking and cooking during the production of the meat products, problems can occur during the extraction of DNA from meat products. This could have an effect on the integrity of the DNA and lead to DNA degradation. Generally, heating has major effects on the quality of DNA, while seasoning and smoking have the least effect [14]. In the present study, mitochondrial DNA was used to detect animal species, which has several excellent properties. Several properties increase its survival chances under processing conditions including high mitochondrial DNA copy number, small size, being double-stranded, and the circular shape in the cells. Also these properties help the preservation and survival of DNA fragments to be adequately amplified by PCR, even in very small amounts of raw or processed meats. Because of its unique properties such as high maternal inheritance, high mutation rate, and the rare possibility of this gene being involved in recombination, mitochondrial DNA polymorphism is widely used to detect different species. These characteristics have made them ideal for identifying animal species in processed meat products [39]. Complete and incomplete mitochondrial DNA sequences are traceable in many animals [40].
Primers used in this study were chosen from different genes of mitochondrial DNA. Binding sites of primers to the amplification of specific fragments were chosen with less than 300 bp because the DNA of the samples could have been severely damaged by the heating process, causing some problems in amplification with the PCR [41].
Recently, concerns about possible fraud in meat products have increased. In this paper, we have tried to introduce an accurate and reliable method for detecting the kind of meat species added to compound processed meat products. In this study, simplex PCR and multiplex PCRs were performed on the samples, and the same results were obtained with both methods. Species-specific simplex PCR confirmed the authenticity of the multiplex PCR method.
In the multiplex PCR method, several specific primers are used for amplification of the target fragments. Selection of primers in the multiplex PCR method is extremely important. In this technique, the specificity of primers and T m temperature are more important than in ordinary PCR. In Iran, the highest possibility of fraud in meat products is with the use of cheaper meats such as goat, camel, donkey and chicken. In order to distinguish between ruminant animals such as goat, camel and cattle, their species-specific primers were used and by optimizing the amount of primers and PCR conditions, each of the primers was able to amplify target fragments without cross-reaction. Multiplex PCR was performed on the DNA of control species (raw meat) and ten sausage brands. With no cross-reactions, the poultry-specific primer for the 12S rRNA gene amplified 183-bp fragment was present in all of the sausage samples, and the cattle specific primer of the cytochrome b gene caused amplification of the 274-bp fragment in samples A, C, D, G and I, therefore given that 55 % beef was written on the label of all of these samples, fraud was detected in all of the samples. It is worth noting that in five of the samples, in addition to chicken, beef meat was also confirmed by PCR.
In a similar study, Ghovati et al. [42] used three types of primers consisting of; poultry, ruminant animals and pork, while in present study, five types of species-specific primers were used including goat, donkey, camel, poultry and cattle, as these meats were more likely to be added to the sausages produced in Iran. A specific primer of ruminant animals used by Ghovati et al. was unable to distinguish between camel, goat or cattle. However, the optimized multiplex PCR in this study was able to detect the different meats of goat, donkey, camel, chicken and cattle, in the sausage samples.
Matsunaga et al. applied a quick and simple method to detect animal species in meat products using PCR. To identify six kinds of meat consisting of beef, pork, chicken, lamb, goat and horse, the raw materials for the products were used by mixing them with seven primers designed for the cytochrome b region. They were able to identify DNA fragments of specific species by multiplex PCR [38].
Convenient, sensitive and specific real-time PCR assay was described for specific identification of horse and donkey by Chisholm et al. This method was applicable to the detection of low levels of horse or donkey meat in commercial products [43] .
Four types of primers (ruminant animals, poultry, fish and pork) were used by Dalmasso et al. In fact, by using one primer for ruminant animals and fish, they could not distinguish between different species in one stage of PCR [32].
Three types of primers were developed by Kesmen et al. They mixed meat samples and prepared the sausages themselves. In fact, they carried out the multiplex PCR test, knowing the type of meat species in the sausages. However, in the present study, industrial sausages were used and based on their labels, stating that they were beef, the samples were assumed to contain beef [18].
Daiming Zha et al. developed a multiplex PCR method for detecting deer meat. Their results showed that this method is highly reliable, accurate and sensitive for detecting deer products [44]. Primer design is extremely important in the multiplex PCR method because specificity and melting temperature (T m) in each primer is more important than compared to ordinary PCR, and the melting temperature of the different primers must be optimized so that different species can be detected in one stage of PCR [45]. In this present study, five pairs of different primers were used, and the amount of each primer was optimized for the amplification of all target sequences.
Some methods such as PCR–RFLP and sequencing, are time consuming and expensive. They cannot detect a mixture of samples and they are not suitable for routine species detection. Multiplex PCR can identify all species in a one reaction stage, as simplex PCRs can detect only one species at a time, and it is also simpler, quicker and cheaper than PCR–RFLP or sequencing [39, 46].
The multiplex PCR described in this study is highly sensitive, specific and its specificity is not influenced by the meat processing temperature. This method is able to amplify mitochondrial DNA in every cell, which increases the chance of survival under severe production conditions. This method has been developed as an ideal method to detect meat species in processed products [28].
Results of this study showed that this method was able to identify fraud in all of the samples without cross-reaction. In addition, sensitivity, specificity and repeatability of multiplex PCR have turned it into a quick method for analyzing fraud in mixed meat products such as sausages. Furthermore, it seems that this method is an accurate technique for detecting various raw materials used in meat products because in a single reaction, it is able to detect different raw materials added to food products using their specific primers.
Conclusion
Results obtained from simplex PCR and multiplex PCR of species-specific meats in this study showed that despite stating a content of 55 % beef on the label of all samples, and although none of the species of goat, camel and donkey were identified in these products, chicken residuals were detected in all samples, which is indicative of fraud in these products. Given the cheap price of poultry components, especially chicken paste, the use of chicken paste in these products is considered to be fraud. According to the second revision of Iran’s national standard guidelines, Article 2303 sausage and meat products, properties and test methods in the 961st session of the National Committee of Food and Agriculture Products dated August 7, 2010, it is illegal to use mechanically produced red or white meat (meat and chicken pastes) in any type of sausage or cold meat.
Recently, in order to increase the protein content of these products, the use of chicken paste, which is in fact chicken residuals, has increased in meat products. It is anticipated that by applying the proposed multiplex PCR method for identifying chicken paste in sausages and meat products, this kind of fraud can be inspected and effectively prevented.
References
Mahajan MV, Gadekar YP, Dighe VD, Kokane RD, Bannalikar AS (2010) Molecular detection of meat animal species targeting MT 12S rRNA gene. Meat Sci 88:23–27
Culbertson JD, Duncan S, Guerrero-Legarreta I, Li-Chan ECY, Ma CY, Manley CH, McMeekin TA, Nip WK, Nollet LML, Rahman MS, Toldr F, Xiong YL (2006) Handbook of food science technology and engineering, vol 1. CRC Press, New York
Pattersan RLS (1985) Biochemical identification of meat species. Elsevier Applied Science, London
Mafra I, Ferreira LVOP, Beatriz MP, Oliveira P (2008) Food authentication by PCR-based methods. Eur Food Res Technol 227:649–665
Ciampolini R, Leveziel H, Mazzanti E, Grohs C, Cianci D (2000) Genomic identification of the breed of an individual or its tissue. Meat Sci 54:35–40
Goffaux F, China B, Dams L, Clinquart A, Daube G (2005) Development of a genetic traceability test in pig based on single nucleotide polymorphism detection. Forensic Sci Int 151:239–247
Zerifi A, Labie C, Benard G (1992) SDS-PAGE technique for the species identification of cooked meat. Fleiswirtschaft 1:54–59
Amstrong SG, Leach DN (1992) The use of HPLC protein profiles in fish species identification. Food Chem 44:147–155
Hsieh YH, Sheu SC, Bridgman RC (1998) Development of a monoclonal antibody specific to cooked mammalian meat mixtures. Meat Sci 15:1–13
Kang’ethe EK, Gathuma JM, Landqvist KJ (1986) Identification of the species of origin of fresh, cooked and canned meat on meat products using antisera to thermostable muscle antigens by Ouctherlony’s double diffusion test. J Sci Food Agric 37:157–162
Patterson RL, Jones SJ (1990) Review of current technique for verification of the species origin of meat. Analyst 115:501–506
Calvo JH, Zaragoza P, Osta R (2001) A quick and more sensitive method to identify pork in processed and unprocessed food by PCR amplification of a new specific DNA fragment. J Anim Sci 79:2108–2112
Koh MC, Lim CH, Chua SB, Chew ST, Phang ST (1998) Random amplified polymorphic DNA (RAPD) fingerprints for identification of red meat animal species. Meat Sci 48:275–285
Saez R, Sanz Y, Toldrá F (2004) PCR-based fingerprinting techniques for rapid detection of animal species in meat products. Meat Sci 66:659–665
Asensio L (2007) PCR-based methods for fish and fishery products authentication. Trends Food Sci Technol 18:558–566
Dalvit C, De Marchi M, Cassandro M (2007) Genetic traceability of livestock products: a review. Meat Sci 77:437–449
Arslan A, Ilhak OI, Calicioglu M (2006) Effect of method of cooking on identification of heat processed beef using polymerase chain reaction (PCR) technique. Meat Sci 72:326–330
Kesmen Z, Sahin F, Yetim H (2007) PCR assay for the identification of animal species in cooked sausages. Meat Sci 77:649–653
Arslan A, Ilhak OI, Calicioglu M (2005) Identification of meats using random amplified polymorphic DNA (RAPD) technique. J Muscle Foods 16:37–45
Chikuni K, Ozutsume K, HoishiKawa T, Kato S (1990) Species identification of cooked meats by DNA hybridization assay. Meat Sci 27:119–128
Ebbehoj KF, Thomsen PD (1991) Species differentiation of heated meat products by DNA hybridization. Meat Sci 30:221–234
Ebbehoj KF, Thomsen PD (1991) Differentiation of closely related species by DNA hybridization. Meat Sci 30:359–366
Hunt DJ, Parkes HC, Lumley ID (1997) Identification of the species of origin of raw and cooked meat products using oligonucleotide probes. Food Chem 60:437–442
Bartlett SE, Davidso WS (1991) Identification of Thunnus tuna species by the polymerase chain reaction and direct sequence analysis of their mitochondrial cytochrome b genes. Can J Fish Aquat Sci 48:309–317
Chikuni K, Tabat T, Saito M, Monma M (1994) Sequencing of mitochondrial cytochrome b genes for the identification of meat species. Anim Sci Technol 65(6):571–579
Meyer R, Hoefelein C, Luethy J, Candrian U (1995) Polymerase chain reaction–restriction fragment length polymorphism analysis: a simple method for species identification in food. J AOAC Int 78(6):1542–1551
Desjardins P, Morais R (1999) Sequence and gene organization of the chicken mitochondrial genome. J Mol Biol 212:599–634
Partis L, Croan D, Guo Z, Clark R, Coldham T, Murby J (2000) Evaluation of a DNA fingerprinting method for determining the species origin of meats. Meat Sci 54:369–376
Cespedes A, Garcia T, Carrera E, Gonzalez I, Fernandez A, Hernandez PE (1999) Application of polymerase chain reaction–single strand conformational polymorphism (PCR–SSCP) to identification of flatfish species. J AOAC Int 82:903–907
Rehbein H, Kress G, Schmidt T (1997) Application of PCR SSCP to species identification of fishery products. J Sci Food Agric 74:35–41
Asensio L (2008) Application of multiplex PCR for the identification of grouper meals in the restaurant industry. Food Control 19(11):1096–1099
Dalmasso A, Fontanella E, Piatti P, Civera T, Rosati S, Bottero M (2004) A multiplex PCR assay for the identification of animal species in feedstuffs. Mol Cell Probe 18:81–87
Di Pindo A, Forte VT, Conversano MC, Tantillo G (2004) Duplex polymerase chain reaction for detection of pork meat in horse meat fresh sausages from Italian retail sources. Food Control 16:391–394
Che Man YB, Aida AA, Raha AR, Son R (2007) Identification of pork derivatives in food products by species-specific polymerase chain reaction (PCR) for halal verification. Food Control 18:885–889
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Sambrook J, Fritch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold spring Habor Laboratory, New York
Chen Y, Wu Y, Xu BL, Wan J, Qian ZM (2005) Species-specific polymerase chain reaction amplification of camel (camelus) DNA extracts. J AOAC Int 88(5):1394–1398
Matsunga T, Chikuni K, Tanabe R, Muroya S, Shibata K, Yamada J, Shinmura Y (1999) A quick and simple method for the identification of meat species and meat products by PCR assay. Meat Sci 51:143–148
Girish PS, Anjaneyulu ASR, Viswas KN, Anand M, Rajkumar N, Shivakumar BM, Bhaskar S (2003) Sequence analysis of mitochondrial 12S rRNA gene can identify meat species. Meat Sci 66:551–556
Umetsu K, Yuasa I (2005) Recent progress in mitochondrial DNA analysis. J Legal Med 7:259–262
Bottero MT, Dalmasso IA, Nucera D, Turi RM, Rosati S, Squadrone S, Goria M, Civera T (2003) Development of a PCR assay for the detection of animal tissues in ruminant feeds. J Food Prot 66:2307–2312
Ghovvati S, Nassiri MR, Mirhoseini SZ, Heravi Moussavi A, Javadmanesh A (2008) Fraud identification in industrial meat products by multiplex PCR assay. Food Control 20:696–699
Chisholm J, Conyers C, Booth C, Lawley W, Hird H (2005) The detection of horse and donkey using real-time PCR. Meat Sci 70:727–732
Zha D, Xing X, Yang F (2010) A multiplex PCR assay for fraud identification of deer products. Food Control 21:1402–1407
Bai W, Xu W, Huang K, Yanfang Y, Cao S, Luo Y (2009) A novel common primer multiplex PCR (CP-M-PCR) method for the simultaneous detection of meat species. Food Control 20:366–370
Lin WF, Hwang DF (2008) A multiplex PCR assay for species identification of raw and cooked bonito. Food Control 19:879–885
Conflict of interest
None.
Compliance with Ethics Requirements
All institutional and national guidelines for the care and use of laboratory animals were followed.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Parchami Nejad, F., Tafvizi, F., Tajabadi Ebrahimi, M. et al. Optimization of multiplex PCR for the identification of animal species using mitochondrial genes in sausages. Eur Food Res Technol 239, 533–541 (2014). https://doi.org/10.1007/s00217-014-2249-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-014-2249-1