Abstract
A facultatively anaerobic sulfur-oxidizing bacterium, strain skT11T, was isolated from anoxic lake water of a stratified freshwater lake. As electron donor for chemolithoautotrophic growth, strain skT11T oxidized thiosulfate, tetrathionate, and elemental sulfur under nitrate-reducing conditions. Oxygen-dependent growth was observed under microoxic conditions, but not under fully oxygenated conditions. Growth was observed at a temperature range of 5–37 °C, with optimum growth at 28 °C. Strain skT11T grew at a pH range of 5.1–7.5, with optimum growth at pH 6.5–6.9. Heterotrophic growth was not observed. Major components in the cellular fatty acid profile were C16:1 and C16:0. The complete genome of strain skT11T consisted of a circular chromosome with a size of 3.8 Mbp and G + C content of 60.2 mol%. Phylogenetic analysis based on the 16S rRNA gene sequences indicated that the strain skT11T is related to sulfur-oxidizing bacteria of the genera Sulfuricella, Sulfurirhabdus, and Sulfuriferula, with sequence identities of 95.4% or lower. The analysis also indicated that these three genera should be excluded from the family Gallionellaceae, as members of another family. On the basis of its genomic and phenotypic properties, strain skT11T (= DSM 110711 T = NBRC 114323 T) is proposed as the type strain of a new species in a new genus, Sulfurimicrobium lacus gen. nov., sp. nov. In addition, emended descriptions of the families Gallionellaceae and Sulfuricellaceae are proposed to declare that Sulfuricellaceae is not a later synonym of Gallionellaceae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The order Nitrosomonadales originally encompassed three families, Nitrosomonadaceae, Gallionellaceae, and Spirillaceae (Garrity et al. 2005). A restructuring of the order was made by Boden et al. 2017, along with reclassification of some taxa at levels ranging from class to genus (Boden et al. 2017). As a result, the order Nitrosomonadales currently includes Methylophilaceae, Thiobacillaceae, and Sterolibacteriaceae, in addition to the original three families. They also emended the description of the family Gallionellaceae, to encompass the genera Sulfuricella, Sulfurirhabdus, and Sulfuriferula (Boden et al. 2017). These three genera formerly constituted the family Sulfuricellaceae (Watanabe et al. 2014, 2015), which is currently regarded as a later synonym of the family Gallionellaceae. The families Sulfuricellaceae and Gallionellaceae were judged to be the same family, on the basis of analysis of the 16S rRNA gene sequences (Boden et al. 2017). The 16S rRNA gene analysis, however, included only one sequence as representative of the family Gallionellaceae. The sole sequence representing Gallionellaceae was that of Ferriphaselus amnicola, a stalk-forming iron-oxidizing bacterium (Kato et al. 2014). After the reclassification in 2017, the genus Ferrigenium was added to the family Gallionellaceae (Khalifa et al. 2018). This genus is represented by Ferrigenium kumadai, an iron-oxidizing bacterium which does not form stalks during iron-dependent growth (Khalifa et al. 2018).
In the genera which once belonged to the family Sulfuricellaceae, there are six species with validly published names. Sulfuricella denitrificans and Sulfurirhabdus autotrophica represent monospecific genera (Kojima and Fukui 2010; Watanabe et al. 2016a). The genus Sulfuriferula encompasses Sulfuriferula multivorans, Sulfuriferula plumbiphila, Sulfuriferula thiophila, and Sulfuriferula nivalis (Watanabe et al. 2015, 2016b; Kojima et al. 2020). All these species grow chemolithoautotrophically by oxidizing inorganic sulfur compounds. As an alternative electron donor, Sulfuriferula plumbiphila can use hydrogen, and Sulfuriferula multivorans can grow heterotrophically. Among these species, considerable variations in sulfur oxidation pathways have been identified (Watanabe et al. 2019). In the present study, a novel sulfur-oxidizing bacterium related to these bacteria was isolated and characterized. In addition, the taxonomic status of Sulfuricellaceae was reassessed to indicate that it is not a later synonym of Gallionellaceae.
Materials and methods
Isolation of novel bacterial strain
Strain skT11T was isolated from an enrichment culture growing on thiosulfate disproportionation, described in a previous study (Watanabe et al. 2016c). The enrichment culture was established from anoxic lake water of a stratified freshwater lake, Lake Mizugaki in Japan (35° 51′ 35" N, 138° 29′ 58" E), and maintained by periodical transfer. From the enrichment culture, a small portion was transferred to the medium S5 (Kojima et al. 2017), supplemented with NaNO3 (10 mM). As described previously, the medium S5 was buffered with 30 mM bicarbonate and contained 10 mM thiosulfate as the sole electron donor. The culture was incubated at 28 °C under anoxic condition. The resulting nitrate-reducing culture was further transferred to the same medium for three times. After that, pure culture of strain skT11T was obtained with repeated agar shake dilution using the same medium, by picking up white colonies (approx. 0.5 mm in diameter) appeared after 1–2-week incubation. Purity of the culture was culture was routinely checked by microscopy and sequencing of PCR-amplified fragments of the 16S rRNA gene.
Analysis of the 16S rRNA gene sequences
For strain skT11T, nearly full length of the 16S rRNA gene sequence was determined as described previously (Kojima et al. 2015). The obtained sequence was subjected to BLASTN search against the public database nr at the NCBI to identify its close relatives. Based on the results obtained, a custom database was created with genomes of the type strains in the genera Sulfuricella, Sulfurirhabdus, and Sulfuriferula. The 16S rRNA gene sequence identities were obtained by BLASTN search against the custom database, for strain skT11T and representatives of the family Gallionellaceae. Further phylogenetic analysis was conducted with the program MEGA version X (Kumar et al. 2018). The sequence of skT11T was aligned with reference sequences, using the MUSCLE algorithm. The references were selected from type strains of species with validly published names in the order Nitrosomonadales. Additionally, sequences of Gallionella ferruginea (lacking a pure culture of the type strain) and its close relatives (without validly published names) were also included in the analysis for a close inspection of the family Gallionellaceae. Based on the resulting alignment, models for genetic distance calculation were evaluated using the model selection tool in MEGA X. With the selected models, genetic distances were calculated by excluding positions with gaps.
Phenotypic characterization
For phenotypic characterization, the medium used for the isolation (S5 supplemented with nitrate) was used as basal medium. Unless otherwise specified, headspace of the culturing bottles was filled with mixture of N2 and CO2 (80:20; v/v), and the cultures were incubated at 28 °C in the dark. Tests of Gram-stain, catalase activity, and oxidase activity were performed as described previously (Kojima et al. 2015).
Utilization of electron donors was tested under nitrate-reducing conditions, by excluding thiosulfate from the basal medium. Oxygen-dependent growth was tested with S5 medium without nitrate, under different oxygen concentrations in the headspace, 0% (as negative control), 1%, 2%, and 20% v/v. The ability for thiosulfate disproportionation was assessed by inoculating the isolate to the medium used for the original enrichment culture (Watanabe et al. 2016c). Temperature dependence of growth was tested by culturing at 0, 5, 8, 10, 13, 15, 18, 22, 25, 28, 30, 32, 35, 37, 42, 45, and 50 °C. Effect of NaCl on growth was examined with the basal medium supplemented with different concentrations of NaCl (0, 10, 50, 100, 250, and 500 mM). Effects of pH on growth were examined under nitrate-reducing conditions, with a method modified from that previously described (Kojima et al. 2015). Briefly, the composition of the medium was changed as follows (l−1): 2.5 g Na2S2O3.5H2O, 0.85 g NaNO3, 0.3 g MgSO4.7H2O, 0.1 g CaCl2. 2H2O, 0.1 g NH4Cl, 0.1 g KH2PO4, and 0.1 g KCl. It also contained solutions of trace elements, selenite-tungstate, and vitamin mixture as same in the original medium. As a buffering agent, MES or MOPS was used at a final concentration of 20 mM, as follows; pH 3.4, 4.2, 4.4, 4.7, 5.1, 5.5, 5.8, 6.0, and 6.5 with MES; pH 6.4, 6.6, 6.8, 6.9, 7.2, 7.5, 7.7, 8.1, 8.3, and 8.6 with MOPS. The ingredients were mixed and then sterilized by filtration, after pH adjustment with HCl or NaOH. The sterilized media were dispensed in closed culture bottles whose headspace was filled with N2 gas.
For fatty acid analysis, cells were grown at 28 °C in a modified version of the basal medium, which contained increased thiosulfate and nitrate (20 mM each). The cellular fatty acid profile was analyzed using the Sherlock Microbial Identification System Version 6.0 (MIDI) with database TSBA6.
Genomic analysis
The genome sequencing was performed using the Illumina NextSeq and Nanopore GridION platforms. A hybrid assembly was performed using Unicycler (Ver 0.4.7), to generate a circular contig with coverage of 861-fold. The genome sequence was annotated with DFAST (Tanizawa et al. 2017), and genes involved in sulfur oxidation were identified as described previously (Watanabe et al. 2019).
A genome-based taxonomic classification of the strain skT11T was conducted with the Genome Taxonomy Database (GTDB), based on 120 proteins coded in the genome (Parks et al. 2018). Taxonomic position of the strain skT11T in the GTDB (release 89) was identified using GTDB-Tk (Chaumeil et al. 2020).
Results and discussion
Basic characteristics of the novel isolate
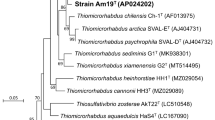
The fundamental characteristics of skT11T are summarized in Table 1 and the species description. Cells of strain skT11T were motile, rod-shaped (Fig. S1), and Gram-staining-negative. In the BLASTN analysis of the 16S rRNA gene sequence of strain skT11T (1461 bp), the highest sequence identity of 95.4% to Sulfuriferula multivorans TTNT was indicated, followed by 95.2% to Sulfuricella denitrificans skB26T (Table S1). Phylogenetic relationships between strain skT11T and these strains are shown in Fig. 1.
Phylogenetic position of strain skT11T within the order Nitrosomonadales. This unrooted phylogenetic tree was constructed with the maximum likelihood method, based on Kimura 2-parameter model with gamma distribution and invariant sites. All positions containing gaps were eliminated and there were a total of 1279 positions in the final dataset. Bar represents substitutions per site. Numbers on nodes represent percentage values of 1000 bootstrap resampling (values lower than 50 are not shown). Names of all strains included in the analysis are shown in full version of the tree provided as Figure S1
Chemolithoautotrophic growth of strain skT11T under nitrate-reducing condition was supported by thiosulfate (10 and 20 mM), tetrathionate (10 mM), and elemental sulfur (0.5 g l−1). Sulfide (2 mM) and hydrogen (H2:N2:CO2, 5:4:1 v/v/v; 200 kPa total pressure) did not support autotrophic growth. The following organic substrates did not support growth of strain skT11T: lactate (5 mM), acetate (5 mM), propionate (2.5 mM), fumarate (2.5 mM), malate (2.5 mM), butyrate (2.5 mM), isobutyrate (2.5 mM), benzoate (2.5 m M), methanol (5 mM), ethanol (2.5 mM), formate (5 mM), citrate (5 mM), and glucose (2.5 mM). In the medium without nitrate, growth of skT11T was observed under microaerobic conditions (1–2% v/v O2), but not under atmospheric oxygen tension (20%). Although strain skT11T was isolated from the enrichment culture growing on thiosulfate disproportionation, it did not disproportionate thiosulfate. A negative effect of NaCl on growth was observed at 50 mM or higher concentrations, and strain skT11T did not grow in the medium supplemented with 250 mM or 500 mM NaCl.
In the cellular fatty acid profile of strain skT11T, summed feature 3 (C16:1ω7c and/or C16:1ω6c; 51.7%) was the most dominant, followed by C16:0 (26.2%) and summed feature 8 (C18: 1ω7c and/or C18:1ω6c; 11.8%). These three major components accounted for nearly 90% of the total. As minor components, C10:0 3-OH (3.4%), C12:0 (2.2%), C16:1ω5c (1.3%), and anteiso-C17:1 ω9c (0.6%) were detected. The other fatty acids detected (< 0.5% of total) were C18:1ω5c, C17:1ω5c, C18:1ω9c, C17:0, C18:0 C10:0, C14:0, C17: 1ω6c, iso-C20:0, C17: 1ω8c, and C16:0 3-OH. In fatty acid profiles of the related genera (Sulfuricella, Sulfurirhabdus, and Sulfuriferula), summed feature 3 and C16:0 were detected as the most and second most dominant components [3, 4, 8, 9, 10].
Genomic characters of the strain skT11T
The genome of strain skT11T consists of a circular chromosome with a size of 3,826,324 bp and G + C content of 60.2 mol%, respectively. In the complete genome, 3678 protein-coding sequences were predicted. It harbors two copies of the 16S rRNA gene with the identical sequence. As the genetic basis for sulfur oxidation, gene encoding proteins involved in the Sox–Dsr–Soe pathway were identified. This pathway is one of the three core pathways defined in a previous study (Watanabe et al. 2019), which revealed coexistence of the Sox–Dsr–Soe and Sox–Hdr–Soe pathways in Sulfurirhabdus autotrophica BiS0T and Sulfuriferula multivorans TTNT. The gene set for Sox–Hdr–Soe pathway, conserved in the genera Sulfurirhabdus and Sulfuriferula, was not identified in the genome of strain skT11T (Table 1). Besides the genes involved in the core sulfur oxidation pathway, strain skT11T has the aprBA genes encoding adenylylsulfate reductase (Apr). The aprBA genes in genomes of sulfur oxidizers are known to be divided into two groups, Apr lineage I and lineage II (Meyer and Kuver 2007; Watanabe et al. 2016c). In the genome of strain skT11T, two copies of the aprBA genes were identified, and they belonged to the Apr lineage I and lineage II, respectively (Table 1).
Reassessment of the families Gallionellaceae and Sulfuricellaceae
Phylogenetic position of strain skT11T within the order Nitrosomonadales was examined by constructing trees with three different methods, based on the 16S rRNA gene sequences (Fig. 1, Figs. S2–S4). Different branching patterns were obtained with different methods, but robust clusters corresponding to the families Nitrosomonadaceae, Methylophilaceae, Spirillaceae, Thiobacillaceae, and Sterolibacteriaceae were formed in all trees. On the other hand, none of the trees supported the monophyly of the family Gallionellaceae. All the trees indicated that members of the family belong to two distinct phylogenetic groups. One of the groups (hereafter referred to as group 1) included the genera Gallionella, Ferriphaselus, and Ferrigenium. The other group (hereafter referred to as group 2) consisted of the genera Sulfuricella, Sulfurirhabdus, and Sulfuriferula, along with strain skT11T (Fig. 1). The phylogenetic dissociation of these two groups has also been observed in phylogenetic trees constructed in the previous studies (Watanabe et al. 2014, 2016c, 2019). These results suggest that the group 2 should be excluded from the family Gallionellaceae corresponding to the group 1. In the previous study which merged the Gallionellaceae and Sulfuricellaceae, sequence identity of 92.25% was used as a “guide” to define the families based on the 16S rRNA gene sequences (Boden et al. 2017). The previous analysis included Ferriphaselus as the solo representative of the Gallionellaceae, and the sequence identity between Ferriphaselus amnicola OYT1T and Sulfuricella denitrificans skB26T (> 92.5%) is greater than this guide value. However, those between Ferriphaselus amnicola OYT1T and the other type strains belonging to the group 2 are lower than 92.2% (Table S1). Ferrigenium kumadai An22T, the other type strain in the group 1, gave similar results in which identity greater than the guide value was observed only with Sulfuricella denitrificans skB26T (Table S1). Although there is no isolated type strain of Gallionella species at present, the 16S rRNA gene sequences of Gallionella are available in the public database. They showed sequence identities lower than the guide value, to all type strains of the group 2 (Table S1). To obtain more evidence for the dissociation of two groups, some sequences closely related to the species of group 1 were additionally analyzed in the same manner. Even in the expanded analysis including uncultured bacteria, sequence identities greater than 92.25% were observed only in limited number of combinations across the two groups (Table S1). These results indicate that the group 2 should be regarded as independent family, and thus, Sulfuricellaceae is not later synonym of Gallionellaceae. Accordingly, description of Gallionellaceae should be emended to exclude the genera belonging to the group 2, i.e., Sulfuricellaceae.
Taxonomic position of the strain skT11T within the family Sulfuricellaceae
The phylogenetic trees of 16S rRNA gene indicate that strain skT11T is member of the family Sulfuricellaceae, but does not belong to any existing genera (Fig. 1, Figs. S2–S4). It was also shown that strain skT11T is most closely related to Sulfuricella and Sulfurirhabdus. To draw conclusion about genus-level classification based on whole genomic data, the genome sequences of strain skT11T and Sulfurirhabdus autotrophica BiS0T were analyzed using the GTDB-Tk. As a result, these organisms and Sulfuricella denitrificans skB26T (included in the GTDB release 89) were classified into three different genera. This means that a novel genus should be created to accommodate strain skT11T. As shown in Table 1, strain skT11T has some characteristics distinct from those of the type strains of species in the genera Sulfuricella, Sulfurirhabdus, and Sulfuriferula. On the basis of these results, strain skT11T is proposed as the type strain of a novel species of a new genus in the family Sulfuricellaceae, with the name Sulfurimicrobium lacus gen. nov., sp. nov.
Description of Sulfurimicrobium gen. nov.
Sulfurimicrobium (Sul.fu.ri.mi.cro’bi.um. L. neut. n. sulfur sulfur; N.L. neut. n. microbium, a microbe; N.L. neut. n. Sulfurimicrobium sulfur-oxidizing microbe).
Grows by oxidation of inorganic sulfur compounds; microaerophilic and neutrophilic; Gram-stain-negative. Major cellular fatty acids are C16:1 and C16:0. Belongs to the family Sulfuricellaceae. The type species is Sulfurimicrobium lacus.
Description of Sulfurimicrobium lacus sp. nov.
Sulfurimicrobium lacus (la’cus. L. gen. n. lacus, of a lake).
In addition to the properties listed for the genus, cells are motile, rod-shaped, 1.1–3.7 µm long and 0.5–0.7 µm wide. Catalase-negative and oxidase-positive. Grows chemolithoautotrophically by oxidation of thiosulfate, tetrathionate, and elemental sulfur. Uses oxygen and nitrate as electron acceptor. Oxygen-dependent growth occurs only under microoxic conditions. Temperature range for growth is 5–37 °C, with an optimum of 28 °C. Growth occurs at pH 5.1–7.5, with an optimum of pH 6.5–6.9. G + C content of genomic DNA of the type strain is 60.2 mol%.
The type strain skT11T (= DSM 110711 T = NBRC 114323 T) was isolated from anoxic lake water of a stratified freshwater lake in Japan. The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene and complete genome sequence of strain skT11T are LC533074 and AP022853, respectively.
Emended description of Sulfuricellaceae (Watanabe et al. 2015)
Sulfuricellaceae (Sul.fu.ri.cel.la.ce’ae. N.L. fem. n. Sulfuricella type genus of the family; -aceae ending to denote family; N.L. fem. pl. n. Sulfuricellaceae the family of the genus Sulfuricella).
Delineation is primarily based on phylogenetic information from 16S rRNA gene sequences. Belongs to the order Nitrosomonadales. Encompasses Gram-stain-negative bacteria isolated from freshwater environments. At the time of writing, the family contains the genera Sulfuricella, Sulfurirhabdus, Sulfuriferula, and Sulfurimicrobium gen. nov. All members utilize inorganic sulfur compounds as their energy source. Uses oxygen or nitrate as terminal electron acceptors for respiration. The type genus is Sulfuricella.
Emended description of Gallionellaceae (Henrici and Johnson 1935 (Approved Lists 1980) emend. Boden et al. 2017)
Gallionellaceae (Gal.li.o.nel.la.ce’ae. N.L. fem. n. Gallionella type genus; -aceae suffix to denote family; N.L. fem. pl. n. Gallionellaceae the Gallionella family).
Delineation is primarily based on phylogenetic information from 16S rRNA gene sequences. Comprises iron-oxidizing bacteria of the genera Gallionella, Ferriphaselus,, and Ferrigenium. Microaerophilic and neutrophilic. Motile with one polar flagellum. Cells are curved-rod or reniform. Reduced sulfur or ferrous iron is used as electron donors for autotrophic and/or mixotrophic growth. Stalk formation is observed during the iron-dependent growth of some species. The type genus is Gallionella.
References
Boden R, Hutt LP, Rae AW (2017) Reclassification of Thiobacillus aquaesulis (Wood & Kelly, 1995) as Annwoodia aquaesulis gen. nov., comb. nov., transfer of Thiobacillus (Beijerinck, 1904) from the Hydrogenophilales to the Nitrosomonadales, proposal of Hydrogenophilalia class. nov. within the 'Proteobacteria', and four new families within the orders Nitrosomonadales and Rhodocyclales. Int J Syst Evol Microbiol 67:1191–1205. https://doi.org/10.1099/ijsem.0.001927
Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH (2020) GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinfomatics 36:1925–1927. https://doi.org/10.1093/bioinformatics/btz848
Drobner E, Huber H, Rachel R, Stetter KO (1992) Thiobacillus plumbophilus spec. nov., a novel galena and hydrogen oxidizer. Arch Microbiol 157:213–217. https://doi.org/10.1007/BF00245152
Garrity GM, Bell JA, Lilburn T (2005) Order VI. Nitrosomonadales ord. nov. In: Brenner DJ, Krieg NR, Staley JT and Garrity GM (editors). Bergey’s Manual of Systematic Bacteriology, 2nd ed, vol. 2, (The Proteobacteria), part C (The Alpha-, Beta-, Delta-, and Epsilonproteobacteria). New York: Springer. p. 863
Khalifa A, Nakasuji Y, Saka N, Honjo H, Asakawa S, Watanabe T (2018) Ferrigenium kumadai gen. nov., sp. nov., a microaerophilic iron-oxidizing bacterium isolated from a paddy field soil. Int J Syst Evol Microbiol 68:2587–2592. https://doi.org/10.1099/ijsem.0.002882
Kojima H, Fukui M (2010) Sulfuricella denitrificans gen. nov., sp. nov., a sulfur-oxidizing autotroph isolated from a freshwater lake. Int J Syst Evol Microbiol 60:2862–2866. https://doi.org/10.1099/ijs.0.016980-0
Kojima H, Shinohara A, Fukui M (2015) Sulfurifustis variabilis gen. nov., sp. nov., a sulfur oxidizer isolated from a lake, and proposal of Acidiferrobacteraceae fam. nov. and Acidiferrobacterales ord. nov. Int J Syst Evol Microbiol 65:3709–3713. https://doi.org/10.1099/ijsem.0.000479
Kojima H, Watanabe M, Fukui M (2017) Sulfurivermis fontis gen. nov., sp. nov., a sulfur-oxidizing autotroph, and proposal of Thioprofundaceae fam. nov. Int J Syst Evol Microbiol 67:3458–3461. https://doi.org/10.1099/ijsem.0.002137
Kojima H, Mochizuki J, Fukui M (2020) Sulfuriferula nivalis sp. nov. and emended description of Sulfuriferula plumbiphila. Int J Syst Evol Microbiol 70:3273–3277. https://doi.org/10.1099/ijsem.0.004166
Kato S, Krepski S, Chan C, Itoh T, Ohkuma M (2014) Ferriphaselus amnicola gen. nov., sp. nov., a neutrophilic, stalk-forming, iron-oxidizing bacterium isolated from an iron-rich groundwater seep. Int J Syst Evol Microbiol 64:921–925. https://doi.org/10.1099/ijs.0.058487-0
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Meyer B, Kuever J (2007) Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5’-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology 153:3478–3498. https://doi.org/10.1099/mic.0.2007/008250-0
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 3:996–1004. https://doi.org/10.1038/nbt.4229
Tanizawa Y, Fujisawa T, Nakamura Y (2017) DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 35:1037–1039. https://doi.org/10.1093/bioinformatics/btx713
Watanabe T, Kojima H, Fukui M (2014) Complete genomes of freshwater sulfur oxidizers Sulfuricella denitrificans skB26 and Sulfuritalea hydrogenivorans sk43H: genetic insights into the sulfur oxidation pathway of betaproteobacteria. Syst Appl Microbiol 37:387–395. https://doi.org/10.1016/j.syapm.2014.05.010
Watanabe T, Kojima H, Fukui M (2015) Sulfuriferula multivorans gen. nov., sp. nov., isolated from a freshwater lake, reclassification of 'Thiobacillus plumbophilus' as Sulfuriferula plumbophilus sp. nov., and description of Sulfuricellaceae fam. nov. and Sulfuricellales ord. nov. Int J Syst Evol Microbiol 65:1504–1508. https://doi.org/10.1099/ijs.0.000129
Watanabe T, Kojima H, Shinohara A, Fukui M (2016a) Sulfurirhabdus autotrophica gen. nov., sp. nov., isolated from a freshwater lake. Int J Syst Evol Microbiol 66:113–117. https://doi.org/10.1099/ijsem.0.000679
Watanabe T, Kojima H, Fukui M (2016b) Sulfuriferula thiophila sp nov., a chemolithoautotrophic sulfur-oxidizing bacterium, and correction of the name Sulfuriferula plumbophilus Watanabe, Kojima and Fukui 2015 to Sulfuriferula plumbiphila corrig. Int J Syst Evol Microbiol 66:2041–2045. https://doi.org/10.1099/ijsem.0.000988
Watanabe T, Kojima H, Fukui M (2016c) Identity of major sulfur-cycle prokaryotes in freshwater lake ecosystems revealed by a comprehensive phylogenetic study of the dissimilatory adenylylsulfate reductase. Sci Rep 6:36262. https://doi.org/10.1038/srep36262
Watanabe T, Kojima H, Umezawa K, Hori C, Takasuka ET et al (2019) Genomes of neutrophilic sulfur-oxidizing chemolithoautotrophs representing 9 proteobacterial species from 8 genera. Front Microbiol 10:316. https://doi.org/10.3389/fmicb.2019.00316
Acknowledgements
We would like to thank A. Shinohara for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that there are no conflicts of interest.
Additional information
Communicated by Erko stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene and the complete genome of strain skT11T are LC533074 and AP022853, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kojima, H., Kanda, M., Umezawa, K. et al. Sulfurimicrobium lacus gen. nov., sp. nov., a sulfur oxidizer isolated from lake water, and review of the family Sulfuricellaceae to show that it is not a later synonym of Gallionellaceae. Arch Microbiol 203, 317–323 (2021). https://doi.org/10.1007/s00203-020-02029-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-02029-0