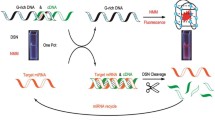
Abstract
Analysis of microRNAs (miRNAs) is important in cancer diagnostics and therapy. Conventional methods used to extract miRNA for analysis are generally time-consuming. A novel approach for rapid and sensitive extraction of miRNAs is urgently need for clinical applications. Herein, a novel strategy based on electrical potential-assisted DNA-RNA hybridization was designed for miRNA extraction. The entire extraction process was accomplished in approximately 3 min, which is much shorter than the commercial adsorption column method, at more than 60 min, or the TRIzol method, at more than 90 min. Additionally, the method offered the advantages of simplicity and specificity during the extraction process by electrical potential-assisted hybridization of single-stranded DNA and RNA. Taking let-7a as an example, satisfactory results were achieved for miRNA extraction in serum, demonstrating the applicability in miRNA nucleic acid amplification.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
RNAs are associated with various physiological and pathological processes, including metabolism, apoptosis, necrocytosis, proliferation, differentiation and development [1,2,3]. MicroRNAs (miRNAs) are noncoding RNAs with length less than 100 base pairs (bp). With the increasing interest in miRNA therapeutics and targeting of miRNAs to treat disease, there is a need for tools to enable the fast and sensitive detection of miRNAs to determine their function [4,5,6]. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) is a particularly attractive technology for detection of miRNA due to its specificity, sensitivity and relatively rapid time-to-answer [7,8,9,10]. A rapid approach for obtaining pure and abundant miRNA molecules is vital in nucleic acid-based analysis.
The most widely implemented approaches reported for miRNA extraction include conventional solvent methods or commercial kit reagents. TRIzol reagent is the most commonly used solvent due to good consistency in recovery [11]. However, the method relies on a set of time-consuming reagents and enrichment steps, with over 4 h of complex operation to collect and process miRNAs, hindering its application for rapid detection preparation. The adsorption column method has been extensively harnessed for numerous commercial kit reagents owing to the simplicity of direct physical adsorption. However, more than 60 min of time consumption and low recovery have impeded widespread application of this method because of the difficulty in separation. Rapid extraction of miRNA remains a great challenge because of degradation and forming co-precipitation [12, 13]. These challenges require the development of novel strategies for rapid, simple and efficient miRNA extraction.
Owing to operational simplicity, remarkable sensitivity and the potential for automation, electrochemical-based strategies have drawn tremendous interest, and have achieved significant progress in the early clinical analysis of diseases. Electrochemical detection of small nucleic acid molecules in particular has become a research hotspot in the field of bioengineering [14,15,16,17]. In the utilization of oriented DNA immobilized on electrodes, diverse electrochemical devices and platforms have been employed to detect miRNAs in the development of electrochemical assay to achieve high sensitivity and selectivity in terms of the electrochemical signal transduction, which also provides the possibility for the extraction of miRNAs owing to the combination of specific DNA-RNA recognition [18,19,20]. However, the current process of the specific hybridization between the DNA capture and the target miRNA all need hours of reaction [21], and the miRNA specifically captured can only be detected by electrochemical methods, and cannot be isolated [22]. The complex modifications commonly used in electrochemical methods undoubtedly increase the total time of the detection process, and the reproducibility of the detection of low-abundance targets is low [23]. For the same low concentration of targets, RT-qPCR increases its concentration through exponential amplification, which greatly improves the sensitivity and accuracy of detection [24, 25]. However, the complicated sample extraction steps such as in the TRIzol method still make RT-qPCR detection of miRNA time-consuming [26]. Controlled electric fields can be used to regulate transport and hybridization of single-stranded oligonucleotides [27]. Heller et al. [28] first used electric fields to enhance DNA hybridization. Conde et al. [29] subsequently demonstrated that the use of microsecond voltage pulses increased the speed of hybridization by at least 108 times compared with the passive control reaction without an electric field. In addition, Gebala et al. [18] proposed a method for assisting DNA hybridization by applying an external voltage. However, although much has been done for extension or adaptation of nucleic acids, the overall detection process for miRNA still takes several hours, and there is no report based on electrical potential-assisted hybridization for miRNA extraction.
The expression of let-7a is closely related to a variety of cancers [30], so it is considered an important biomarker for accurate tumor diagnosis and prognosis [31]. Several methods have been developed for let-7a detection, including sequencing, electrochemical and amplification methods. Next-generation sequencing achieves high-throughput profiling, but cannot reliably quantify low-abundance analytes [32]. Electrochemical methods and standard assays based on amplification such as PCR are sensitive but often require complex sensor fabrication and time-consuming extraction [32,33,34]. At present, a combination electrochemical and amplification protocol has been proposed to achieve more sensitive and specific detection of let-7a, but complex modifications and rigorous protocol design are still required [35]. Herein, a rapid, simple and efficient method for miRNA extraction was investigated based on electrical potential-assisted DNA-RNA hybridization. Taking miRNAs of let-7a as an example, the region of the single-stranded DNA responsible for let-7a hybridization was designed and immobilized on the electrode. With the assistance of the electric field, let-7a quickly formed an enrichment on the electrode surface, thereby increasing the chance of collision, and greatly increasing the nucleic acid hybridization rate. The entire let-7a isolation process was completed in 150 s, including 90 s capture and 60 s rapid elution steps. The extraction time was significantly shortened. The performance characteristics of the method for RNA extraction were investigated by RT-qPCR and accelerated strand exchange amplification (ASEA) [36], and its possible application in real samples was evaluated for early cancer screening.
Materials and methods
Materials and reagents
DNA oligonucleotides were purchased from Sangon Biotech (Shanghai, China). DNA capture probe (15-mer): 5′CAA CCT ACT ACC TCA–SH 3′; complementary let-7a target: 5′ UGA GGU AGU AGG UUG UAU AGU U 3′ (The underlined portion was reverse complement with DNA capture probe). TaqMan™ MicroRNA Reverse Transcription Kit was purchased from Applied Biosystems (USA). PerfectStart™ II Probe qPCR SuperMix was purchased from TransGen Biotech Co., Ltd. (Beijing, China). The ASEA detection kit was provided by Navid Biotech Co., Ltd. (Qingdao, China). The miRcute miRNA extraction and isolation kit was provided by Tiangen Biotech Co., Ltd. (Beijing, China). TRIzol was provided by Thermo Fisher Scientific Co., Ltd. (Shanghai, China). Ethanol, potassium ferricyanide (K3Fe(CN)6), potassium ferricyanide trihydrate [K4Fe(CN)6·3H2O], chloroform and isopropanol were obtained from Sinopharm Chemical Reagent Co., Ltd. (Jinan, China). Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 6-mercaptohexanol (MCH), RNase-free water, KCl and NaCl were purchased from Sigma. All reagents were of analytical grade and were used as received. Human serum was provided by Qingdao Central Hospital.
Preparation of electrodes and DNA immobilization
Gold electrodes (2 mm diameter, Wuhan Goslink Technology Co., Ltd., China) were polished with wet alumina slurries with particle size of 0.3 μm (Shanghai Chenhua Instrument Co., Ltd., China) with a motor sander (GU-100) for 1 min each. Then they were rinsed ultrasonically for 1 min with ethanol and deionized water, respectively, and finally dried with nitrogen. The electrodes were further electrochemically characterized by means of cyclic voltammetry (CV) in a real-time prepared 0.5 M H2SO4 solution in a potential range from 0 V to +1.2 V vs. Ag/AgCl (3.0 M KCl) until a stable voltammogram was obtained. Finally, the electrodes were rinsed with water and dried under a nitrogen stream.
Electrical potential-assisted DNA-RNA hybridization for miRNA extraction
The prepared electrode was soaked in 20 μL of immobilization solution for 8 h. The immobilization solution contains 1 μM capture DNA probe and 10 mM TCEP. In addition, the electrode incubated in the 10 μL 1 mM MCH for 30 min to block the sites not occupied by the ligated capture. The modified electrode was put into 200 μL of hybridization solution containing the target strand, two potential pulses of 100 mV for 0.01 s and −200 mV for 0.01 s were applied for a total of 4500 cycles, and electrical potential-assisted DNA-RNA hybridization was used to hybridize the target miRNA to the capture probe. The hybridization solution contained 10 mM phosphate-buffered saline (PBS) and 1 M NaCl, pH 7.4. The electrode was then rinsed with RNase free water, and a potential pulse of −1 V was applied for 60 s to elute the DNA-miRNA hybrid into 50 μL of RNase-free water to complete the isolation of the target miRNA.
Electrochemical measurement
CV and electrochemical impedance spectroscopy (EIS) were performed with an electrochemical workstation (CHI600E, CH Instruments, Inc.). The three-electrode system consisting of a gold working electrode, a platinum wire auxiliary electrode and an Ag/AgCl (3 M KCl) reference electrode was used. The CV was carried out at a scan rate of 100 mV/s. The EIS was measured in potassium ferri/ferrocyanide solution (5.0 mM [Fe(CN)6]3−/4−, 0.1 M KCl), the voltage parameter was equilibrium potential, and the frequency was from 0.1 Hz to 100,000 Hz.
RT-qPCR and ASEA amplification
The miRNAs were reverse transcribed into complementary DNA (cDNA) according to the procedure described by Chen et al. [34]. Reactions of 7.5 μL were incubated in the CFX96 Connect™ Real-Time PCR System (Bio-Rad, CA, USA) for 30 min at 16 °C, 30 min at 42 °C, 5 min at 85 °C and then held at 4 °C. Real-time PCR was performed using a PerfectStart™ II Probe qPCR SuperMix. The fluorescence signal was detected by the CFX96 Connect™ Real-Time PCR System at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. ASEA amplification for 30-bp target was performed using the ASEA detection kits. The fluorescence signal was detected by the CFX96 Connect™ Real-Time PCR System at 40 cycles of 76 °C for 1 s and 60 °C for 1 s. Sequences of the primers and probes of RT-qPCR and ASEA are shown in Table S1.
Results and discussion
Scheme of the electrical potential-assisted hybridization to extract miRNA
The principle of the electrical potential-assisted hybridization for miRNA extraction is illustrated in Fig. 1. The construction of the miRNA extraction platform was based on electrical potential-assisted DNA/RNA hybridization. The three terminally modified sulfhydryl DNA capture probes were immobilized on the surface of the gold electrode by forming gold−sulfur bonds consisting of a highly reproducible recognition interface. In the presence of target miRNAs, hybridization between the DNA capture and miRNA target occurs, and acceleration of hybridization is achieved by electrical potential assistance. The extraction process only includes a binding step and isolation step. In the binding step, the repetitive electric fields of the two potential pulses were applied within 90 s to hybridize the target miRNA with the DNA capture. In the isolation step, the target miRNA was separated from the surface of the gold electrode by applying a continuous potential pulse of 60 s. The potential pulse condition was optimized, and the results are shown in Fig. S1.
Feasibility of electrical potential-assisted DNA-RNA hybridization to extract miRNA
To obtain insight into the interface properties of the electrode surface of electrodes, EIS was employed to test the detailed information of surface-modified impedances changes before and after DNA modification. In the Nyquist diagram, semicircle diameters represent the charge transfer resistance (Rct). Figure 2A shows the Nyquist diagram of gradual modification processes measured in PBS buffer (pH 7) containing 5 mmol/L [Fe(CN)6]3−/4− and 0.1 mol/L KCl. Compared with the bare Au electrode (curve a), the Rct of the Au electrode modified with capture probe (b) was obviously increased, which proved the successful modification of the capture probe. After the hybridization with 100 nM of the target sequence, there was an obvious improvement in the charge transfer rate over the electronic surface, confirmed by a smaller semicircle (curve c). The decrease in Rct after hybridization was attributed to the new conformation of the secondary structure of the double-layer [37, 38]. After applying a negative electric field, smaller semicircles were observed in the electrode surface (curve d) due to the breakage of the gold–sulfur bond for RNA recovery. The above EIS results suggest the feasibility of the proposed method for miRNA extraction. To further investigate the miRNA extraction process, RT-qPCR was chosen to determine the amount of miRNA (Fig. 2B). As expected, the fluorescence signal based on the electrical potential-assisted method for RNA extraction (a) was significantly increased compared with the no-template control (curve c). The samples eluted directly after 90 s passive hybridization did not exhibit any changes in fluorescence signal (curve b), indicating that the electrical potential-assisted DNA-RNA hybridization was able to effectively extract miRNA. In addition, this strategy relies on a specific complementary nucleic acid base to achieve hybridization and extraction of specific targets. Therefore, the specificity of the proposed miRNA extraction strategy was evaluated by extracting various sequences contained in the let-7 families and detecting using RT-qPCR. As shown in Fig. 2C, only let-7a was amplified, while non-target sequences had no signal during the amplification process, which proved that let-7a was successfully extracted, and indicated the high specificity of the strategy for let-7a extraction.
The feasibility of potential-assisted hybridization to extract miRNA. (A) Electrochemical impedance spectroscopy for the gold electrode. a = EIS of an unmodified gold electrode; b = EIS of a single-stranded (ss)DNA/MCH-modified gold electrode; c = 60 s electrical potential-assisted hybridization with let-7a; d = the gold electrode at a constant potential of −1 V for 60 s. (B) Amplification of let-7a. a = let-7a extracted by electrical potential-assisted hybridization; d = let-7a eluted directly after 90 s passive hybridization; c = no-template control. (C) RT-qPCR result corresponding to some nucleic acid sequences of let-7 family. a–d = let-7a, let-7b, let 7-c and let-7 i, respectively. e = no-template control
The volume of the hybridization system for the extraction of miRNA
The volume of hybridization buffer is crucial for the electrochemical performance response to the binding target. During the hybridization step, a known potential pulse was applied to the electrode. The larger the hybridization volume, the farther the edge of the solution is from the position where the electric field is applied, so that the influence of the electric potential becomes smaller [39]. Therefore, the parameter of reaction volume was optimized here. The different volumes of the hybridization system with the same number of 106 copies (Fig. 3A) or same concentration of 105 copies/μL (Fig. 3B) were prepared for electrical potential-assisted extraction. Subsequently, RT-qPCR was investigated for quantification. As shown in Fig. 3A, a lower volume of the hybridization system decreased the cycle threshold (Ct) value, suggesting higher efficiency. The difference in Ct value between different volumes was within one cycle, when the sample concentration was constant (Fig. 3B). Therefore, the difference in performance was related to the volume, and the extraction effect in 200 μL was the best. Consequently, the volume of the hybridization system was determined in 200 μL.
RT-qPCR of miRNA isolated from different sample volumes. (A) The different volumes of the hybrid system with 106 copies target. a–d = hybridization in 200 μL, 500 μL, 1 mL and 2 mL hybridization buffer, respectively, and e = no-template control. (B) The different volumes with the same concentration of 105 copies/μL target. a–d = hybridization in 200 μL, 500 μL, 1 mL and 2 mL hybridization buffer, respectively, and e = no-template control
Extraction of targets of different lengths using electrical potential-assisted hybridization
In order to evaluate the universal capability of the developed strategy for miRNA extraction, simulated samples with three lengths of 22 bp (Fig. 4a), 39 bp (Fig. 4b) and 60 bp (Fig. 4c) were extracted by the electrical potential-assisted method. The target concentrations were 103 copies/μL to 106 copies/μL, and they were quantified by RT-qPCR (22 bp) and ASEA (39 bp and 60 bp). Although there were some differences in the extraction effects of different concentrations and different lengths of samples, all the simulated samples were successfully extracted, which provided evidence for the universal application of the proposed strategy.
Extraction of the targets with length of (a) 22 bp, (b) 39 bp and (C) 60 bp using electrical potential-assisted hybridization. Relationship between the Ct values of RT-qPCR and the logarithmic values of the target concentration. Black lines represent the synthetic targets used directly for RT-qPCR; red lines represent the targets extracted by electrical potential-assisted hybridization before RT-PCR. Error bars show mean standard deviations of three determinations. The illustrations represent RT-qPCR quantitative values
The application under a biological environment
To access the ability of the proposed assay method for diagnostic purposes, miRNA samples of 108 copies/μL let-7a were extracted from different concentration of human serum based on the electrical potential-assisted method. The quality of miRNA was determined based on the Ct value obtained by RT-qPCR analysis. Although the Ct value gradually increased with the increase in serum concentration due to the interfering substances (Fig. 5B), the quantitative values of miRNA extracted by the electrical potential-assisted method were sufficient for RT-qPCR (Fig. 5C). In particular, at a high concentration of serum, there was still detectable signal appearing for the RT-qPCR (Fig. 5A), indicating that the potential assisted-based method provided a simple and effective tool for miRNA extraction in complex samples. The possible reasons for the inhibitory effect of serum were as follows: On the one hand, serum could not provide the most suitable ionic strength for the hybridization of capture DNA and target miRNA like a hybridization solution. On the other hand, complex biological components such as proteins in the serum migrated to the vicinity of the electrode under the influence of the electric field; the steric hindrance of nucleic acid hybridization was caused by these components. Therefore, the efficiency of nucleic acid hybridization was reduced.
The anti-interference ability of electrical potential-assisted hybridization to extract miRNA. (A) RT-PCR of different volume percentages of hybridization solution and interference serum. a = 100% hybridization buffer, b = 75% hybridization buffer + 25% serum, c = 50% hybridization buffer + 50% serum, d = 25% hybridization buffer + 75% serum, and e = 100% serum. f = no-template control. (B) The Ct value of RT-qPCR corresponding to different percentages of hybridization solution and interfering serum in the hybridization system. (C) The quantitative values of RT-qPCR. 1 = 100% hybridization buffer, 2 = 75% hybridization buffer + 25% serum, 3 = 50% hybridization buffer + 50% serum, 4 = 25% hybridization buffer + 75% serum, and 5 = 100% serum
Comparison of electrical potential-assisted extraction method with adsorption column and TRIzol method
To assess the performance of the proposed method for miRNA extraction, different copies of miRNA were extracted using the electrical potential-assisted method, adsorption column method and TRIzol method, and RT-qPCR analysis was performed. The operation steps of the adsorption column method and TRIzol method are shown in the supplementary material. The quality of RNA samples was determined by the Ct value based on RT-qPCR. As shown in Fig. 6, the amount of RNA strongly affected the efficiency of RT-qPCR. The Ct value of RNA samples increased with the decreasing concentration of RNA samples from 105 copies/μL to 10 copies/μL for the three methods. The RNA sample with lower concentration extracted using the electrical potential-assisted method showed a faster Ct value than that of the adsorption column and TRIzol methods, indicating higher efficiency for low-abundance RNA extraction. In particular, the acceptable Ct value obtained in 10 copies/μL of RNA sample with the electrical potential-assisted method is important for low-abundance miRNA-based diagnostics. These results suggest the universality of the new strategy based on the electrical potential-assisted method focusing on RNA extraction, which enhances the sensitivity of miRNA-based pathogenic diagnostics.
Comparison of electrical potential-assisted hybridization with adsorption column method and TRIzol method. The concentration of RNA samples ranged from 105 copies/μL to 10 copies/μL. The abscissa represents the logarithm of the target concentration, and the ordinate represents the average Ct value (a) and quantitative value (b) of RT-PCR. Each value represents 10 parallel experiments
As mentioned earlier, the TRIzol and adsorption column methods are the most commonly used for RNA extraction. Compared to time consumption of more than 90 min for the traditional TRIzol method and more than 60 min for the adsorption column method, the potential-assisted method needed only 3 min because of the electric field on the electrode-accelerated hybridization and desorption, which effectively protected RNA from enzymatic digestion (Table 1) [40, 41]. Moreover, the proposed method demonstrated good specificity and sensitivity due to the highly efficient hybridization reaction between the DNA recognition probe and RNA. These features will allow the method to be used for relative RNA quantification in diagnostics.
Versatility of miRNA extraction
Based on specific base complementation, the capture probe sequence can be replaced according to the sequence of the detection target, in order to realize the extraction of various miRNAs. miRNA 21 is considered a diagnostic and prognostic biomarker of various human malignancies [42]. Therefore, the capture probe was designed according to the miRNA 21 sequence. A concentration of miRNA 21 with 106 copies/μL was diluted in artificial serum, and then the extraction was assisted by electric potential. As shown in Fig. 7, miRNA 21 was successfully amplified by RT-qPCR, proving that miRNA 21 isolated by potential-assisted hybridization can be used for amplification detection. This demonstrates the miRNA extraction versatility of the proposed electrical potential-assisted extraction method.
Conclusion
An ultrafast electrical potential-assisted miRNA extraction scheme is reported here for the first time. This work has demonstrated that electrical potential-assisted DNA-RNA hybridization is capable of miRNA recovery within a shorter time than with commercial methods. miRNA extraction was achieved within 3 min, much shorter than that of traditional methods such as the adsorption column method (>60 min) or TRIzol method (>90 min). The method of electrical potential-assisted DNA-RNA hybridization for miRNA extraction was simple, rapid and efficient compared with traditional methods. The resulting preparation of miRNAs in serum was highly enriched in the target miRNA, supporting the applicability of the method in real samples, which is effectively ready for RT-qPCR analysis. It is worth mentioning that this system was able to detect miRNA in concentrations as low as 10 copies/μL combined with RT-qPCR, which is much lower than the conventional concentration of miRNA in cancer cells. The high extraction efficiency of low-concentration miRNA in this work will aid in accurate quantitation of low-abundance miRNAs, providing an effective tool in the development of miRNA extraction for clinical diagnosis.
Data availability
All data generated or analyzed during this study are included in this manuscript and its supplementary information files.
Abbreviations
- miRNAs:
-
microRNAs
- RT-qPCR:
-
Reverse transcription quantitative polymerase chain reaction
- ASEA:
-
Accelerated strand exchange amplification
- TCEP:
-
Tris(2-carboxyethyl)phosphine hydrochloride
- MCH:
-
6-mercaptohexanol
- CV:
-
Cyclic voltammetry
- EIS:
-
Electrochemical impedance spectroscopy
References
Ban E, Chae D-K, Yoo YS, Song EJ. An improvement of miRNA extraction efficiency in human plasma. Anal Bioanal Chem. 2017;409(27):6397–404. https://doi.org/10.1007/s00216-017-0580-7.
Gu J, Qiao Z, He X, Yu Y, Lei Y, Tang J, Shi H, He D, Wang K. Enzyme-free amplified detection of miRNA based on target-catalyzed hairpin assembly and DNA-stabilized fluorescent silver nanoclusters. Analyst. 2020;145(15):5194–9. https://doi.org/10.1039/D0AN00545B.
Liu H, Tian T, Ji D, Ren N, Ge S, Yan M, Yu J. A graphene-enhanced imaging of microRNA with enzyme-free signal amplification of catalyzed hairpin assembly in living cells. Biosens Bioelectron. 2016;85:909–14. https://doi.org/10.1016/j.bios.2016.06.015.
Lavaee P, Taghdisi SM, Abnous K, Danesh NM, Khayyat LH, Jalalian SH. Fluorescent sensor for detection of miR-141 based on target-induced fluorescence enhancement and PicoGreen. Talanta. 2019;202:349–53. https://doi.org/10.1016/j.talanta.2019.04.084.
Li D, Luo Z, An H, Yang E, Wu M, Huang Z, Duan Y. Poly-adenine regulated DNA density on AuNPs to construct efficient DNA walker for microRNA-21 detection. Talanta. 2020;217:121056. https://doi.org/10.1016/j.talanta.2020.121056.
Kim H, Kang S, Park KS, Park HG. Enzyme-free and label-free miRNA detection based on target-triggered catalytic hairpin assembly and fluorescence enhancement of DNA-silver nanoclusters. Sens Actuators B: Chem. 2018;260:140–5. https://doi.org/10.1016/j.snb.2017.12.137.
Cury JA, Koo H. Extraction and purification of total RNA from Sreptococcus mutans biofilms. Anal Biochem. 2007;365(2):208–14. https://doi.org/10.1016/j.ab.2007.03.021.
Burgess KA, Workman VL, Elsawy MA, Miller AF, Oceandy D, Saiani A. RNA extraction from self-assembling peptide hydrogels to allow qPCR analysis of encapsulated cells. PLoS One. 2018;13(6):e0197517. https://doi.org/10.1371/journal.pone.0197517.
Zhao F, Lee EY, Shin Y. Improved reversible cross-linking-based solid-phase RNA extraction for pathogen diagnostics. Anal Chem. 2018;90(3):1725–33. https://doi.org/10.1021/acs.analchem.7b03493.
Yang H, Liu J, Huang S, Guo T, Deng L, Hua W. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene. 2014;538(1):113–22. https://doi.org/10.1016/j.gene.2013.12.057.
Duy J, Koehler JW, Honko AN, Minogue TD. Optimized microRNA purification from TRIzol-treated plasma. BMC Genomics. 2015;16(1):95. https://doi.org/10.1186/s12864-015-1299-5.
Zhu C, Varona M, Anderson JL. Magnetic ionic liquids as solvents for RNA extraction and preservation. ACS Omega. 2020;5(19):11151–9. https://doi.org/10.1021/acsomega.0c01098.
Drula R, Ott LF, Berindan-Neagoe I, Pantel K, Calin GA. MicroRNAs from liquid biopsy derived extracellular vesicles: recent advances in detection and characterization methods. Cancers. 2020;12(8). https://doi.org/10.3390/cancers12082009.
LdS FA, Amorim RG, Scopel WL, Scheicher RH. Controlled current confinement in interfaced 2D nanosensor for electrical identification of DNA. Phys Chem Chem Phys. 2019;21(45):24884–90. https://doi.org/10.1039/c9cp03950c.
Jambrec D, Gebala M, La Mantia F, Schuhmann W. Potential-assisted DNA immobilization as a prerequisite for fast and controlled formation of DNA monolayers. Angew Chem Int Ed Engl. 2015;54(50):15064–8. https://doi.org/10.1002/anie.201506672.
Miranda-Castro R, Palchetti I, de-Los-Santos-Alvarez N (2020) The translational potential of electrochemical DNA-based liquid biopsy. Front Chem 8:143. https://doi.org/10.3389/fchem.2020.00143.
Sanchez-Salcedo R, Miranda-Castro R, de-Los-Santos-Alvarez N, Lobo-Castanon MJ (2021) Dual electrochemical genosensor for early diagnosis of prostate cancer through lncRNAs detection. Biosens Bioelectron 192:113520. https://doi.org/10.1016/j.bios.2021.113520.
Tymoczko J, Schuhmann W, Gebala M. Electrical potential-assisted DNA hybridization. How to mitigate electrostatics for surface DNA hybridization. ACS Appl Mater Interfaces. 2014;6(24):21851–8. https://doi.org/10.1021/am5027902.
Cai W, Peck JR, van der Weide DW, Hamers RJ. Direct electrical detection of hybridization at DNA-modified silicon surfaces. Biosens Bioelectron. 2004;19(9):1013–9. https://doi.org/10.1016/j.bios.2003.09.009.
Liu Q, Ma C, Liu X-P, Wei Y-P, Mao C-J, Zhu J-J. A novel electrochemiluminescence biosensor for the detection of microRNAs based on a DNA functionalized nitrogen doped carbon quantum dots as signal enhancers. Biosens Bioelectron. 2017;92:273–9. https://doi.org/10.1016/j.bios.2017.02.027.
Miao P, Wang B, Meng F, Yin J, Tang Y. Ultrasensitive detection of microRNA through rolling circle amplification on a DNA tetrahedron decorated electrode. Bioconjug Chem. 2015;26(3):602–7. https://doi.org/10.1021/acs.bioconjchem.5b00064.
Tavallaie R, McCarroll J, Le Grand M, Ariotti N, Schuhmann W, Bakker E, Tilley RD, Hibbert DB, Kavallaris M, Gooding JJ. Nucleic acid hybridization on an electrically reconfigurable network of gold-coated magnetic nanoparticles enables microRNA detection in blood. Nat Nanotechnol. 2018;13(11):1066–71. https://doi.org/10.1038/s41565-018-0232-x.
Rafiee-Pour HA, Behpour M, Keshavarz M. A novel label-free electrochemical miRNA biosensor using methylene blue as redox indicator: application to breast cancer biomarker miRNA-21. Biosens Bioelectron. 2016;77:202–7. https://doi.org/10.1016/j.bios.2015.09.025.
Dong J, Chen G, Wang W, Huang X, Peng H, Pu Q, Du F, Cui X, Deng Y, Tang Z. Colorimetric PCR-based microRNA detection method based on small organic dye and single enzyme. Anal Chem. 2018;90(12):7107–11. https://doi.org/10.1021/acs.analchem.8b01111.
Xu H, Zhang S, Ouyang C, Wang Z, Wu D, Liu Y, Jiang Y, Wu ZS. DNA nanostructures from palindromic rolling circle amplification for the fluorescent detection of cancer-related microRNAs. Talanta. 2019;192:175–81. https://doi.org/10.1016/j.talanta.2018.07.090.
Jimenez LA, Gionet-Gonzales MA, Sedano S, Carballo JG, Mendez Y, Zhong W. Extraction of microRNAs from biological matrices with titanium dioxide nanofibers. Anal Bioanal Chem. 2018;410(3):1053–60. https://doi.org/10.1007/s00216-017-0649-3.
Sosnowski RG, Tu E, Butler WF, O'Connell JP, Heller MJ. Rapid determination of single base mismatch mutations in DNA hybrids by direct electric field control. Proc Natl Acad Sci U S A. 1997;94(4):1119–23. https://doi.org/10.1073/pnas.94.4.1119.
Edman CF, Raymond DE, Wu DJ, Tu E, Sosnowski RG, Butler WF, Nerenberg M, Heller MJ. Electric field directed nucleic acid hybridization on microchips. Nucleic Acids Res. 1997;25(24):4907–14. https://doi.org/10.1093/nar/25.24.4907.
Fixe F, Branz HM, Louro N, Chu V, Prazeres DM, Conde JP. Electric-field assisted immobilization and hybridization of DNA oligomers on thin-film microchips. Nanotechnology. 2005;16(10):2061–71. https://doi.org/10.1088/0957-4484/16/10/014.
Yao M, Lv X, Deng Y, Rasheed M. Specific and simultaneous detection of micro RNA 21 and let-7a by rolling circle amplification combined with lateral flow strip. Anal Chim Acta. 2019;1055:115–25. https://doi.org/10.1016/j.aca.2018.12.040.
Wang ZH, Xu CJ. Research Progress of MicroRNA in early detection of ovarian Cancer. Chin Med J. 2015;128(24):3363–70. https://doi.org/10.4103/0366-6999.171459.
Johnson-Buck A, Su X, Giraldez MD, Zhao M, Tewari M, Walter NG. Kinetic fingerprinting to identify and count single nucleic acids. Nat Biotechnol. 2015;33(7):730–2. https://doi.org/10.1038/nbt.3246.
Daneshpour M, Karimi B, Omidfar K. Simultaneous detection of gastric cancer-involved miR-106a and let-7a through a dual-signal-marked electrochemical nanobiosensor. Biosens Bioelectron. 2018;109:197–205. https://doi.org/10.1016/j.bios.2018.03.022.
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. https://doi.org/10.1093/nar/gni178.
Zhang Z, Zhang L, Wang Y, Yao J, Wang T, Weng Z, Yang L, Xie G. Ultrasensitive electrochemical biosensor for attomolar level detection of let 7a based on toehold mediated strand displacement reaction circuits and molecular beacon mediated circular strand displacement polymerization. Anal Chim Acta. 2021;1147:108–15. https://doi.org/10.1016/j.aca.2020.12.057.
Li M, Liu M, Ma C, Shi C. Rapid DNA detection and one-step RNA detection catalyzed by Bst DNA polymerase and narrow-thermal-cycling. Analyst. 2020;145(15):5118–22. https://doi.org/10.1039/D0AN00975J.
Faria HAM, Zucolotto V. Label-free electrochemical DNA biosensor for zika virus identification. Biosens Bioelectron. 2019;131:149–55. https://doi.org/10.1016/j.bios.2019.02.018.
Rant U, Arinaga K, Tornow M, Kim YW, Netz RR, Fujita S, Yokoyama N, Abstreiter G. Dissimilar kinetic behavior of electrically manipulated single- and double-stranded DNA tethered to a gold surface. Biophys J. 2006;90(10):3666–71. https://doi.org/10.1529/biophysj.105.078857.
Deraney RN, Schneider L, Tripathi A. Synergistic use of electroosmotic flow and magnetic forces for nucleic acid extraction. Analyst. 2020;145(6):2412–9. https://doi.org/10.1039/c9an02191d.
Guo Y, Bosompem A, Zhong X, Clark T, Shyr Y, Kim AS. A comparison of microRNA sequencing reproducibility and noise reduction using mirVana and TRIzol isolation methods. Int J Comput Biol Drug Design. 2014;7(2-3):102–12. https://doi.org/10.1504/ijcbdd.2014.061642.
Kim Y-K, Yeo J, Kim B, Ha M, Kim VN. Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol Cell. 2012;46(6):893–5. https://doi.org/10.1016/j.molcel.2012.05.036.
Juan D, Alexe G, Antes T, Liu H, Madabhushi A, Delisi C, Ganesan S, Bhanot G, Liou LS. Identification of a microRNA panel for clear-cell kidney cancer. Urology. 2010;75(4):835–41. https://doi.org/10.1016/j.urology.2009.10.033.
Acknowledgements
The study was supported by grants from the National Key Research and Development Programs of China (2018YFE0113300), National Natural Science Foundation of China (81801264) and Key Project of Shandong Provincial Natural Science Foundation (ZR2020KH030).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The studies were approved by the appropriate ethics committee and were performed in accordance with ethical standards.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 429 kb)
Rights and permissions
About this article
Cite this article
Zhao, X., Li, Y., Sun, R. et al. Electrical potential-assisted DNA-RNA hybridization for rapid microRNA extraction. Anal Bioanal Chem 414, 3529–3539 (2022). https://doi.org/10.1007/s00216-022-03979-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-03979-8