Key message
Fine mapping of the barley leaf rust resistance locus Rph13 on chromosome 3HL facilitates its use in breeding programs through marker-assisted selection.
Abstract
Barley leaf rust (BLR—caused by Puccinia hordei) is a widespread fungal disease that can be effectively controlled by genetic resistance. There is an ongoing need to both diversify and genetically characterise resistance loci to provide effective and durable control given the ongoing threat of rapidly evolving P. hordei populations. Here, we report on the molecular genetic characterisation of the Rph13 locus, originally derived from wild barley and transferred to barley accession Berac (then referred to as PI 531849). The 2017 reference genome of cv. Morex was used as a road map to rapidly narrow both a genetic and physical intervals around the Rph13 resistance locus. Using recombination-based mapping, we narrowed the physical interval to 116.6 kb on chromosome 3H in a segregating population of a cross of the Rph13 carrying resistant line PI 531849 with the leaf rust-susceptible cultivar Gus. We identified two nucleotide-binding leucine-rich repeat genes as likely candidates for the Rph13 resistance. Sequences from the candidate genes enabled the development of a KASP marker that distinguished resistant and susceptible progeny and was found to be predictive and useful for MAS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Barley is the fourth most important cereal crop in global production. Barley production is, however, affected by numerous diseases that reduce both yield and grain quality. Spreading from host to host by wind- or human-aided spore migration, rust pathogens (Puccinia spp.) cause some of the most damaging diseases, especially in cereals including: wheat, barley and oat. Almost all of Australia’s devastating rust epidemics have followed unpredictable exotic rust incursions. Barley leaf rust (BLR) is a disease caused by the basidiomycete fungal pathogen Puccinia hordei Otth., which has been reported to significantly reduce grain quality and yield (up to 62%) given suitable environmental conditions (Cotterill et al. 1992). The fungus mostly propagates via asexual modes of reproduction, but in the presence of the alternate host (Ornithogalum spp.) can also undergo sexual recombination that gives rise to new diverse pathogenic variants that threaten crop production (Wallwork et al. 1992). Genetic resistance is the most economically and environmentally responsible method to control rust diseases including BLR. One of the biggest challenges in disease resistance breeding is the breakdown of resistance genes due to mutations in effector (avirulence genes) genes in prevailing pathogen populations, resulting in new virulent races that can cause disease on previously resistant barley varieties. One approach that has proved highly effective in breeding for durable resistance to stem rust of wheat in Australia is to pyramid multiple resistance genes in cultivars (Park 2007).
Over the past century, 26 Rph (Resistance to Pucciniahordei) genes have been formally designated for BLR resistance in additional to numerous QTLs conferring partial resistance (Niks et al. 2015; Park et al. 2015; Kavanagh et al. 2017; Yu et al. 2018). Of these, most confer resistance expressed at all stages of plant development (all stage resistance—ASR), while three confer adult plant resistances (APRs) that are often minor in effect. Among the designated Rph genes, Rph10, Rph11, Rph13, Rph15 and Rph16 are derived from wild barley (H. spontaneum); however, none of these genes have yet been deployed in cultivated barley (Park et al. 2015). PI 531849 is an experimental barley line from the UK that exhibited a high level of resistance (hypersensitive response) based on phenotypic testing with a panel of isolates virulent for Rph1-12 (Jin and Steffenson 1994). In Australia, virulence has been detected in a few P. hordei pathotypes (Park 2003). However, because virulence is rare, Rph13 remains a valuable source of resistance in combination with other effective seedling genes and especially with adult plant resistance genes such as Rph20. When PI 531849 was crossed with the leaf rust-susceptible barley cv. Bowman, the F2 population segregated for a single dominant gene (Jin and Steffenson, 1994). Tests for allelism with different Rph resistance donors (Rph1-12) produced either a two or three gene segregation ratios demonstrating that the resistance in accession PI 531849 (RphPI531849) was independent from all other previously described Rph genes. Jin et al. (1996) did, however, observe a deficiency of susceptible F2 plants in a cross between PI 531849 and an Rph9 donor (HOR 2596) located on the long arm of chromosome 5H, suggesting possible linkage with Rph9. RphPI531849 was subsequently designated Rph13 in accordance with the gene nomenclature for Rph genes.
Molecular markers have been used extensively in barley breeding, mainly for genetic mapping, marker-assisted selection (MAS) and positional gene cloning efforts. Despite the rapid evolution of molecular marker technologies, single-nucleotide polymorphisms (SNPs) are the marker of choice due to their abundance, reproducibility and their ease of application. The availability of the first complete reference genome of barley has increased the efficacy of marker design in targeted genomic regions and the exploitation of putative candidate genes (Mascher et al. 2017). In a recent study, high-throughput SNP genotyping was performed for 55 different Bowman introgression lines carrying different Rph genes (Martin 2018). SNP differences between the respective Rph13 NIL (carrying Rph13 from PI 531849) and the Bowman recurrent parent revealed two introgressed segments on 3HL. The aims of the present study were to (i) develop a high-resolution mapping population for the Rph13 resistance gene, (ii) use the barley reference genome to design KASP markers to saturate the genetic interval harbouring Rph13, (iii) relate the genetic map to the barley reference and (iv) explore the use of sequences from candidate genes predicted at the Rph13 locus to develop a diagnostic marker for the efficient selection of Rph13.
Materials and methods
Plant and pathogen materials
The line PI 531849 carrying the Rph13 resistance allele (Rph13.x) was originally generated by introgressing leaf rust resistance from Hordeum vulgare subsp. spontaneum (Hs2986) to the barley cultivar Berac (Berac*3/HS 2986). PI 531849 was crossed to the leaf rust-susceptible barley cv. Gus (Franckowiak 1998; Jin et al. 1996; Jin and Steffenson 1994) to develop a segregating F2 mapping population comprising 719 plants. A panel of Australian cultivars was used for marker validation (Table S1).
The Puccinia hordei isolates used in this study are maintained and stored in liquid nitrogen at the Plant Breeding Institute (PBI), University of Sydney. Pathotype 5457 P + (PBI accession number 090017 = 612) used for phenotyping PI 531849 × Gus population is avirulent on resistance genes Rph5, Rph7, Rph8, Rph11, Rph13, Rph14, Rph15 and Rph21; and virulent on resistance genes Rph1, Rph2, Rph3, Rph4, Rph6, Rph9, Rph10, Rph12 and Rph19 (Singh et al. 2018). The Rph13-virulent pathotype 5653 P + (+Rph13) (PBI accession number 010116 = 569) was also used to confirm the specificity of the resistance segregating in the PI 531849 x Gus population.
Plant growth and pathogen infection
F2 plants were inoculated with P. hordei pathotype 5457 P+ at the primary leaf stage. P. hordei urediniospores were heat-shocked at 42 °C for two minutes, mixed with Isopar L and sprayed over plants. Seedlings of the mapping population and controls were inoculated as described by Dracatos et al. (2019). Infection types were recorded 10 d post-inoculation using the “0” to “4” scale (Park and Karakousis 2002). Infection types of test lines of each population were compared with those displayed by the parents and by barley differential lines to assure accurate classification into resistant and susceptible classes
Marker development
We designed oligonucleotides based on the Morex reference sequence (Mascher et al. 2017) to amplify DNA fragments by PCR from the two parents PI 531849 and Gus, sequenced them by SANGER sequencing and converted identified polymorphisms into KASP or CAPS markers (Table S2). Primer design for PCR and KASP marker assays was conducted with the Primer3 online tool (http://bioinfo.ut.ee/primer3-0.4.0//). All PCRs were performed on Bio-Rad T100 thermal cycler. Short fragments (< 2 kb) were amplified with HotStarTaq DNA polymerase (Qiagen) and larger fragments by Phusion high-fidelity DNA polymerase (New England Biolabs) according to manufacture guidelines.
The codominant KASP marker HvKASP_Rph13: resistant allele (HvKASP_Rph13plus) ‘GAAGGTCGGAGTCAACGGATTGATGCTCTGGAAGAAGTGGTTC,’ wild-type allele (Gus, HvKASP_Rph13minus) ‘GAAGGTGACCAAGTTCATGCTGATGCTTTGGAAGAAGTGGTTG’ and common reverse primer ‘ATCATCACCCCCAATCAACA’ (HvKASP_Rph13reverse) was initially tested across segregating leaf material of five F3 recombinant families (8 plants per family, see table S6). Once optimised, the KASP marker was further validated using 180 individuals of the F2 mapping population and a reference set of Australian cultivars detailed in Table S1.
Candidate gene analysis
We amplified the whole genomic sequences of candidate genes by PCR using gene-specific primers (Table S2). PCR products were cloned into TOPO XL2 vector (Invitrogen) using manufacturer’s instructions. At least six colonies per cloned PCR amplicon were preceded for plasmid isolation using ISOLATE II Plasmid Mini Kit (Bioline) and sent for Sanger sequencing.
KASP marker analysis
Standard oligos were designed with Hex and FAM overhangs, and reaction mix using KASP version 4.0 2x Master mix (LGC) was prepared as previously described (Gao et al. 2016). KASP analysis was performed on CFX96 Real-Time PCR Detection System (Bio-Rad) by two cycling steps: initial denaturation for 15 min at 95 °C, followed by 9 cycles of denaturation at 94 °C for 20 s, annealing at 65 °C for 1 min (decreasing by 0.8 °C per cycle); this was followed by 32 cycles (up to 40 cycles, depending on marker) of denaturation at 94 °C for 20 s, annealing at 57 °C for 1 min, followed by 5 min at 25 °C with image taken for end point fluorescence discrimination.
NLR annotation
The sequence from 666,516,273 to 667,693,961 bp of chromosome 3H of the Morex reference genome (Mascher et al. 2017) was extracted by SAMtools version 1.9.0. The sequences were chopped to fragments of 20 kb length with 5 kb overlap, and the NLR prediction was performed based as previously described (https://github.com/steuernb/NLR-Annotator (Steuernagel et al. 2018).
Phylogeny
A maximum likelihood tree was developed with MEGA10 (https://www.megasoftware.net) based on nucleotide sequences using ClustalW alignment and following previously published protocol (Hall 2013). The tree was constructed using HKY + G model and Bootstrap with 1000 replicates,
Data availability
The sequences of described genes in this study can be accessed from NCBI database (https://www.ncbi.nlm.nih.gov/) under the following accession number: NLR1_Rph13 (MN601345), NLR1_Gus (MN601346), NLR2_Rph13_v1 (MN601347), NLR2_Gus (MN601348) and NLR2_Rph13_v2 (MN601349).
Results
Genetic mapping
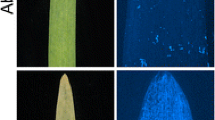
In a previous study, high-density marker analysis of the Rph13 near isogenic line (NIL) and its recurrent parent Bowman revealed two segments with polymorphic markers with sizes of ~ 30.6 Mb (640.3 to 670.9 Mb) and 14 Mb (684.6 to 698.5 Mbp), respectively (Martin 2018). We firstly confirmed the presence of the Rph13-mediated resistance in the reported introgression/s on the long arm of chromosome 3H through correlating the phenotype and genotype of 160 PI 531849 x Gus F2 plants. Seedlings of the PI 531849 x Gus F2 population segregated for a single resistance gene (121 R:39 S) when inoculated with P. hordei pathotype 5457 P+. We observed distinct phenotypic classes within the mapping population characterised either by a hypersensitive response (HR necrotic flecks and chlorosis) for resistance or by large sporulating pustules for susceptibility, respectively (Fig. 1). Despite this apparent ease of phenotypic classification, a majority of resistant F2 plants were more intermediate relative to the PI 531849 parent, suggesting incomplete dominance (Fig. 1). Kompetitive allele-specific PCR (KASP) markers spanning the last 70 Mbp of the distal end of 3HL were concurrently designed based on sequence information from the 2017 Morex assembly (Table S2). The compiled genotypic and phenotypic data enabled mapping of the Rph13 locus to a genetic interval of 3.4 cM translating to a physical region of ~ 6 Mbp based on anchoring the polymorphic KASP markers to the Morex reference genome (Mascher et al. 2017) (Fig. 2).
Variation of disease response in leaf rust infected seedlings. The susceptible parent Gus (left) shows large sporulating pustules (uredinia) compared to the small non-sporulating pustules restricted by a hypersensitive response induced in the Rph13 line PI 531849 (far right). Between the parental lines, representatives from the segregating population showing the range of resistant and susceptible infection responses phenotyped in the PI 531849 × Gus F2-F3 population
Low-resolution genetic map. Genetic map based on leaf rust phenotyping and genotyping with seven markers of 160 F2 plants distributed along the 70 Mb target area on chromosome 3HL. The F2 phenotypic screen allowed mapping of the Rph13 locus (red) between markers K6 and K5 in a 3.4-cM interval (color figure online)
Based on this initial map, we selected markers K5 and K6 (Fig. 2, Table S2) as flanking markers to genotype a larger number of F2 plants to increase the genetic resolution at the Rph13 locus. In total, 719 F2 plants (including the 160 specified above) were phenotyped (164 susceptible and 555 resistant plants) and then genotyped, leading to the identification of 38 plants with recombination events between K5 and K6. To further refine the genetic and physical interval of the Rph13 locus, we developed 10 polymorphic KASP markers between K5 and K6 and genotyped the 38 recombinants (Fig. 3). We identified one co-segregating marker (K34) flanked by K69 and K49 that delimited a 116.6 kb physical interval (Fig. 3) according to the genome reference of Morex (Mascher et al. 2017).
High-resolution mapping: a fine mapping of the Rph13 locus in 719 F2 plants revealed 38 recombinants between the flanking markers K6 and K5, which were further saturated with markers. Numbers between the markers are according to identified recombinants. b Schematic drawing of the physical map including BAC clones spanning the interval (Table 1) and high-confidence genes predicted based on the Barley Ref v1
Candidate gene analysis at the Rph13 locus
We explored the narrowed 116.6-kb interval for putative candidate genes implicated in host response to pathogen. In total, three high-confidence (HC) genes (Table 1) were predicted based on the reference genome sequence of Morex (Mascher et al. 2017): HORVU3Hr1G105110.1, a short protein of unknown function; HORVU3Hr1G105070.7, described as ankyrin repeat family protein; and HORVU3Hr1G105020.39 as disease resistance protein. The disease resistance protein contained a predicted nucleotide-binding (NB) domain and multiple leucine-rich repeats (LRRs). The disease resistance gene showed strong nucleotide similarity (94%) to the gene HORVU3Hr1G104770.5, which has 1, 237 additional nucleotides upstream of the predicted ATG from compared to HORVU3Hr1G105020.39. Interestingly, the additional sequence was predicted to encode a coiled-coil (CC) domain, suggesting HORVU3Hr1G104770.5 was likely a CC-NBS-LRR (NLR) encoding gene. In the Morex reference, HORVU3Hr1G104770.5 was located 414 kb outside of the interval based on the genome reference representing the genetically defined interval harbouring the Rph13 locus. Further sequence comparison of the full-length CC-NBS-LRR gene (HORVU3Hr1G104770.5) with HORVU3Hr1G105020.39 within the Rph13 interval determined they shared 98% nucleotide similarity and confirmed the presence of a CC domain in HORVU3Hr1G105020.39 that was missing from the gene model prediction (Mascher et al. 2017).
To minimise the risk of overlooking possible gene candidates, we decided to re-annotate the gene space at the Rph13 locus to search for NLR candidates not present based on the current Morex reference gene annotation. We used NLR prediction software, which enables an automatic high-throughput analysis for the NLR-specific conserved motif combinations (Steuernagel et al. 2018). We interrogated 1.18 Mbp of Morex sequence (666,516,273–667,693,961 Mbp of Chr3H) spanning the co-segregating region and the highly related CC-NBS-LRR (NLR) second gene copy outside the mapping interval. The software predicted the same two genes (HORVU3Hr1G104770.5 and HORVU3Hr1G105020.39) as the only full-length NLR genes as described in Table S3. This confirms the presence of all motifs for a complete NLR within the mapping interval. However, if the reference sequence is accurate, the predicted extended open reading frame (ORF) leads to a truncated protein (lacking the last 532 amino acids compared to the paralog) and hence putatively represents a pseudogene in Morex.
The absence of either a genome sequence or physical contigs spanning the locus from the parental genotypes limits any exhaustive assessment of the gene content of the Rph13 locus. To gain further understanding of possible structural variation at the Rph13 locus, we sequenced (start to stop codon) the NLR candidates HORVU3Hr1G105020.39 (NLR1) and HORVU3Hr1G104770.5 (NLR2) from both parents PI 531849 and Gus using the Morex reference as a template for primer design (Table S4). Gus had a similar haplotype to Morex as only a single copy each of NLR1 and NLR2 was amplified using this approach. However, in the Rph13 donor PI 531849 we amplified a single copy of NLR1 and two apparent paralogs of NLR2 based on sequencing multiple (8) colonies per sub-cloned PCR amplicon. We compared the nucleotide sequences of all identified NLRs with the versions from Morex and observed a mixed clustering of NLRs originating from amplification of NLR1 and NLR2 (Fig. 4). All amplified variants from both parents could be transcribed using an in silico approach to infer functional proteins. Interestingly, the NLR1 homolog amplified from Gus contained a 991-bp insertion within an intron, resulting in the encoded protein from this variant being highly divergent relevant to all other NLRs from PI 531849 and Gus (Figure S1).
Homology to the wheat stem rust Sr35 locus
To improve our understanding of the syntenic relationship between the candidate genes for Rph13 identified in the present study and other characterised NLRs that map to the same chromosomal region, we searched for putative orthologues within closely related species in the Triticeae. We took the predicted translated protein sequences of the detected NLR candidates at the Rph13 locus amplified from donor accession PI 531849 and used them as BLAST P queries against the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Interestingly, the predicted protein sequences of PI 531849-derived NLRs at the Rph13 locus showed strong homology and coverage (99%) to numerous closely related NLRs [CNL9 (81%), CNL4 (81%), CNL6 (77%), CNL2 (79%) and CNL1 (74%)] from Triticum monococcum subsp. monococcum corresponding to the Sr35 wheat stem rust locus (Saintenac et al. 2013). The given percentage of identity refers to the protein sequence of NLR1 as query for the BLAST as representative of the NLRs detected in PI 531849 (Rph13), but the alignment of all protein sequences to Sr35 (CNL9) shows strong conserved sequence among all detected NLRs in this study (Figure S1). The third functional annotated segregating gene HORVU3Hr1G105070.7 showed homology to APGG1 and also a reported member of the Sr35 gene cluster (Saintenac et al. 2013). Furthermore, the KASP markers used for genetic mapping showed perfect collinearity with the Sr35 locus when used for comparative mapping with the recent release of the wheat reference genome (Appels et al. 2018), further supporting the syntenic relationship between Rph13 and Sr35 (Table S5).
Marker development for breeding
We aimed to develop a marker which co-segregates with the phenotype and is specific to the Rph13 resistance locus. We followed a two-step validation approach: (i) assess co-segregation by genotyping critical recombinants flanking the Rph13 locus and (ii) genotype a representative set of cultivars to assess the specificity of the SNP haplotype among barley germplasm. Firstly we used a SNP from HORVU3Hr1G105070.7 (PGG/ANK) and confirmed co-segregation using DNA from our segregating mapping population. Unexpectedly, screening the markers across various Australian cultivars (Table S1) identified numerous false positives in cultivars that were either susceptible to Rph13-avirulent pathotypes or were known not to carry the Rph13 resistance based on detailed multi-pathotype testing (RF Park & D Singh, unpublished), suggesting that these KASP markers were not predictive of Rph13 only (Fig. 5).
Predictive marker for Rph13a observed segregation in F3 families as illustrated by the distinct clustering of susceptible (orange) heterozygote (green) and resistance (blue). b Marker screening of Australian cultivars showed that Rph13 is absent. Heterozygote and resistant accessions represent controls in the experiment (color figure online)
We PCR-amplified, sub-cloned and sequenced NLR paralogs from both PI 531849 and Gus to permit the development and optimisation of gene copy-specific KASP markers for each of the NLRs. We aimed to: (i) obtain a marker that is predictive for Rph13 and (ii) determine the most plausible gene candidate to facilitate sequence comparison between mutant versus wild type in future studies. In order to develop a Rph13 predictive marker, we interrogated SNPs for KASP marker design within the NLR1 and NLR2 genes and tested several KASP markers of different NLR combinations for co-segregation (data not shown). Some of the designed markers failed possibly influenced by other undetected NLR paralogs at the locus or from the massive amplification of other members of the large NLR gene family in the genome. However, the KASP marker developed for NLR1_Gus versus NLR2_Rh13_v1 did not co-segregate with the phenotype of F3 families of critical recombinants flanking the Rph13 locus (Table S6), suggesting NLR2_Rph13v2 is not a candidate. We were not able to map NLR2_Rph13v2; thus, it remains as a candidate. One KASP marker (referred to as HvKASP_Rph13, Table S2) was designed from a SNP in Exon 1 G385C between Gus (NLR1_Gus) and PI 531849 (NLR1_Rph13) and was predictive for the Rph13 resistance. HvKASP_Rph13 once optimised was validated on a panel of 72 diverse barley genotypes with known phenotypic responses to leaf rust including Australian, Chinese and European accessions and numerous resistant and susceptible experimental control genotypes. All accessions with the exception of HOR 1063 and the positive controls carried the Gus (susceptibility) allele, suggesting this marker is suitable for the marker-assisted selection of Rph13.
Discussion
The effectiveness of commonly used R genes is often short-lived due to rapidly evolving pathogen populations and/or exotic pathogen incursions. One approach is to pyramid multiple disease resistances from diverse origins in cultivars and/or source resistance from wild relatives. The efficacy of gene pyramiding in crops especially in barley relative to wheat is reduced by lack of detailed molecular genetic studies and robust molecular markers. Numerous sources of leaf rust resistance have been identified in both bulbous barley (Hordeum bulbosum) and wild barley (H. spontaneum), but few have been genetically characterised to a point that they can be utilised effectively for crop improvement. The Rph13 allele (Rph13.x) was originally transferred from Hordeum vulgare subsp. spontaneum into the cultivar (cv.) Berac. In this study, we genetically characterised and fine-mapped the Rph13 locus from PI 513849 to a narrow genetic and physical interval on the long arm of chromosome 3H (3HL).
Rph13 was concluded to be located on chromosome 5H based on genetic linkage observed when PI 513849 was crossed with the donor for Rph9 (HOR 2596) (Jin et al. 1996). In a very recent study by Martin (2018), polymorphisms identified using a genotyping-by-sequencing (GBS) approach revealed two introgressed segments from the Rph13 donor, PI 513849 in the long arm of chromosome 3HL in a line near isogenic line with Bowman background (Martin 2018). Despite this, there is still the possibility of additional smaller introgressions that were not detected by the genotypic analysis. Our mapping data revealed that Rph13 is located in the larger introgression, and we narrowed the 30.6-Mb physical interval to 116.5 kb using a recombination-based mapping approach. A QTL from a wild barley introgressed segment into cultivar Scarlett has been previously allocated to the same region on 3HL (von Korff et al. 2005). The reported associated marker of the detected QTL on 3HL in Scarlett maps to the same physical position in the Morex reference (667 Mbp) as identified for Rph13 in the present study (Mascher et al. 2017). Given the fact that both resistances were derived from wild barley, they might both carry Rph13; however, further test of allelism and Rph13-virulent pathotype testing would be required to confirm this hypothesis.
Based on the syntenic relationship between Triticum and Hordeum spp. in the Triticeae and the infection type observed in PI 531849, Rph13 from H. spontaneum likely encodes an NLR immune receptor orthologous to an NLR at the wheat stem rust resistance Sr35 locus. We identified two NLR copies derived from PI 531849 in our 116.5-kb mapping interval that were highly similar to Sr35 from T. monococcum. Both are progenitors of today’s cultivated barley and wheats, respectively, indicating a putative common ancient resistance locus within the common ancestor predating the division of the Triticeae tribe. Gene Sr35 was localised within a complex cluster of NLR genes that comprises five complete, two partial and additional fragments of NLR genes (Saintenac et al. 2013). The effectiveness of the stem rust resistance gene Sr35 from T. monococcum in barley was previously assessed for resistance to the wheat stem rust pathogen P. graminis f. sp. tritici (Pgt) and P. hordei using isolates that were avirulent with respect to Sr35 and Rph13, respectively. When transformed into the Golden Promise background, Sr35 conferred resistance to Pgt, but did not provide resistance to P. hordei (M. Asyraf Md. Hatta et al. 2018). This suggests that sequence diversification between the Sr35 gene and NLR members at the Rph13 locus generates resistance specificity between Pgt and P. hordei. Alternatively, Sr35 may not be the true orthologue of Rph13. Putatively, an orthologue of another NLR gene within the cluster could be responsible for Rph13-mediated leaf rust resistance in PI 531849. The current lack of a sequence scaffold at the Rph13 locus from the resistance donor prevents an accurate depiction of the number of NLRs in the Rph13 donor and further comparison with the wheat Sr35 NLR cluster.
Due to evolutionary forces such as gene duplication driving the diversity of NLRs in crop plants, pan genomic variation should be expected especially in the case of disease resistance loci. A common disadvantage of the map-based cloning strategy is the underestimation of pan genomic variation between the resistance donor and the reference genome. For example, the causal gene of Yr15 conferring stripe rust resistance in common wheat was not actually present in the Chinese Spring reference genome (Klymiuk et al. 2018). The recent release of the Morex genome reference sequence in 2017 allowed improvements to gene order and annotation in barley. This resource, along with the high rate of polymorphism between cultivated and wild barley, unquestionably fast-tracked the fine mapping of Rph13 in our study to three possible candidate genes. An empirical comparison between Morex, Gus and the Rph13 donor PI 531849 was made through PCR amplification of each of the three candidate genes in the Rph13 interval. Our data suggest that both Morex (rph13) and Gus (rph13) carry a single copy each of NLR1 and NLR2. In contrast, PI 531849 (Rph13) carried two copies of NLR2 indicative of pan genome variation at the Rph13 locus, suggesting possible gene duplication. However, further sequence data from PI 531849 are required to confirm this hypothesis.
To develop and validate molecular markers with high utility in terms of their reliability and prediction for the presence and absence of Rph13 resistance, we had to satisfy two main criteria. Firstly, we needed to interrogate gene copy-specific SNPs between the parents and produce a reproducible KASP assay with clear groupings to distinguish homozygotes from heterozygotes. Secondly, once developed we needed to validate the efficacy of the marker using a panel of barley genotypes that were accurately phenotyped for the presence/absence of the Rph13 resistance. Our initial attempt to design predictive KASP markers to the non-NLR genes in the mapping interval was based on the assumption that the likelihood of amplifying paralogous gene copies would be reduced. Despite this, KASP markers designed to HORVU3Hr1G105070.7 were not predictive for the Rph13 resistance based on false positives in numerous Australian cultivars that lacked the Rph13 resistance based on detailed testing with P. hordei isolates with diverse virulence profiles. This may suggest possible recombination, which may not be overly surprising given the proximity of the Rph13 locus to the telomere on the long arm of chromosome 3H. Furthermore, KASP markers designed to SNPs within NLR2 were also not predictive for the presence of Rph13. In the reference genome, NLR2 is located 400 kb upstream of NLR1, consistent with our data, suggesting the gene order in Morex is likely correct. In contrast, a KASP marker designed to a SNP in NLR1 co-segregated perfectly with the Rph13 resistance when validated on the PI 531849 x Gus mapping population and 72 barley accessions with known leaf rust responses, suggesting this marker is highly predictive for Rph13 and can be used for marker-assisted selection in breeding. The German landrace HOR1063 was screened twice with the NLR KASP marker resulting in the Rph13 and heterozygous marker haplotype, respectively, suggesting it may be heterogeneous for Rph13. This, however, contradicts previous studies, suggesting the presence of leaf rust resistance on chromosome 2H in HOR1063 that is allelic with Rph15/Rph16 (Derevnina et al. 2015).
Future prospects in pan genomics will unravel critical structural, copy number and cultivar-specific sequence variations and contribute critical resources to fast-track the understanding of disease resistance and other traits in barley (Monat et al. 2019). For example, the availability of a whole genome sequence of PI 531849 would unravel the molecular basis at the Rph13 locus and determine the presence and permit the validation of candidate genes not present in the reference. Furthermore, recently developed approaches in mutational genomics such as MutRenSeq and MutChromSeq have proved effective for rapidly isolating rust resistance genes in crops species (Dracatos et al. 2019; Sanchez-Martin et al. 2016; Steuernagel et al. 2016, 2017). Future work will involve the development of further pan genome information and identification of chemically induced rph13 knockout mutants in the PI 531849 background to functionally verify the gene candidates for Rph13 identified in the current study.
References
Appels R et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. https://doi.org/10.1126/science.aar7191
Cotterill PJ, Rees RG, Platz GJ, Dillmacky R (1992) Effects of leaf rust on selected Australian barleys. Aust J Exp Agric 32:747–751. https://doi.org/10.1071/Ea9920747
Derevnina L, Singh D, Park RF (2015) The genetic relationship between barley leaf rust resistance genes located on chromosome 2HS. Euphytica 203:211–220. https://doi.org/10.1007/s10681-014-1315-x
Dracatos PM et al (2019) The coiled-coil NLR Rph1, confers leaf rust resistance in barley cultivar Sudan. Plant Physiol 179:1362–1372. https://doi.org/10.1104/pp.18.01052
Franckowiak JD (1998) BGS 590, reaction to Puccinia hordei 13 (barley leaf rust), Rph13. Barley Genet Newslett 28:31
Gao L, Jia J, Kong X (2016) A SNP-based molecular barcode for characterization of common wheat. PLoS ONE 11:e0150947. https://doi.org/10.1371/journal.pone.0150947
Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30:1229–1235. https://doi.org/10.1093/molbev/mst012
Hatta MAM, Johnson R, Matny O, Smedley MA, Yu G, Chakraborty S, Bhatt D, Xia X, Arora S, Steuernagel B, Richardson T (2018) The wheat Sr22, Sr33, Sr35 and Sr45 genes confer resistance against stem rust in barley. BioRxiv. https://doi.org/10.1101/374637
Jin Y, Steffenson BJ (1994) Inheritance of resistance to Puccinia-Hordei in cultivated and wild barley. J Hered 85:451–454. https://doi.org/10.1093/oxfordjournals.jhered.a111500
Jin Y, Cui GH, Steffenson BJ, Franckowiak JD (1996) New leaf rust resistance genes in barley and their allelic and linkage relationships with other Rph genes. Phytopathology 86:887–890. https://doi.org/10.1094/Phyto-86-887
Kavanagh PJ, Singh D, Bansal UK, Park RF (2017) Inheritance and characterization of the new and rare gene Rph25 conferring seedling resistance in Hordeum vulgare against Puccinia hordei. Plant Breed 136:908–912. https://doi.org/10.1111/pbr.12535
Klymiuk V et al (2018) Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat Commun 9:3735. https://doi.org/10.1038/s41467-018-06138-9
Martin M (2018) Development of barley introgression lines carrying the leaf rust resistance genes Rph1 To Rph15. University of Minnesota, Minneapolis
Mascher M et al (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544:426. https://doi.org/10.1038/nature22043
Monat C, Schreiber M, Stein N, Mascher M (2019) Prospects of pan-genomics in barley. Theor Appl Genet 132:785–796. https://doi.org/10.1007/s00122-018-3234-z
Niks RE, Qi X, Marcel TC (2015) Quantitative resistance to biotrophic filamentous plant pathogens: concepts, misconceptions, and mechanisms. Annu Rev Phytopathol 53:445–470. https://doi.org/10.1146/annurev-phyto-080614-115928
Park RF (2003) Pathogenic specialization and pathotype distribution of Puccinia hordei in Australia, 1992 to 2001. Plant Dis 87:1311–1316. https://doi.org/10.1094/Pdis.2003.87.11.1311
Park RF (2007) Stem rust of wheat in Australia. Aust J Agric Res 58:558–566. https://doi.org/10.1071/Ar07117
Park RF, Karakousis A (2002) Characterization and mapping of gene Rph19 conferring resistance to Puccinia hordei in the cultivar ‘Reka 1’ and several Australian barleys. Plant Breed 121:232–236. https://doi.org/10.1046/j.1439-0523.2002.00717.x
Park RF et al (2015) Leaf rust of cultivated barley: pathology and control. Annu Rev Phytopathol 53:565–589. https://doi.org/10.1146/annurev-phyto-080614-120324
Saintenac C, Zhang WJ, Salcedo A, Rouse MN, Trick HN, Akhunov E, Dubcovsky J (2013) Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 341:783–786. https://doi.org/10.1126/science.1239022
Sanchez-Martin J et al (2016) Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol 17:221. https://doi.org/10.1186/s13059-016-1082-1
Singh D et al (2018) Genome-wide association studies provide insights on genetic architecture of resistance to leaf rust in a worldwide barley collection. Mol Breed. https://doi.org/10.1007/s11032-018-0803-4
Steuernagel B et al (2016) Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol 34:652–655. https://doi.org/10.1038/nbt.3543
Steuernagel B, Witek K, Jones JDG, Wulff BBH (2017) MutRenSeq: a method for rapid cloning of plant disease resistance genes methods. Mol Biol 1659:215–229. https://doi.org/10.1007/978-1-4939-7249-4_19
Steuernagel B, Witek K, Krattinger SG, Ramirez Gonzalez RH, Schoonbeek H-J, Yu G, Baggs E, Witek A, Yadav I, Krasileva KV, Jones JDG, Keller B, Ridout C (2018) Physical and transcriptional organisation of the bread wheat intracellular immune receptor repertoire. BioRxiv 5:2. https://doi.org/10.1101/339424
von Korff M, Wang H, Leon J, Pillen K (2005) AB-QTL analysis in spring barley. I. Detection of resistance genes against powdery mildew, leaf rust and scald introgressed from wild barley. Theor Appl Genet 111:583–590. https://doi.org/10.1007/s00122-005-2049-x
Wallwork H, Preece P, Cotterill P (1992) Puccinia hordei on barley and Omithogalum umbellatum in South Australia. Australas Plant Pathol 21:95–97
Yu X et al (2018) Genetic mapping of a barley leaf rust resistance gene Rph26 introgressed from Hordeum bulbosum. Theor Appl Genet 131:2567–2580. https://doi.org/10.1007/s00122-018-3173-8
Acknowledgements
The authors would like to thank Ms Bethany Clark, Mr Matthew Williams and Mr Gary Standen for technical assistance and the Grains Research Development Corporation for funding this project (US00074).
Author information
Authors and Affiliations
Contributions
MJ and PD designed and performed all experiments, analysed and interpreted results. DS contributed plant material for the study. MJ and PD wrote the manuscript. All authors reviewed and approved the manuscript. RP and EL conceptualised the research and received funding.
Corresponding author
Additional information
Communicated by Albrecht E. Melchinger.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jost, M., Singh, D., Lagudah, E. et al. Fine mapping of leaf rust resistance gene Rph13 from wild barley. Theor Appl Genet 133, 1887–1895 (2020). https://doi.org/10.1007/s00122-020-03564-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03564-6