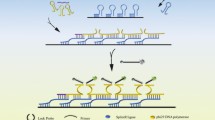
Abstract
A CRISPR/Cas12a-coupled multiplexed strand displacement amplification (CMSDA) for the detection of miR155 has been developed. Non-specific amplification was avoided by designing a single-stranded DNA template with a hairpin structure. The detection target miR155 was used as a primer to initiate a multiple-strand displacement reaction to produce abundant ssDNA. ssDNA was recognized by the Cas12a/CrRNA binary complex, activating the trans-cleaving activity of Cas12a. The multiple-strand displacement reaction is more efficiently detected compared with a single-strand displacement reaction. The detection range is from 250 pM to 1 nM, and the limit of the detection is 6.5 pM. The proposed method showed a good applicability in complex serum environments, indicating that the method has a broad prospect for disease detection and clinical application. In addition, we designed a dual-cavity PCR tube, which realized one-tube detection of miRNA155 and avoided open-cap contamination.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are a class of non-coding single-stranded RNA molecules of approximately 22 nucleotides in length encoded by endogenous genes [1]. miRNAs are involved in the regulation of gene expression at different time and space in the human body and are closely related to the occurrence of many human diseases. The function of miR155 is broad, as it is involved in several biological processes such as cell development and differentiation [2], inflammatory response [3, 4], and immune response [5], and presents high expression in various cancer tissues or cell lines such as liver cancer [6], pancreatic cancer [7], salivary gland adenoid cystic carcinoma [8], breast cancer [9], lymphoma, and lung cancer [10]. It has also been closely associated with other diseases such as multiple sclerosis [11] and leukemia [12]. With continuous research, miR155 has emerged as a tumor marker and a target for oncogene therapy and is considered one of the important blood biomarkers.
The impact of miR155 on human health and its importance in the field of medical detection has fostered the rapid development of miR155 detection methods. In recent years, miR155 detection methods have emerged, including electrochemical sensors [13, 14], fluorescent sensors [15, 16], nanomaterial luminescence [17, 18], nanoprobes [19], colorimetric methods [20, 21], surface-enhanced Raman spectroscopy [22, 23], and enzyme-catalyzed molecular reactions [24]. Among them, the fluorescence methods have the advantage of stability, convenience, and visualization. However, the current fluorescence detection methods have the defects of cumbersome steps, the introduction of additional nanomaterials, and the prolonged detection time, which restrict the detection effect and detection application scenarios of miR155 greatly and make the detection process more time-consuming and labor-intensive. Therefore, a more convenient, concise, sensitive, and time-efficient fluorescence detection method has become an urgent need for current miR155 detection.
Nowadays, the CRISPR/Cas systems and isothermal amplification technologies are becoming two new favorites in molecular detection. The programmability, the formal cleavage activity for target strands, and the trans-cleavage activity for non-target strands of the CRISPR/Cas system make it a convenient tool and with unlimited possibility for gene editing and molecular detection. In particular, Cas12a, Cas13a, and Cas14f, which possess excellent trans-cleavage activity, have been popularly employed in molecular detection. Isothermal amplification technology has the advantages of non-reliance on thermal cycling instruments, no need for high-temperature resistant enzymes, and mild reaction, and is comparable to traditional PCR methods in terms of detection performance. Therefore, the question of how to effectively integrate CRISPR/Cas technology and isothermal amplification technology to make them better applied to molecular detection has recently attracted extensive discussion and research by many researchers.
Zhou [25] fused Cas12a and rolling circling amplification to achieve ultrasensitive detection of miR155, but its steps were cumbersome. Huang [26] combined the advantages of RTF-EXPAR and CRISPR-Cas12a and greatly improved the sensitivity of SARS-CoV-2 RNA detection, but the different reaction temperatures of the enzymes (37 °C and 60 ℃) prevented them from satisfying one-tube detection. Zhang [27] accomplished the detection of multiple miRNAs in vivo using CRISR/Cas12a and loop-mediated isothermal amplification (LAMP), but with a high background signal. Xiong [28] used recombinase polymerase amplification and Cas12a to realize one-pot detection of viral DNA, but the CRISPR/Cas reaction system dripped onto the PCR tube lid was less stable and not suitable for large-volume reaction system detection. Therefore, the molecular detection system combining CRISPR/Cas12a and isothermal amplification technology has to be further explored in terms of stability, specificity, simplicity, and timeliness of detection.
Based on the above needs, in this work, we coupled multiplex strand displacement amplification (MSDA) with the CRISPR/Cas12a system to detect miR155. We used a DNA template to achieve multiplex signal amplification based on strand displacement reaction and designed a self-hairpin structure on the template to eliminate as much as possible the non-specific amplification caused by the complementation of other miRNAs with the DNA template in vivo. In the process, we identified a single-base difference in Cas12a/CrRNA recognizing non-completely complementary paired single-stranded DNA. We believe that this will provide a new idea for the application of the CRISPR/Cas12a system to single-base mutations of single-stranded nucleic acids. The MSDA is performed at 37 °C and the reaction of MSDA requires only one step of sample addition, which simplifies the reaction. However, due to the process of Cas12a/CrRNA recognition of the activation strand, the whole experiment is still divided into two steps, and the resulting operation of opening the cap of the tube twice is likely to cause cross-disturbance. To solve this problem, we designed a dual-cavity PCR tube inspired by the Chinese traditional yin-yang jug, which successfully realized the spatial separation of the CRISPR/Cas12a reaction from the MSDA reaction in a single tube, which truly enables a one-tube detection while avoiding open-cap contamination.
Material
Reagents
Klenow Fragment (3′– > 5′ exo-), Nb.BbvCI, and EnGen ® Lba Cas12a (Cpf1) were available from New England Biolabs (New England, USA). Deoxynucleotide triphosphates (dNTPs), DNA marker, DNA loading buffer (6 ×), Gel Red nucleic acid dye, Water-DEPC Treated Water, TE-buffer, Acryl/Bis 30% Solution (29:1), and TBE Buffer (5 ×) were purchased from Sangon Biotech Inc. (Shanghai, China). Human serum was purchased from Solarbio (Beijing, China). The oligonucleotides used in this work were synthesized by Sangon Biotech Inc. (Shanghai, China) with HPLC purification (listed in Table 1). Fluorescence signals and gel images were performed on a fluorescence spectrophotometer (PerkinElmer LS-55) and an electrophoresis instrument (YENA UV solo Imager), respectively.
Multiplexed strand displacement amplification system
A total of 20 µl reaction system containing 2 µL template (200 nM), 2 µL miRNA 155 in different concentrations, 2 µL Klenow Fragment (1 U), 2 µL 10 × NEBuffer 2 (500 mM NaCl,100 mM Tris–HCl, 100 mM MgCl2, 10 mM DTT (pH 7.9 @ 25 °C)), 2 µL Nb.BbvCI (1 U), 2 µL 10 × NEB rCutSmart buffer (500 mM potassium acetate, 200 mM Tris–acetate, 100 mM magnesium acetate, 1000 µg/ml recombinant albumin, (pH 7.9 @ 25 ℃)), and 7 µL TE buffer were reacted at 37 ℃ for 2 h.
CRISPR/Ca12a reaction system and fluorescence detection
Nine microliters of TE buffer, 2 μL Cas12a (1U), 2 μL 10 × NEBuffer 2.1 (100 mM Tris − HCl + 500 mM NaCl + 1000 μg/mL BSA + 100 mM MgCl2, pH 7.9), 2 μL CrRNA (200 nM), 4 μL SDA product, and 1 μL FQ reporter (10 μM) reacted at 37 ℃ for 35 min. We added 80 μL ddH2O to the above 20-μL solution and mixed and then recorded the fluorescence signal by a fluorescence spectrometer (PerkinElmer) at 490 nm excitation wavelength.
PAGE analysis
A 10% polyacrylamide gel was used to analyze amplification products. After electrophoresis in 1 × TBE buffer (2 mM EDTA and 89 mM Tris-boric acid, pH 8.3) for 1 h, images were collected by a gel electrophoresis analyzer.
Results and discussion
The principle of CMSDA for miRNA155 detection
The current strand displacement and exponential amplification templates are mostly 40–100 nt single-stranded nucleic acids, and excessively long single-stranded templates may cause non-specific amplification of non-target RNAs in vivo. To solve this problem and achieve specific amplification of miR155, we designed a single-stranded DNA T with a self-hairpin structure as the amplification template (Scheme 1A). The end of the T is a toehold sequence complemented by miRNA155, and the middle sequence of T is a 20 nt CrRNA recognition sequence in which the hairpin structure is contained. The hairpin structure consists of seven complementary base pairs (stem) and one single base (loop), which can effectively close the T and eliminate non-specific amplification caused by other DNA/RNA in real serum samples. In addition, two Nb.BbvcI digest site sequences were designed between the toehold sequence and the CrRNA recognition sequence.
According to the kinetic analysis, miR155 preferentially pairs with the 3′ end of T to form an RNA/DNA hybrid duplex (Scheme 1B). In the presence of KF, miR155 acts as a primer to sequentially amplify the enzyme cut site sequence, the CrRNA recognition sequence (P2), and the DNA sequence (P1) whose sequence is the same as the miR155 (U > T). Subsequently, Nb.BbvcI cuts an inscribed gap at the enzyme cut site, and then KF continues to amplify the new sequence at the gap and displaces single-stranded P1 and P2, forming the reaction strand H1 and the first signal amplification.
Since the P1 sequence is identical to the miR155 sequence (U > T), P1 also serve as a primer to trigger a second round of amplification by KF and Nb.BbvcI. This process forms H2 and is identical to the first round of amplification. Since P2 is fully complementary to the CrRNA recognition sequence (Blue), P2 likewise acts as a primer to unlock the hairpin of T1 and initiates a third round of strand displacement reaction under the action of the two enzymes, a process that forms H3 and releases the new P1. the above three cycles ensure the production of large amounts of P1 and P2.
In addition, since the 3′ end of H3 is exposed, P1 can pair with the 3′ end of H3 to form H2 again and start the fourth cycle. The P2 released during the whole process would be recognized by CrRNA to activate the trans-cleavage activity of Cas12a to perform random cleavage to the FQ probe, releasing a strong fluorescent signal. In the absence of miR155, strand displacement cannot proceed, no P2 is produced, Cas12a cannot be activated, and no fluorescent signal is generated. The whole process is divided into two steps at 37 ℃: multiplexed strand displacement amplification and Cas12a recognition.
Feasibility exploration
The clear bands in lanes 1 and 2 correspond to 23 bp of miR155 and 73 bp of template T, respectively (Fig. 1A). Lane 3 is a blank set containing only template T and the enzyme reaction system, so the lane of the blank set shows only the band corresponding to T. In contrast, compared with lane 3, lane 4 showed two additional bands in addition to the T band. They corresponded to P1 (25 bp) and P2 (27 bP) which were displaced after KF amplification and Nb.BbvCI cleavage, respectively. The PAGE results indicated that MSDA was carried out successfully. The successful coupling of Cas12a and MSDA is the key step to generating the signal. As shown in Fig. 1B, under UV light irradiation, no fluorescent signal was observed in the control group by the naked eye, while the experimental group showed a strong fluorescent signal. Further analysis by a fluorometer showed a fluorescence value of 128 in the control group and that of 820 in the experimental group. The result demonstrated that CMSDA for miR155 detection is fully feasible.
Optimization of experimental conditions
The study showed that CrRNA with more than 7 bases pair to the 3′ end of ssDNA may activate the activity of Cas12a, and the mismatching effect at the near PAM site (5′) end is greater than that at the 3′ end. The self-hairpin structure makes T 13 bases pair to CrRNA, and there is a theoretical risk of signal leakage due to the direct activation of Cas12a/CrRNA by template T. Therefore, the sequence of T was first explored. We designed two templates T1 and T2 with the same hairpin structure. The difference is that compared to T2, an extra C base was added between the recognition sequence of CrRNA of T1 and the P2 sequence. We simulated the pairing of CrRNA with T1 and T2, respectively. As shown in Fig. 2A, in addition to the 13 bases at the hairpin structure, there are three bases complementary to CrRNA at the 5′ end of T2, while for T1, only 13 bases at the hairpin are complementary to CrRNA. The efficiency of the recognition and cleavage of Cas12a to T1 and T2 was compared by fluorescence spectroscopy (Fig. 2B). After 30 min shearing, the fluorescence signal of T2 was about eight times that of T1, and almost no background signal was generated by T1. This indicated that T2 successfully activated the trans-activity of Cas12a, while T1 did not. This result also indicated that pairing near the 5′ end has an effect on the recognition of single-stranded recognition sites by CrRNA. Thus, T1 was selected for subsequent experiments.
Annealing affected the pairing effect of miR155 with the template in the system, and we further explored the effect of annealing on the results. In Fig. 2C, the annealed fluorescence results were less than half of the unannealed fluorescence values (F = the fluorescence value of miR155, F0 = the fluorescence value of the blank group). We analyzed that annealing may cause dimer formation between the single-stranded templates themselves, which would hinder the polymerization process of KF. The unannealed results perfectly satisfied the assay requirements and also made the experiment free of the dependence of the PCR instrument so that the whole experiment could be completed at 37 °C. Finally, the template concentration (Fig. 2D), amplification time (Fig. 2E), and the reaction time of Cas12a (Fig. 2F) were optimized. The optimal results were 200 nM, 2 h, and 35 min, respectively.
Sensitivity exploration
The sensitivity of the sensor was investigated under the optimal conditions described above. Different concentrations of miR155 from 250 pM to 1 nM were detected. Figure 3A shows that the fluorescence signals of miR155 were all higher than those of the control group, and the signals increased with the increase of miR155 concentration. The fluorescence signal of miRNA155 from 250 pM to 1 nM showed a faster upward trend and the fluorescence values from 250 pM to 1 nM were linearly fitted as shown in Fig. 3B. The data showed a good linearity with a linear equation of F–F0 = 16.93lgC + 60.03 (C is the concentration of miR155, and R is the correlation coefficient, R2 = 0.971). The limit of detection (LOD) is 6.5 pM calculated based on 3 σ/K (σ is the standard deviation value of the blank, and K is the slope of the standard curve). Compared CMSDA with other methods for miR155 detection (Table 2), CMSDA showed good advantages in the limit of detection and the detection step [29,30,31,32].
Selectivity and repeatability
Specificity and stability are important indicators to evaluate the sensor. We first examined 5 RNAs at 500 pM to assess the resistance of the fluorescent sensor. Among 5 RNAs (Fig. 4A), the fluorescent signals of miR15, miR21, miR141, and miRLet 7f were slightly higher than those of the blank group, whereas the signal of miR21 was the highest, only 80 higher than the blank group. The signal of miR16 was even lower than the blank group, which was much lower than the signal of miR155, demonstrating the excellent selectivity of the sensor. Next, five groups of parallel samples were tested using 500 pM miR155 (Fig. 4B), the signals of all five experimental groups were in the range of 590–610, the signals of all five control groups were in the range of 85–120, and the differences of F–F0 of the five groups were stable in the range of 490–510. The result demonstrated that the fluorescence sensor has a good reproducibility and stability.
Real sample detection
The serum supplementation of miRNA155 at 250 pM, 400 pM, and 500 pM was performed with human serum diluted ten times. As shown in Table 3, the detected results for the concentrations of 250 pM, 400 pM, and 500 pM were 247.25 pM, 427.28 pM, and 519.00 pM, with recoveries of 98.90%, 106.82%, and 103.80%, respectively. The results showed that the method had a good ability of application to complex samples.
The design of the dual-cavity tube
Despite the simplicity of the amplification step, the whole reaction system is still divided into two processes including MSDA and Cas12a activation. The additional step of the Cas12a system not only adds to the tediousness, but the possible cross-contamination caused by the open-cap operation is an issue of concern. To implement the one-step method, the reaction order of the two systems in time should be considered first as the Cas12a system can only be introduced after the end of MSDA. This requires that the two systems are separated from each other spatially even in a single tube so that they do not interfere with each other during amplification. In order to achieve a one-tube detection to avoid contamination caused by opening the cap, it is also necessary to ensure that the two systems can finally be mixed without opening the cap after the amplification is completed.
To achieve the above two conditions simultaneously, thanks to the inspiration of the dual-cavity wine jug in traditional Chinese culture, we designed a dual-cavity PCR tube, which successfully realized the one-step method and one-tube detection of miR155. As shown in Fig. 5A, B, we designed a 0.8-mm-thick section in the conventional PCR tube, which can spatially divide the tube into two spaces on the left and right. The height of the upper end of the section from the surface of the tube is 4 mm. In order to verify the performance of the dual-cavity tube in separating the liquid, we added 20 uL of the blue staining solution to the left and right spaces of the tube for observation. As shown in Fig. 5C, the liquid was successfully separated when the tube was upright. Considering that the tube turning sideways during the assay might cause premature mixing of the two systems, we inverted the tube by 180°. The results showed (Fig. 5D) that the liquids in the two chambers were still separated from each other, which was due to the dense reaction system having a viscous force with the tube wall greater than its own gravity. This showed that the dual-cavity tube separated the different reaction systems very well, with excellent resistance to vibration and side-turning, which was more advantageous compared to the method of dropping the Cas12a system inside the PCR lid [28].
Next, we explored whether the dual-cavity tube could achieve an effective fusion of the two reaction systems. We manually flung the two chambers of liquid from the bottom to the top simultaneously to mix the two well first and then to one side for reaction. As shown in Fig. 5E, the liquids in the two chambers were successfully mixed in one chamber after two to three flings. This proved that the dual-cavity tube could be successfully applied for the separation and mixing of different systems. We added the MSDA system and Cas12a recognition system into two lumens of the dual-cavity tube for miR155 detection, respectively. In Fig. 5F, the two systems in a were not manually mixed, while the b was. The Cas12a systems in the a and b lumens were aspirated for detection. As shown in Fig. 5F, no fluorescence signal was generated in a, and b showed intense fluorescence. This indicated that with the help of the dual-cavity tube, the CMSDA truly achieved one-step and one-tube detection for miR155. This demonstrated that dual-cavity tubes successfully reduced the testing steps and solved the problem of cross-contamination, which greatly contributed to the level of molecular testing and promoted the practical application of clinical research.
Conclusion
In this study, we developed a multiple-strand displacement amplification coupled with CRISPR/Cas12a system for miR155 detection. The sensitivity and selectivity were improved by the single-stranded DNA template with a hairpin structure. The ssDNA released from the first strand displacement reaction triggered multiple rounds of strand displacement reactions and finally activated the trans-cutting activity of Cas12a, which efficiently amplified the biological signal. Therefore, CMSDA has a good detection performance with a limit of detection of 6.5 pM. The dual-lumen PCR tube allows for complete separation of the Cas12a system from the MSDA system in a single tube, which truly enables one-tube detection and avoids open-cap contamination. In this process, we also discovered the effect of single-base differences of ssDNA on the activation of Cas12a activity, which provided a new idea for the application of the CRISPR/Cas12a system to identify single-base mutations in single-stranded DNA. Finally, based on the programmability of the CRISPR/Cas12a system, the method is expected to be used for the detection of other miRNAs.
Data Availability
The data that support the findings of this study are available from the corresponding author, Changjun Hou, upon reasonable request.
References
Dong HF, Lei JP, Ding L, Wen YQ, Ju HX, Zhang XJ (2013) MicroRNA: function, detection, and bioanalysis. Chem Rev 113(8):6207–6233
Kempinska-Podhorodecka A, Milkiewicz M, Wasik U, Ligocka J, Zawadzki M, Krawczyk M, Milkiewicz P (2017) Decreased expression of vitamin D receptor affects an immune response in primary biliary cholangitis via the VDR-miRNA155-SOCS1 pathway. Int J Mol Sci 18(2):289
O’Rourke M, Trenkmann M, Connolly M, Fearon U, Murphy CC (2019) Novel gene targets for miRNA146a and miRNA155 in anterior uveitis. Brit J Ophthalmol 103(2):279–285
Chen DF, Gong BD, Xie Q, Ben QW, Liu J, Yuan YZ (2010) MicroRNA155 is induced in activated CD4(+) T cells of TNBS-induced colitis in mice. World J Gastroenterol 16(7):854–861
Solomou EE, Kalyvioti E, Verigou E, Katsanaki K, Gogos C, Hahalis G, Alexopoulos D (2016) platelet-induced T cell activation in patients with myocardial infarction; the role of miR155. Blood 128(22):4913
Ezzat WM, Amr KS, Raouf HA, Elhosary YA, Hegazy AE, Fahim HH, Kamel RR (2016) Relationship between serum microRNA155 and telomerase expression in hepatocellular carcinoma. Arch Med Res 47(5):349–355
Novais PC, Moscatel MB, Rosa MS, Tirapelli D, Cirino ML, Kemp R, Santos JS (2022) Expression profile of miR-148a and miR-155 associated with apoptosis mechanism in pancreatic cancer. Faseb J 36(S1):L8056
Liu L, Hu YJ, Fu JY, Yang XH, Zhang ZY (2013) MicroRNA155 in the growth and invasion of salivary adenoid cystic carcinoma. J Oral Pathol Med 42(2):140–147
Roth C, Rack B, Muller V, Janni W, Pantel K, Schwarzenbach H (2010) Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res 12(6):R90
Ren SX, Rivard CJ, Yu H, Genova C, Rozenboom L, Gao DX, Hinz TK, Rikke BA, Wynes MW, Caldwell C, Agustoni F, Suda K, Jiang T, Zhou CC, Heasley LE, Hirsch FR (2018) A miRNA panel predicts sensitivity of FGFR inhibitor in lung cancer cell lines. Clin Lung Cancer 19(5):450–456
Shademan B, Nourazarian A, Nikanfar M, Avci CB, Hasanpour M, Isazadeh A, (n.d.), (2020) Investigation of the miRNA146a and miRNA155 gene expression levels in patients with multiple sclerosis. J Clin Neurosci 78:189–193
Schneider E, Staffas A, Rohner L, Malmberg ED, Ashouri A, Krowiorz K, Pochert N, Miller C, Wei SY, Arabanian L, Buske C, Dohner H, Bullinger L, Fogelstrand L, Heuser M, Dohner K, Xiang P, Ruschmann J, Petriv OI, Heravi-Moussavi A, Hansen CL, Hirst M, Humphries RK, Rouhi A, Palmqvist L, Kuchenbauer F (2018) Micro-ribonucleic acid-155 is a direct target of Meis1, but not a driver in acute myeloid leukemia. Haematologica 103(2):246–255
Fu P, Xing S, Xu MJ, Zhao Y, Zhao C (2020) Peptide nucleic acid-based electrochemical biosensor for simultaneous detection of multiple microRNAs from cancer cells with catalytic hairpin assembly amplification. Sensor Actuat B-Chem 305:127545
Shen ZP, He LY, Wang WH, Tan L, Gan N (2020) Highly sensitive and simultaneous detection of microRNAs in serum using stir-bar assisted magnetic DNA nanospheres-encoded probes. Biosens Bioelectron 148:111831
Nemati F, Hosseini M (2022) Fluorescence turn-on detection of miRNA-155 based on hybrid Ce-MOF/PtNPs/graphene oxide serving as fluorescence quencher. J Photoch Photobio A 429:113943
Shahsavar K, Shokri E, Hosseini M (2020) A fluorescence-readout method for miRNA-155 detection with double-hairpin molecular beacon based on quadruplex DNA structure. Microchem J 158:105277
Huang R, Liao YH, Zhou XM, Fu Y, Xing D (2017) Multiplexed detection of microRNA biomarkers from tumor cells and tissues with a homogeneous nano-photon switch. Sensor Actuat B-Chem 247:505–513
Lu YY, Wang L, Chen HQ (2019) Turn-on detection of MicroRNA155 based on simple UCNPs-DNA-AuNPs luminescence energy transfer probe and duplex-specific nuclease signal amplification. Spectrochim Acta A 223:117345
Wu Y, Cheng H, Zhu M, Zhang L, Mao Z, Wang C, Liu Z (2023) Monitoring subtle changes of blood-brain barrier permeability via detection of MiRNA-155 in brain microvasculature. ACS Appl Mater Interfaces 15(18):21893–21903
Shahsavar K, Shokri E, Hosseini M (2022) Sensitive colorimetric detection of miRNA-155 via G-quadruplex DNAzyme decorated spherical nucleic acid. Microchim Acta 189(9):357
Liu L, Zhu SY, Wei YM, Liu XL, Jiao SL, Yang JJ (2019) Ultrasensitive detection of miRNA-155 based on controlled fabrication of AuNPs@MoS2 nanostructures by atomic layer deposition. Biosens Bioelectron 144:111660
Wu CJ, Huang SQ, Wang YY, Chai YQ, Yuan R, Yang X (2021) DNA structure-stabilized liquid-liquid self-assembled ordered Au nanoparticle interface for sensitive detection of MiRNA 155. Anal Chem 93(31):11019–11024
He Y, Yang X, Yuan R, Chai YQ (2017) “Off” to “on” surface-enhanced raman spectroscopy platform with padlock probe-based exponential rolling circle amplification for ultrasensitive detection of microRNA 155. Anal Chem 89(5):2866–2872
Fu P, Xu MJ, Xing S, Zhao Y, Zhao C (2021) Dual cascade isothermal amplification reaction based glucometer sensors for point-of-care diagnostics of cancer-related microRNAs. Analyst 146(10):3242–3250
Zhou S, Sun H, Dong J, Lu P, Deng L, Liu Y, Yang M, Huo D, Hou C (2023) Highly sensitive and facile microRNA detection based on target triggered exponential rolling-circle amplification coupling with CRISPR/Cas12a. Anal Chim Acta 1265:341278
Hang XM, Wang HY, Liu PF, Zhao KR, Wang L (2022) Cas12a-assisted RTF-EXPAR for accurate, rapid and simple detection of SARS-CoV-2 RNA. Biosens Bioelectron 216:114683
Zhang M, Wang HH, Wang H, Wang FF, Li ZP (2021) CRISPR/Cas12a-assisted ligation-initiated loop-mediated isothermal amplification (CAL-LAMP) for highly specific detection of microRNAs. Anal Chem 93(22):7942–7948
Xiong YF, Cao GH, Chen XL, Yang J, Shi MM, Wang Y, Nie FP, Huo DQ, Hou CJ (2022) One-pot platform for rapid detecting virus utilizing recombinase polymerase amplification and CRISPR/Cas12a. Appl Microbiol Biot 106(12):4607–4616
Li PX, Wei M, Zhang F, Su J, Wei W, Zhang YJ, Liu SQ (2018) A novel fluorescence switch for MicroRNA imaging in living cells based on DNAzyme amplification strategy. ACS Appl Mater Interfaces 10(50):43405–43410
Yu JY, Liu QY, Qi LJ, Fang Q, Shang XD, Zhang XJ, Du Y (2024) Fluorophore and nanozyme-functionalized DNA walking: a dual-mode DNA logic biocomputing platform for microRNA sensing in clinical samples. Biosens Bioelectron 252:116137
Lu YY, Wang L, Chen HQ (2019) Turn-on detection of MicroRNA155 based on simple UCNPs-DNA-AuNPs luminescence energy transfer probe and duplex-specific nuclease signal amplification. Spectrochim Acta A: Mol Biomol Spectrosc 223(12):117345
Zhong Y, Li B, Xin H, Wang CY (2024) Endogenous mRNA-driven “one-to-more” signal amplification of DNA probe for intracellular miR155 sensing. Chem Asian J e202400401
Acknowledgements
This work was Supported by the Open Research Fund of State Key Laboratory of Digital Medical Engineering (2023-K08), Chongqing Natural Science Foundation (CSTB2022NSCO-BHX0727), Fundamental Research Founds for the Central Universities (2022CDJYGRH-013), and the sharing fund of Chongqing University’s large equipment.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical approval
This research did not involve human or animal samples.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, X., Deng, L., Zhou, S. et al. CRISPR/Cas12a-coupled multiplexed strand displacement amplification for miRNA155 one-tube detection: via a dual-cavity PCR tube. Microchim Acta 191, 470 (2024). https://doi.org/10.1007/s00604-024-06538-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-024-06538-3