Abstract
A novel chemolithoautotrophic bacterium, strain H1576T, was isolated from water of a brackish lake. Strain H1576T grew aerobically on inorganic sulfur compounds. Hydrogen gas did not support autotrophic growth, and heterotrophic growth was not observed. Cells were rod shaped, motile, 1.5–2.7 μm in length and 0.6–0.7 μm in width. Growth was observed at 3–22 °C with an optimum growth temperature of 13–15 °C. The pH range for growth was 6.0–7.4 with an optimum pH of 6.6–6.8. Major fatty acids were summed feature 3 (C16: 1ω7c and/or C16: 1ω6c). The complete genome of strain H1576T consists of a circular chromosome and a plasmid, with total length of 2.8 Mbp and G+C content of 46.4 mol%. Phylogenetic analyses indicated that strain H1576T belongs to the genus Sulfurimonas but distinct from representatives of existing species. On the basis of genomic and phenotypic characteristics, a new species named Sulfurimonas aquatica sp. nov. is proposed with the type strain of strain H1576T (= BCRC 81254T = JCM 35004T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to the List of Prokaryotic Names with Standing in Nomenclature, LPSN (Parte et al. 2020), the genus Sulfurimonas belongs to the family Helicobacteraceae and currently includes eight species with validly published names (as of 26 July 2022). They grow chemolithoautotrophically by oxidizing inorganic sulfur compounds, with oxygen as electron acceptor. In some species, anaerobic growth and H2 gas oxidation are observed. As chemotaxonomic feature, they share major fatty acids of C16: 1, C18: 1 and C16: 0. Besides these eight species, three other species and two Candidatus species have been proposed in this genus, on the basis of genomic and phenotypic characterizations of isolated strains (Table 1).
As reviewed previously (Han and Perner 2015), members of the genus Sulfurimonas have been repeatedly detected by 16S rRNA gene sequence analysis, in various ecosystems represented by hydrothermal vents, marine sediments and water columns. In addition, Sulfurimonas is known to be a dominating bacterial genus in some engineered microbial systems, as shown in recent studies employing 16S rRNA gene amplicon sequencing (Fu et al. 2020; Wu et al. 2020; Haosagul et al. 2021). With the same approach, a dominance of Sulfurimonas species at specific water depths of a stratified brackish lake was recently reported (Watanabe et al. 2022). This shallow eutrophic lake, Lake Harutori in Japan, is characterized by steep chemocline and high concentration of sulfide in bottom water (Kubo et al. 2014; Watanabe et al. 2022). In this study, a novel sulfur-oxidizing autotroph was isolated from anoxic water of Lake Harutori, and characterized as a representative of a new species in the genus Sulfurimonas.
Materials and methods
Sampling of water from Lake Harutori was conducted on 16 Feb 2016. A sample of anoxic bottom water was collected from 5 m depth, at a site where previous studies were conducted (Kubo et al. 2014; Watanabe et al. 2022). A portion of the sample (0.3 ml) was inoculated into 30 ml of a medium for aerobic thiosulfate oxidizers. The medium (hereafter referred to as basal medium) was prepared as described below. First, the following salts (g l−1) were dissolved in distilled water and then sterilized by autoclaving: NaCl (20), Na2S2O3·5H2O (5), MgCl2·6H2O (3), MgSO4·7H2O (0.3), CaCl2·2H2O (0.1), NH4Cl (0.1), KH2PO4 (0.1) and KCl (0.1). To the autoclaved and cooled salt solution, the following stock solutions (ml l−1) were aseptically added: trace element solution (1), selenite-tungstate solution (1), vitamin mixture solution (1) and 1 M NaHCO3 solution (30). The vitamin mixture solution consisted of the followings (mg l−1): biotin (20), folic acid (20), pyridoxine–HCl (100), thiamine–HCl・2H2O (50), riboflavin (50), nicotinic acid (50), calcium d( +) pantothenate (50), 4-Aminobenzoic acid (50), lipoic acid (50) and cyanocobalamine (1). The other stock solutions were prepared as described previously (Widdel and Bak 1992). Finally, pH of the medium was adjusted to 7.0–7.2 with HCl. From the enrichment culture established, pure culture of strain H1576T was obtained by repeated serial dilution with the basal medium. The enrichment and isolation were performed at 15 °C in the dark.
Phenotypic characteristics of strain H1576T were investigate by culturing the strain at 15 °C in the basal medium, unless otherwise specified. Cell morphology was observed with phase-contrast light microscopy, and Gram stain test was conducted with a kit (Fluka). Cellular fatty acid profile was obtained with the Sherlock Microbial Identification System (MIDI) version 6.0 (database; TSBA6).
To determine upper and lower limits of temperature for growth, strain H1576T was inoculated into the basal medium and incubated at 0, 3, 5, 8, 13, 15, 18, 22, 25, 28, 30 and 32 °C. Effect of NaCl concentration on growth was examined by using media modified from the basal medium, with lowered concentration of MgCl2·6H2O (0.2 g l−1) and varying concentrations of NaCl (0.0, 0.5, 1.0, 2.0, 3.0, 4.0, 5.0% and 6.0 w/v). Effect of pH on growth was tested with media of various pH which were prepared as below. The media commonly contained the following constituents (l− 1): 20 g NaCl, 5 g Na2S2O3·5H2O, 1 g NaHCO3, 0.2 g MgCl2·6H2O, 0.1 g CaCl2·2H2O, 0.1 g NH4Cl, 0.1 g KH2PO4, 0.1 g KCl, 1 ml trace element solution, 1 ml selenite-tungstate solution and 1 ml vitamin mixture solution. Each medium of varying pH contained one of buffering reagents listed below (at a final concentration of 20 mM), along with NaOH for pH adjustment. Tested pH and buffering reagents were as follows; pH 5.8, 5.9, 6.0, 6.1, 6.2, 6.3, 6.4, 6.5, 6.7, 6.8, 6.9, 7.1, 7.2, 7.4 and 7.7 with MES; pH 6.6, 6.9 and 7.2 with PIPES; pH 7.0, 7.2, 7.3 and 7.6 with MOPS. All ingredients were mixed and then sterilized by filtration.
Utilization of electron donors was tested with the basal medium, by replacing thiosulfate with one of the followings (mM); sulfide (2), pyruvate (5), lactate (5), acetate (5), propionate (2.5), succinate (2.5), fumarate (2.5), malate (2.5), butyrate (2.5), benzoate (2.5), isobutyrate (2.5), methanol (5), ethanol (2.5), formate (5), citrate (5), glucose (2.5), xylose (2.5), phenol (2), m-cresol (1). As insoluble substrates, elemental sulfur (0.5 g l−1) and hydrogen gas (air/H2; 2: 1, v/v; 150 kPa total pressure) were also tested with the thiosulfate-free basal medium. Utilization of electron acceptors was tested with the basal medium supplemented with nitrite (2 mM) or nitrate (5, 10 mM), under atmosphere of N2 and CO2 (80% and 20% in volume, respectively).
The novel isolate was subjected to whole genome sequencing, with the PacBio RS II platform. From linear contigs obtained, circular chromosome and plasmid were manually reconstructed based on sequence alignment. The resulting complete genome sequence was subjected to comparative analysis with the closest relatives, by the TYGS web server (https://tygs.dsmz.de). In the TYGS, the Type (Strain) Genome Server, relatives of the subjected genome were automatically identified for subsequent genome-based phylogenetic analysis and calculation of digital DNA-DNA hybridization (dDDH) values (Meier-Kolthoff and Göker 2019). Phylogenetic analysis was also conducted with the 16S rRNA gene identified in the genome, by using MEGA version 11 (Tamura et al. 2021). The reference sequences of Sulfurimonas species were retrieved from LPSN (accessed on 06 July 2022). The sequences of strain H1576T and references were aligned with the MUSCLE algorithm. As an outgroup, Sulfuricurvum kujiense YK-1T was included in the alignment. The best substitution model with the lowest Bayesian Information Criterion score was selected by the model selection tool in MEGA. Phylogenic tree was constructed with the selected model by excluding positions with gaps. Values of average nucleotide identity (ANI) between strain H1576T and type strains of Sulfurimonas species were computed by ANI calculator available in the EzBioCloud, based on the OrthoANIu algorithm (Yoon et al. 2017).
Results
Cells of the novel isolate, strain H1576T, were Gram stain negative, motile, rod shaped, 1.5–2.7 μm in width, 0.6–0.7 μm in length. The strain grew at 3–22 °C with optimum growth at 13–15 °C. At 15 °C, growth was observed at pH range of 6.0–7.4, with optimum growth at pH of 6.6–6.8. Growth was observed in the presence of 2–5% (w/v) NaCl. The cellular fatty acid profile of strain H1576T is shown in Table S1. In the profile, summed feature 3 (C16: 1ω7c and/or C16: 1 ω6c) and C16: 0 were predominant, accounting for 65.5% and 21.9%, respectively.
Chemolithoautotrophic growth of strain H1576T was supported by thiosulfate, sulfide and elemental sulfur, but not by H2 gas. None of the tested organic substrate supported aerobic growth of the strain. As sole electron acceptor for thiosulfate oxidation, nitrate and nitrite did not support anaerobic growth of strain H1576T.
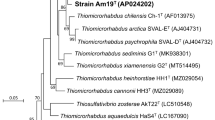
The reconstructed genome of strain H1576T consists of a circular chromosome and a plasmid, with length of 2.76 Mbp and 81.9 kbp, respectively. The G+C contents of the chromosome and plasmid are 34.8% and 32.8%, respectively. By analyzing the genome with the TYGS platform, it was revealed that the closest relatives of strain H1576T are Sulfurimonas species. Genome-based phylogenetic analysis by the TYGS indicated that strain H1576T belongs to the genus Sulfurimonas, but not to any known species (Fig. S1). The calculated values of dDDH and ANI indicated strain H1576T should not be affiliated to any Sulfurimonas species previously proposed (Table 1). Phylogenetic analysis was also conducted with the 16S rRNA gene identified in the genome. The generated phylogenetic tree indicated that strain H1576T is phylogenetically distinct from all type strains of the genus (Fig. 1). The genome of H1576T has been incorporated in the genome taxonomy database (GTDB), which provides genome-based taxonomy framework on the basis of conserved proteins (Parks et al. 2018). In the latest release of the GTDB (07-RS207), strain H1576T is classified into a Sulfurimonas species which encompasses no other organisms. All these analyses consistently indicate that strain H1576T is representative of a new species in the genus Sulfurimonas.
Phylogenetic position of strain H1576T within the genus Sulfurimonas, based on the 16S rRNA gene sequences. This maximum likelihood tree was constructed based on the Kimura 2-parameter model. All positions containing gaps and missing data were eliminated, leaving 1099 positions in the final dataset. A discrete gamma distribution was used to model differences in evolutionary rates among sites (5 categories, parameter = 0.3206). The rate variation model allowed for some sites to be invariable (68.83% sites). Bar, substitutions per site. Numbers on nodes represent percentage values of 1000 bootstrap resampling
Conclusion
The genomic analyses of different approaches consistently indicated that strain H1576T should be classified into a new species of the genus Sulfurimonas. Within the genus, strain H1576T is differentiated from the type strains of the other species by a unique combination of phenotypic characteristics (Table 1). On the basis of these results, H1576T is proposed to be assigned to a new species, with the name Sulfurimonas aquatica sp. nov.
Description of Sulfurimonas aquatica sp. nov
Sulfurimonas aquatica (a.qua’ti.ca. L. fem. adj. aquatica, aquatic)
Cells are rod shaped, motile, 1.5–2.7 μm in length and 0.6–0.7 μm in width. Gram stain negative. Grows chemolithoautotrophically by oxidizing thiosulfate, sulfide and elemental sulfur. Hydrogen gas is not used as electron donor. Aerobic. Nitrate and nitrite do not support anaerobic growth when thiosulfate is provided as the sole electron donor. Grows at 3–22 °C with an optimum growth at 13–15 °C. The pH range for growth is 6.0–7.4, with an optimum pH range of 6.6–6.8. Grows with 2–5% NaCl (optimum 2–3%). Predominant fatty acid is C16: 1. G+C content of genomic DNA of the type strain is 34.7 mol%.
The type strain H1576T (= BCRC 81254T = JCM 35004T) was isolated from water of a brackish lake in Japan.
The GenBank/EMBL/DDBJ accession numbers for the chromosome and plasmid of type strain are CP046072 and CP046073, respectively.
References
Cai L, Shao M-F, Zhang T (2014) Non-contiguous finished genome sequence and description of Sulfurimonas hongkongensis sp. nov., a strictly anaerobic denitrifying, hydrogen- and sulfur-oxidizing chemolithoautotroph isolated from marine sediment. Stand Genomic Sci 9:1302–1310. https://doi.org/10.4056/sigs.4948668
Fu C, Li J, Lv X, Song W, Zhang X (2020) Operation performance and microbial community of sulfur-based autotrophic denitrification sludge with different sulfur sources. Environ Geochem Health 42:1009–1020. https://doi.org/10.1007/s10653-019-00482-5
Han Y, Perner M (2015) The globally widespread genus Sulfurimonas: versatile energy metabolisms and adaptations to redox clines. Front Microbiol 6:989. https://doi.org/10.3389/fmicb.2015.00989
Haosagul S, Oaew S, Prommeenate P, Sawasdee V, Boonyawanich S, Pisutpaisal N (2021) Profile of sulfur oxidizing bacteria in full-scale biotrickling filter to remove H2S in biogas from in cassava starch industry. Energy Rep 7:677–685. https://doi.org/10.1016/j.egyr.2021.07.086
Henkel JV, Vogts A, Werner J, Neu TR, Spröer C, Bunk B, Schulz-Vogt HN (2021) Candidatus Sulfurimonas marisnigri sp. nov. and Candidatus Sulfurimonas baltica sp. Nov., thiotrophic manganese oxide reducing chemolithoautotrophs of the class Campylobacteria isolated from the pelagic redoxclines of the Black Sea and the Baltic Sea. Syst Appl Microbiol 44:126155. https://doi.org/10.1016/j.syapm.2020.126155
Hu Q, Wang S, Lai Q, Shao Z, Jiang L (2021) Sulfurimonas indica sp. nov., a hydrogen-and sulfur-oxidizing chemolithoautotroph isolated from a hydrothermal sulfide chimney in the Northwest Indian Ocean. Int J Syst Evol Microb 71:004575. https://doi.org/10.1099/ijsem.0.004575
Inagaki F, Takai K, Kobayashi H, Nealson KH, Horikoshi K (2003) Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing ε-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int J Syst Evol Microbiol 53:1801–1805. https://doi.org/10.1099/ijs.0.02682-0
Kubo K, Kojima H, Fukui M (2014) Vertical distribution of major sulfate-reducing bacteria in a shallow eutrophic meromictic lake. Syst Appl Microbiol 37:510–519. https://doi.org/10.1016/j.syapm.2014.05.008
Labrenz M, Grote J, Mammitzsch K, Boschker HT, Laue M, Jost G et al (2013) Sulfurimonas gotlandica sp. nov., a chemoautotrophic and psychrotolerant epsilonproteobacterium isolated from a pelagic redoxcline, and an emended description of the genus Sulfurimonas. Int J Syst Evol Microbiol 63:4141–4148. https://doi.org/10.1099/ijs.0.048827-0
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. https://doi.org/10.1038/nbt.4229
Parte AC, SardàCarbasse J, Meier-Kolthoff JP, Reimer LC, Göker M (2020) List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol 70:5607–5612. https://doi.org/10.1099/ijsem.0.004332
Ratnikova NM, Slobodkin AI, Merkel AY, Kopitsyn DS, Kevbrin VV, Bonch-Osmolovskaya EA, Slobodkina GB (2020) Sulfurimonas crateris sp. nov., a facultative anaerobic sulfur-oxidizing chemolithoautotrophic bacterium isolated from a terrestrial mud volcano. Int J Syst Evol Microbiol 70:487–492. https://doi.org/10.1099/ijsem.0.003779
Takai K, Suzuki MT, Nakagawa S, Miyazaki M, Suzuki Y, Inagaki F et al (2006) Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int J Syst Evol Microbiol 56:1725–1733. https://doi.org/10.1099/ijs.0.64255-0
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Timmer-Ten Hoor A (1975) A new type of thiosulphate oxidizing, nitrate reducing microorganism: Thiomicrospira denitrificans sp. nov. Neth J Sea Res 9:344–350. https://doi.org/10.1016/0077-7579(75)90008-3
Wang S, Jiang L, Liu X, Yang S, Shao Z (2020) Sulfurimonas xiamenensis sp. nov. and Sulfurimonas lithotrophica sp. nov., hydrogen and sulfur-oxidizing chemolithoautotrophs within the Epsilonproteobacteria isolated from coastal sediments, and an emended description of the genus. Int J Syst Evol Microbiol 70:2657–2663. https://doi.org/10.1099/ijsem.0.004087
Wang S, Jiang L, Hu Q, Cui L, Zhu B, Fu X, Lai Q, Shao Z, Yang S (2021a) Characterization of Sulfurimonas hydrogeniphila sp. Nov., a novel bacterium predominant in deep-sea hydrothermal vents and comparative genomic analyses of the genus Sulfurimonas. Front Microbiol 12:626705. https://doi.org/10.3389/fmicb.2021a.626705
Wang S, Shao Z, Lai Q, Liu X, Xie S, Jiang L, Yang S (2021b) Sulfurimonas sediminis sp. nov., a novel hydrogen-and sulfur-oxidizing chemolithoautotroph isolated from a hydrothermal vent at the Longqi system, southwestern Indian ocean. Antonie Van Leeuwenhoek 114:813–822. https://doi.org/10.1007/s10482-021-01560-4
Watanabe T, Kubo K, Kamei Y, Kojima H, Fukui M (2022) Dissimilatory microbial sulfur and methane metabolism in the water column of a shallow meromictic lake. Syst Appl Microbiol 45:126320. https://doi.org/10.1016/j.syapm.2022.126320
Widdel F, Bak F (1992) Gram-negative mesophilic sulfate-reducing bacteria. The prokaryotes, vol 4, 2nd edn. Springer, New York, pp 3352–3378
Wu J, Jiang X, Jin Z, Yang S, Zhang J (2020) The performance and microbial community in a slightly alkaline biotrickling filter for the removal of high concentration H2S from biogas. Chemosphere 249:126127. https://doi.org/10.1016/j.chemosphere.2020.126127
Yoon S-H, Ha S-M, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. https://doi.org/10.1007/s10482-017-0844-4
Acknowledgements
This study was and supported by JSPS KAKENHI, Grant Number 15K07209. The authors have no relevant financial or non-financial interests to disclose. The water sample collection was performed with permission of the Agency of Cultural Affairs, Government of Japan.
Funding
This study was funded by JSPS KAKENHI, Grant Number 15K07209.
Author information
Authors and Affiliations
Contributions
MF conducted water sampling. HK, YK and TW performed experiments for phenotypic characterization. HK isolated the strain, conducted genome analysis, wrote manuscript and prepared figures. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kojima, H., Kato, Y., Watanabe, T. et al. Sulfurimonas aquatica sp. nov., a sulfur-oxidizing bacterium isolated from water of a brackish lake. Arch Microbiol 204, 559 (2022). https://doi.org/10.1007/s00203-022-03167-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03167-3